Abstract
This discussion focuses on the cellular dynamics of tRNA transcription, processing, and turnover. Early tRNA biosynthesis steps are shared among most tRNAs, while later ones are often individualized for specific tRNAs. tRNA transcription and early processing occur coordinately in the nucleolus, requiring topological arrangement of ~300 tRNA genes and early processing enzymes to this site; later processing events occur in the nucleoplasm or cytoplasm. tRNA nuclear export requires multiple exporters which function in parallel and the export process is coupled with other cellular events. Nuclear-cytoplasmic tRNA subcellular movement is not unidirectional as a retrograde pathway delivers mature cytoplasmic tRNAs to the nucleus. Despite the long half-lives, there are multiple pathways to turnover damaged tRNAs or normal tRNAs upon cellular stress.
Keywords: tRNA transcription, tRNA processing, tRNA modification, nucleolus, tRNA nuclear export, tRNA retrograde movement, tRNA turnover
Introduction
Eukaryotes have developed multi-layered pathways for modulating the expression of nuclear genes, including the creation of multiple distinct RNA polymerases, highly complex nucleolytic RNA processing pathways, varied subcellular localizations, and multiple turnover pathways. In general, each broad type of RNA has a distinctive complement of components that accomplish and regulate its biosynthesis and turnover, though there is increasing realization that there are components that are adapted to act along the multiple pathways.
Initial synthesis of nuclear RNAs is accomplished by at least three distinct RNA polymerases in eukaryotic nuclei, RNA polymerases I, II and III (Pol I, Pol II, Pol III), although recent evidence suggests plants have evolved two more variants of Pol II termed Pol IV and Pol V to express non-coding RNAs [1,2]. Each of the three ubiquitous RNA polymerases expresses specific classes of RNA, with Pol I specializing in the large ribosomal RNA (rRNA) precursors, Pol II expressing pre-messenger RNAs and a variety of small, functional RNAs, and Pol III exclusively expressing highly conserved, small functional RNAs. This discussion concerns itself with one particular class of functional RNAs expressed by Pol III, transfer RNAs (tRNAs). Transfer RNAs serve as adaptors between information encoded in the genome and proteins synthesis. tRNAs are an ancient form of RNA, having evolved as part of the translation machinery to bridge between the hypothetical “RNA World” of self-replicating RNAs and the more modern world in which proteins mediate reproduction of genetic material (RNA and DNA). tRNAs must be produced in large quantities and coordinately controlled in response to the need for protein synthesis.
tRNAs have a unique set of physical constraints. Their tertiary structure must conform to a narrowly defined structural window to fit into the necessary contacts with the ribosome and other translational machinery, as well as common RNA processing enzymes. On the other hand, dozens to hundreds of different tRNA molecules must differ sufficiently so that they can be aminoacylated with different amino acids and incorporate those amino acids only when directed by sequences in translating mRNA-ribosome assemblies. These seemingly conflicting requirements have led to development of expression pathways that not only coordinate the expression of many different tRNAs in large quantities but also “tweak” the identity of individual tRNAs to allow them to be reliably distinguished from one another.
In bacteria, tRNAs are normally excised from long transcripts by nucleases that recognize their unique structure and then they are enzymatically modified on bases and sugars to allow fine discrimination between tRNA structures. In eukaryotes transcription factors recognize minimal sequence conservation within all tRNA coding regions to mark the genes as individual transcription units through binding of specialized transcription factors and Pol III [3]. The subsequent processing of the precursor tRNA (pre-tRNA) transcripts requires many of the same cleavages and modifications that are required in the bacterial tRNAs. It is important to note that other classes of genes transcribed by Pol III (e.g. 5S rRNA) that do not have the same divergent processing requirements have entirely different genomic organization [4].
The processing pathways for pre-tRNAs have unique constraints in eukaryotic nuclei. The many tRNAs require a huge diversity of different types of processing to refine their structure and identity, though for the most part their gene transcription is coordinately regulated. At the same time, the spatial organization of tRNA expression is subject to pressures that demand a high rate of transcription and common aspects of early tRNA processing that benefit from economies of scale. Thus, spatial coordination of transcription and some aspects of early tRNA processing make sense, whereas there is expected to be considerable divergence of processing pathways to cope with isotype-specific modifications and regulation.
Spatial organization of tRNA genes as a component of expression
Spatial coordination of key components of early tRNA processing is likely to be critical in the efficiency of the overall tRNA biosynthetic pathway. The localization of the Pol I-transcribed ribosomal genes and the Pol III-transcribed 5S genes to the nucleolus appears to facilitate usage of their transcripts in ribosomal assembly and processing. Moreover, the biosynthesis of these ribosomal components at the nucleolus is often co-regulated with expression of tRNAs [5]. Thus, it would not be surprising if the Pol III-transcribed tRNA genes would also be positioned at the nucleolus, where their expression might be more readily co-regulated with ribosomal gene transcription. Yet, until a few years ago, there was no definitive evidence that tRNA genes were preferentially localized to any specific nuclear substructure.
Insight into how organization of the tRNA genes relates to their functional needs was gained from the demonstration in budding yeast that the 274 tRNA genes, which are scattered across the linear map of the genome, are preferentially localized to the nucleolus (Table 1) [6]. More recently, it has been shown that this localization to the nucleolus is a two-part phenomenon, with clustering of the tRNA genes separable from microtubule-dependent positioning of the clusters at the nucleolus, and that this positioning occurs throughout the cell cycle [7]. While three-dimensional clustering of tRNA genes has so far only been shown in yeast, it would not be unexpected if some form of clustering existed in higher eukaryotes, especially in light of evidence that at least one highly conserved protein complex required for the clustering, condensin, is present at tRNA genes in yeast and interacts directly with the Pol III transcription factors [7,8].
Table 1.
Subcellular Location of Biosynthesis/Processing/Turnover Steps for Cytoplasmic tRNAs
| Step | Cellular compartment - yeast |
Cellular sub- compartment - yeast |
Cellular compartment – vertebrate cells |
Cellular sub- compartment – vertebrate cells |
|---|---|---|---|---|
| Transcription | Nucleus, clustered | Nucleolus | Nucleus, clustered | ND |
| 5’ processing | Nucleus | Nucleolus | Nucleus | Unresolved |
| 3’ processing | Nucleus | ND | Nucleus | ND |
| Splicing | Cytoplasm | Mitochondrial surface | Nucleus | Nucleoplasm |
| Nucleoside modifications | Nucleus or cytoplasm | Nucleoplasm, nuclear membrane, ER, cytoplasm | Nucleus and ND | ND |
| TRAMP-mediated 3’>5’ turnover | Nucleus | ND | ND | ND |
| RTD 5’>3’ turnover | Nucleus & cytoplasm | ND | ND | ND |
| Half molecule production | Cytoplasm | Vacuolar enzyme | Cytoplasm | Endocytosed enzyme |
ND – not determined; see text for references supporting locations of the processes
Although nothing definitive is known about the spatial organization of mammalian tRNA genes, they are also scattered throughout the linear genome as in yeast [9]. Limited data on Pol III expression in human cells is consistent with clusters of expression, though not with close association with the nucleolus [10]. Even in yeast, not every individual tRNA gene is likely to be localized to the nucleolus at all times. Mapping of a limited number of tRNA gene loci gave 60%–70% nucleolar association if the tRNA gene was actively expressed versus 5–15% if transcriptionally inactivated, but it is possible that positioning is quite variable among loci, especially if there are other positioning signals from neighboring genes. Still, the predominant clustering of the tRNA genes suggests that this spatial arrangement is not random, and the clustering of the genes is entirely consistent with the localization of most nuclear pre-tRNA transcripts and early processing steps.
Early pre-tRNA processing steps
One important component of the early tRNA processing machinery is RNase P [11,12], an endonuclease that catalyzes the cleavage of the 5’ leader sequence from pre-tRNA transcripts. RNase P is thought to be an ancient enzyme dating back to the “RNA World” and exists as a ribonucleoprotein complex in almost all organisms with a large, catalytic RNA and variable amounts of required protein. In budding yeast both RNase P and early pre-tRNA transcripts, which contain 5’ and 3’ termini and introns, primarily localize to the nucleolus (Table 1) [13]. In addition, there is evidence that RNase P might associate directly with the Pol III transcription apparatus through interactions with the transcription factor TFIIIB, which binds upstream of tRNA genes [14]. Studies of localization patterns of RNase P in metazoans have been more difficult to interpret, with both perinucleolar and nucleoplasmic localizations having been demostrated [15,16], an ambiguity that parallels the lack of knowledge in metazoans concerning localization of the tRNA genes themselves.
In light of the clustering of tRNA genes at the nucleolus in yeast, it is not surprising in retrospect that RNase P is concentrated there. RNase P existed before the development of Pol III and is required for cleavage of virtually all nucleus-encoded pre-tRNAs, providing a possible selection for the enzyme to develop and associate directly with the transcription so as to efficiently capture substrates. Another early processing function common to all pre-tRNA transcripts, cleavage of the 3’ trailing sequences, usually occurs quickly after RNase P cleavage cleavage (see [17] for an exception), and one could imagine that this process might also be spatially coordinated to maximize efficiency. Although multiple nucleases might be involved with this process in vivo, as in bacteria [18], there is an extensive collection of such nucleases in the nucleolus and nucleoplasm. At least one enzyme, RNase Z, known to be involved in processing nuclear and mitochondrial transcripts [19], has been localized to both the nucleus and mitochondria [20].
tRNA modification – distribution throughout cells
tRNAs are highly modified and ~100 different modifications have been described (Review: [21]). Many chemically distinct modifications exist including numerous methylations of all four bases or nucleoside sugars, isomerization of uracil to pseudouracil, conversion of uracil to dihydrouracil, N-acetylation of cytidine, conversion of adenosine to inosine, and elaborate modifications of the anticodon such as isopentylation of adenosine, formation of wybutosine or queuosine from guanosine, and methoxycarbonylmethylation and thiolylation of uracil. Some modifications are restricted to Archaea, bacteria, or eukaryotes, but many are shared. In eukaryotes tRNAs encoded by both the nuclear genome and by organellar genomes are modified. There are numerous cases of a single nuclear gene encoding two or more isoenzymes, one of which is imported into mitochondria to modifiy mitochondrial tRNA and the other of which is responsible for modification of cytoplasmic tRNAs, but particular organellar modifications can be encoded by genes distinct from nonorganellar modifications (Review: [22]).
Genomes encode scores of enzymes responsible for catalyzing the correct modifications at the correct site of particular tRNAs. Yet, until recently the functions of modifications, with the exception of those located at the anticodon and known to participate in decoding, were poorly understood. This is because, where investigated, most tRNA modification enzymes are encoded by unessential genes (Review [23]). For example, even though dihydrouracil is a ubiqitious tRNA modification catalyzed by four separate yeast genes, DUS1–4, yeast deleted for all four family members are viable [24]. However, more recently, it has become appreciated that modifications affect tRNA structure and tRNA stability (see tRNA turnover, below).
Experiments employing Xenopus oocyte injections of tRNA substrates first showed that in eukaryotes tRNA modifications occur in a preferred order [25]. One reason for the preferred order concerns substrate specificity. For example, some modification enzymes employ only pre-tRNAs, such as intron-containing tRNAs, as substrates, whereas others modify only spliced tRNAs [26]. Another reason for ordered modification concerns the subcellular distribution of the modification enzymes. In yeast, some modification enzymes are restricted to the nucleus and particular subnuclear locations, such as the nucleoplasm or inner nuclear membrane, whereas others are restricted to the cytoplasm (Table 1). For example, yeast Trm1 that modifies G26 to m22G26 resides in the nucleus at the inner nuclear membrane, whereas Trm11 that catalyzes methylation of G10 to m2G is a cytoplasmic enzyme [27,28]. Information regarding the subcellular distribution of tRNA modification enzymes for higher eukaryotes is largely unavailable, but if the distribution is similar to yeast, the picture that emerges is that in eukaryotes tRNAs travel about the cell acquiring their modifications at different subcellular sites. This view distinguishes tRNAs from other cellular RNAs whose biogenesis generally is thought to be completed by recruitment of the processing activities to the site of transcription (see above). It will be interesting to learn the function of distributing tRNA modification enzymes throughout the cells and whether there is evolutionary conservation for the location of particular activities.
Pre-tRNA splicing – different subcellular organization among organisms
tRNA genes can be interrupted by introns. In yeast ~20% of the tRNA genes, encoding 10 different tRNAs, contain introns (Review: [23]). The introns range from 14–60 nucleotides and interrupt the anticodon loop 1 nucleotide 3' to the anticodon. There is no conservation of sequence at the splice junctions, although the 3'-splice junction is in a bulged loop. The general features and locations of tRNA introns are conserved, although in Archaea tRNA introns may be located at positions other than the anticodon loop. Most eukaryotic genomes appear to possess a lower percentage intron-containing tRNA genes than does S. cerevisiae [29].
Removal of introns from pre-tRNAs in eukaryotes and Archaea is a protein catalyzed process whereas in bacteria and organelles intron removal proceeds via a group I self-splicing reaction (Review: [30] and references therein). Pre-tRNA splicing has been thoroughly characterized in budding yeast where it has been shown to be a three-step process. Step 1, catalyzed by the heterotetrameric tRNA splicing endonuclease [31], removes introns generating 5' half molecules with a 2'-3’ cyclic phosphate and 3’ half molecules with a 5’ hydroxyl group (Review: [32]). Step 2, catalyzed by the multifunctional monomeric tRNA splicing ligase, Trl1, [33], adds a phosphate to the 3' tRNA half and joins the 5' and 3' halves in a phosphodiester bond. Step 3, catalyzed by tRNA 2' phosphotransferase, removes the extra 2' phosphate from the splice junction and generates a novel byproduct - adenosine diphosphate (ADP)-ribose 1"–2" cyclic phosphate [34]. Although the yeast splicing endonuclease is an αβγδ heterotetamer, Archaeal splicing endonucleases can be αα homodimers, αβ heterodimers, or heterotertramers of αβ heterodimers [35] and references therein). The catalytic splicing endonuclease subunits are conserved from Archaea to human, although the Archaeal enzyme recognizes a helix-bulge-helix structure in pre-tRNAs that the budding yeast and human enzymes do not (Review: [32]). Surprisingly, the ligation and 2’ phosphotransferase steps may not be conserved, as it appears that there are two alternative ligation pathways in animal cells and although the yeast-like tRNA ligase is found in plants and humans, the yeast-like 2’phosphotransferase path appears to be unessential in animal cells [36].
Despite the high conservation of the tRNA splicing endonuclease, there appears to be fundamental differences in the subcellular distribution of pre-tRNA splicing among different organisms (Table 1). For vertebrates, experiments conducted to monitor the fates of particular yeast precursor and mature tRNA species injected into Xenopus oocyte cytoplasm or nuclei, first showed that removal of introns from pre-tRNAs occurred in the nucleoplasm [25]. These results were confirmed by others and extended to human cells [37,38], documenting that pre-tRNA splicing occurs in the nucleus in vertebrate cells. Although initial studies to determine the subcellular location of the tRNA splicing machinery indicated that pre-tRNA splicing in yeast would also occur in the nucleoplasm [39], Yoshihisa et al. ([40] and confirmed by others [41–43]) showed that the tRNA splicing endonuclease subunits are located on the cytoplasmic surface of mitochondria, not in the nucleus as anticipated. The yeast enzymes that function after the endonucleolytic cutting step, Trl1 and Tpt1, also are not nuclear proteins [43]. The location of pre-tRNA splicing in plants has not been entirely resolved. Expression of GFP-tagged recombinant genes from Arabidopsis and Oryza, in Allium (monocot) epidermal or Vicia (dicot) guard cells showed that two of the splicing endonuclease subunits were found in the nucleus and the cytoplasm whereas tRNA ligase co-located with chloroplast and the cytoplasm or nucleus. Tagged proteins encoded by Arabidopsis cDNAs encoding full length 2’ phosphotransferase associated with both mitochondrial and chloroplast organelles as well as the cytoplasm, but mutant constructs designed to initiate translation at internal AUGs produced proteins with a nuclear location in addition to the organellar locations [44]. Since nuclear pools of each of the splicing enzymes could be detected by these studies, the data are consistent with the hypothesis that pre-tRNA splicing in plants occurs in the nucleus. However, genetic studies in Arabidopsis offer a different view as it has been shown that PAUSED mutants, defective in tRNA nuclear export (see below), accumulate intron-containing pre-tRNA, as if the pre-tRNA and splicing machinery do not come into contact when nuclear export is defective, as is the case in budding yeast [45]. Further study is required to resolve the subcellular site for pre-tRNA splicing in Arabidopsis.
In sum, while the biochemistry of pre-tRNA splicing is conserved, curiously, the location of splicing differs in different organisms. Why this is the case is unknown, but one possibility is that the components of the splicing machinery serve additional functions that provide selection for their subcellular distribution (Review: [46]). For example, tRNA ligase in yeast functions in the unfolded protein response pathway that occurs in the cytoplasm which might provide a selection for pre-tRNA splicing to occur in the cytoplasm in yeast; on the other hand, in vertebrate cells splicing endonuclease activity is coupled to mRNA 3’ termini formation that occurs in the nucleus which might provide a selection for the nuclear location of the tRNA splicing endonuclease in vertebrate. It will be interesting to investigate these and other possible reasons underlying why the location of pre-tRNA splicing in eukaryotic cells differs.
Dynamic movement of mature tRNAs within eukaryotic cells
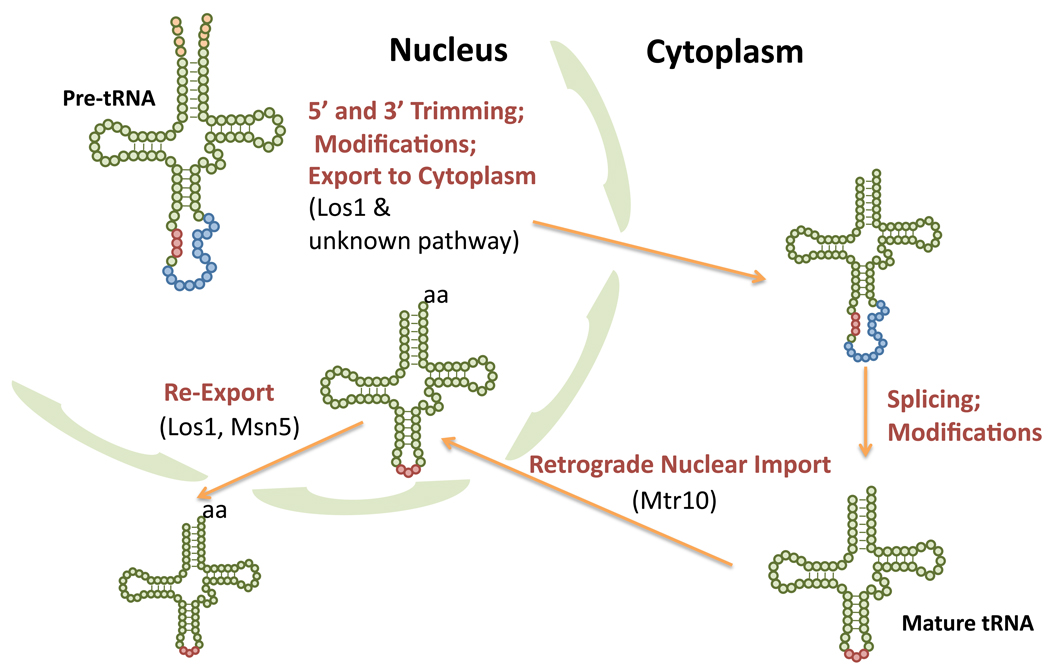
It’s well established that tRNA transcription and numerous pre-tRNA processing steps occur in the nucleus, but that tRNAs function in protein synthesis in the cytoplasm. Moreover, a certain subpopulation of nucleus-encoded tRNAs are delivered to mitochondria for function in protein synthesis within this organelle (Review: [47]). Thus, there must be systems in eukaryotic cells to deliver tRNAs to the correct subcellular locations (Fig. 1). Very surprisingly, nuclear-cytoplasmic tRNA subcellular movement is not unidirectional. A tRNA retrograde pathway that results in cytoplasmic tRNA accumulation in the nucleus under particular conditions has been described for yeast [41,42], vertebrate cells [48], and likely has been usurped by Lenti viruses, like HIV ([49]; Review: [46]).
Figure 1.
Movement of tRNA between the nucleus and cytoplasm in the yeast, S. cerevisiae. tRNA transcription and initial end processing occurs in the nucleolus. Following various modification steps in the nucleoplasm and at the inner nuclear membrane, intron-containing pre-tRNAs are exported to the cytoplasm via the Los1 exportin and at least one unknown pathway. After pre-tRNA splicing on the cytoplasmic surface of mitochondria, mature tRNAs can be constitutively imported into nuclei, directly or indirectly via Mtr10. Re-export to the cytoplasm is mediated by Los1 and Msn5 and is regulated by nutrient status; likely Msn5-dependent re-export requires that the tRNA be appropriately structured and aminoacylated in the nucleus. Symbols: cloverleaf cartoon – tRNA, green and red balls; parts of the tRNA that are maintained in the mature structure; red balls – anticodon triplet; salmon color balls – transcribed 5’ and 3’ extra sequences; blue balls – intron sequence; aa – amino acid. Processing steps are labeled as are the β-importin members that function in the nucleus-cytoplasm import and export steps.
Export of newly synthesized tRNA from the nucleus to the cytoplasm
Movement of macromolecules between the nucleus and the cytoplasm is an active process that generally requires the Ran pathway. Ran, a small GTPase, exists in a GTP-bound form in the nucleus and a GDP-bound form in the cytoplasm. The gradient of high Ran-GTP in the nucleus and low Ran-GTP in the cytoplasm dictates directionality of macromolecular movement across nuclear pores – structures that form transport channels connecting the nucleus and the cytoplasm (Review: [50]). Eukaryotic cells possess a family (~20 members in vertebrates and 14 in yeast) of Ran-binding proteins, referred to as the β-importins. These proteins bind nuclear pore components and particular protein and/or RNA cargoes, delivering the cargoes to the correct subcellular compartment. Generally, β-importin members transport in a single direction and are termed importins if involved in movement of cargo from the cytoplasm to the nucleoplasm or exportins if functioning in movement of cargo from the nucleoplasm to the cytoplasm. Ran-GTP-exportin-nuclear cargo heterotrimeric complexes deliver macromolecules from the nucleus to the cytoplasm where the cargo is released by hydrolysis of Ran-GTP to Ran-GDP.
One well-studied β-importin family member functions in the delivery of nuclear tRNAs to the cytoplasm for most organisms (Fig. 1). This exportin, Exportin-t or Exp-t in vertebrate cells [51,52], and its orthologues - PAUSED in Arabidopsis [53] and Los1 or Xpot in fungi, [54–56], binds tRNA in a Ran-GTP dependent mechanism. Structure probing and use of artificial substrates studies showed that the Exp-t interacts with the backbone of the TψC and acceptor arms of the tRNA cloverleaf structure [51,57]. Recently a 3.1 Å structure of S. pombe Xpot in complex with tRNA and Ran was obtained; as anticipated Xpot makes contacts with the TψC and D loops; it also interacts with both the 5’ and 3’ tRNA termini [56]. The data provide an explanation for how this exportin serves a proofreading role delivering only appropriately structured, properly matured functional tRNAs to the cytoplasm and why Exp-t has higher binding affinity for 5’ and 3’ end-matured tRNAs [51,57]. Exp-t interacts equally well with intron-containing and intron-free pre-tRNA; so the splicing step of processing is not proofread by this export pathway. Lack of interaction of Xpot with the anticodon loop was also verified in the X-ray structure [56].
Inhibition of tRNA export by introduction of antibodies to Exp-t reduced the levels of cytoplasmic tRNA in vertebrate cells >80% [51,57], leading to the conclusion that Exp-t is the major, if not the only, exportin required for tRNA nuclear export in vertebrate cells. In fungi and plants tested to date, this is not the case, because the Exp-t orthologues are unessential genes in S. cerevisiae, S. pombe, and Arabidopsis (Review: [46]).
Despite the fact that Los1 is unessential in yeast, it clearly integrates physiological processes. For example, when yeast cells are exposed to DNA damaging agents, the subcellular distribution of Los1 between the nucleus and the cytoplasm is altered such that tRNA nuclear export is down-regulated. This down-regulation contributes to a cell cycle checkpoint by the delayed translation of the cyclin, Cln2, thus coupling tRNA nuclear export via Los1 to the cell cycle checkpoint for DNA damage [58]. In another example, los1Δ was shown to cause Gcn2-independent induction of the general amino acid regulatory pathway that normally occurs upon amino acid deprivation [59]. Moreover, either a single species of mutant tRNA in the nucleus (even when the wild-type endogenous tRNA gene is present) or inappropriate levels of nuclear tRNA processing intermediates due to altered expression of tRNA processing enzymes induced the Gcn pathway. The data support the existence of uncharacterized communication between nuclear tRNA processing and Los1-dependent nuclear export and events regulating translation in the cytoplasm [59].
Since Los1 and PAUSED are unessential, parallel tRNA nuclear export pathways must exist, as least in some organisms (Fig. 1). The β-importin member, Msn5 in yeast, provides one such parallel pathway. Msn5 was discovered by its ability to export particular phosphorylated transcription regulators from the nucleus to the cytoplasm [60]. However, this exportin also binds tRNA [61] as first demonstrated in studies with its vertebrate orthologue, Exportin-5 [62,63]. Exportin-5 also interacts with short RNA duplexes and its major biological role in vertebrate cells appears to be in pre-microRNA (miRNA) export from the nucleus to the cytoplasm, not tRNA nuclear export [64]. It has been suggested that the ability of Exportin-5 to bind tRNA serves as a means to rid the nucleus of the tRNA binding protein, eEF1A, inadvertently trapped in the nucleus upon nuclear reformation after mitosis [64]. Similarly, the Exportin-5 plant orthologue, HASTY, appears not to affect tRNA levels [45]. In contrast, in Drosophila which has no Exp-t othologue, Exportin-5 functions in both miRNA and tRNA nuclear export and in yeast, S. cerevisiae, which does not encode miRNAs, Msn5 serves to export tRNA from the nucleus to the cytoplasm as deletion of MSN5 causes nuclear accumulation of tRNA ([41,61]; Murthi et al., submitted). Curiously, yeast cells lacking both LOS1 and MSN5 are perfectly healthy [41], likely meaning that there is/are yet additional pathways that export tRNA from the nucleus to the cytoplasm (Fig. 1). In sum, tRNA nuclear export pathways that operate redundantly with Exp-t/Los1 exist in some, but not all, organisms, and although Msn5 serves as one such pathway in yeast, its orthologue may not so function in other organisms. Moreover, it is clear that yeast possesses at least one additional undiscovered tRNA nuclear export pathway.
Since Los1 and Msn5 bind only properly structured tRNAs, they serve a proofreading role to export only functional tRNAs to the cytoplasm. However, there may be at least some level of error prone export by either of these pathways or the pathway(s) yet to be identified because pre-tRNA species lacking introns but still containing 5’ or 3’ extension have been detected [65]. Since splicing occurs in the cytoplasm, these tRNAs must have left the nucleus prior to completion of 5’ and/or 3’ processing. The fate of these tRNAs – turnover in the cytoplasm or return to the nucleus via the tRNA retrograde pathway (see below) for repair or turnover - is unknown.
The tRNA retrograde process
Retrograde nuclear accumulation of cytoplasmic tRNA was predicted because in some mutant yeast strains or under some physiological conditions mature spliced tRNAs accumulate in the nuclei [66,67]. Since splicing occurs in the cytoplasm [40], the spliced tRNAs had to have sampled the cytoplasm prior to accumulation in the nucleus. The prediction that tRNAs move after processing in the cytoplasm to the nucleus was verified by employing heterokaryons with two nuclei, only one of which encoded a reporter tRNA. Results documented movement of the reporter tRNA from the shared cytoplasm to both nuclei in the fused cells [41,42] (Fig. 1). It has since been learned that the tRNA retrograde process is energy-dependent [42], rapid [68], reversible (defining the “re-export” step of this process) [68], and responsive to nutrient availability [42,68,69] and tRNA aminoacylation in the nucleus [68,70,71]. The process is conserved and subject to similar regulation in yeast and vertebrate cells [48]. Moreover, it appears that Lenti retroviruses have usurped the tRNA retrograde process as one mechanism to deliver the reverse transcribed genome from the cytoplasm to the nucleus in nondividing neuronal cells [49].
Nuclear accumulation of cytoplasmic tRNA requires the β-importin family member, Mtr10 [42,69] (Fig. 1); however, it is unknown whether Mtr10 directly functions in tRNA nuclear import. Data support the notion that retrograde nuclear import is constitutive. First, tRNA accumulates in both nuclei of heterokaryon cells for which both nuclei possess LOS1 deletions, indicating that tRNA can access nuclei but accumulate in nuclei when export to the cytoplasm is defective in the mutant heterokaryons. Second, although nuclear accumulation of cytoplasmic tRNA only occurs when wild-type cells are nutrient deprived, accumulation occurs if nuclear export is inhibited, even when all nutrients are supplied. Third, an assay was developed to assess whether tRNAs enter the nucleus when cells are nutrient replete. It employs a tRNA modification, Cm32, catalyzed by Trm7 only after tRNA splicing in the cytoplasm. Tethering of Trm7 inside the nucleus provided a means to determine whether tRNA accesses the nucleus from the cytoplasm when cells are fed as well as when they are starved. Supporting the hypothesis that tRNA nuclear import is constitutive, tRNAPhe Cm32 modification occurred under either condition (Murthi et al., submitted).
Since tRNA nuclear import appears to be constitutive, nuclear accumulation of tRNA upon nutrient deprivation must be due to regulated tRNA re-export (Murthi et al., submitted). The signal pathways that communicate nutrient information to tRNA re-export have not been explored in detail, but it is known that the tRNA retrograde process is not affected by defects in the general amino acid control pathway; in contrast, tRNA nuclear accumulation fails to occur appropriately when cells have defects in the PKA pathway or when the TOR pathway is inhibited by rapamycin [68]. Re-export requires tRNA aminoacylation in the nucleus [67,68,70,71].
The tRNA exportins, Los1 and Msn5, appear to serve somewhat different roles in delivering newly synthesized versus imported tRNA to the cytoplasm. As described above, Exp-t and orthologues function in the initial tRNA nuclear export process delivering newly synthesized tRNA to the cytoplasm. The fact that los1Δ cells accumulate intron-containing pre-tRNAs supports the conclusion that the export of newly synthesized tRNA is defective in reaching the cytoplasmic pools of the tRNA splicing machinery in the absence of Los1 [46]. Two lines of data support the idea that Los1 also functions in tRNA re-export. First, nuclear pools of tRNA that accumulate in los1Δ cells depend upon tRNA retrograde import as los1Δ mtr10Δ cells have significantly less nuclear tRNA than los1Δ cells (Murthi et al., submitted). Second, heterokaryons homozygous for los1Δ mutations accumulate nuclear pools of cytoplasmic tRNA in nuclei that do not encode the reporter tRNA [42]. In contrast, since only spliced tRNA accumulates in nuclei of msn5Δ cells, the vast majority of accumulated nuclear tRNA must have been first spliced in the cytoplasm (Murthi et al. submitted). The data support the hypothesis that Msn5 is dedicated to the tRNA re-export process, at least for tRNAs encoded by intron-containing genes (Fig. 1). Why Msn5 apparently only binds to and re-exports spliced tRNA, but not intron-containing tRNA in the initial export step is unknown; it may involve required cooperation with the translation factor, Tef1 (eEF1A) whose subcellular distribution, like tRNA, is dependent upon Mtr10 and Msn5, and whose function is necessary for the tRNA re-export process ([67]; Murthi et al., submitted).
The biological function(s) of the tRNA retrograde process is unknown, but it has been proposed to serve nonexclusive roles including translation in the nucleus, a means to control translation in response to nutrient deprivation, or tRNA proofreading [46]. A role in regulating protein synthesis is consistent with the fact that the process is responsive to nutrient availability and tRNA aminoacylation in the nucleus [70,72]. Although the data support “hiding” tRNAs from cytoplasmic protein synthesis under conditions when translation should be down-regulated, this hypothesis has not been rigorously tested. Supporting the idea that the retrograde process serves a proofreading function to assure that only functional charged tRNAs encounter the protein synthesis machinery is the fact that mutations or conditions that affect pre-tRNA processing, end-repair, or aminoacylation cause retrograde tRNA nuclear accumulation. Constitutive tRNA nuclear import is consistent with such a proofreading role.
tRNA turnover and endonucleolytic digestion in the nucleus and the cytoplasm
Although tRNAs are highly stable molecules, three separate pathways to turnover damaged tRNAs or normal tRNAs when cells are in stressed conditions recently have been described and reviewed (Reviews: [73–75]). Surprisingly, tRNA turnover is not restricted to a single subcellular compartment; rather, pathways exist in both the nucleus and the cytoplasm and can involve a vacuolar enzyme (Table 1). Individual pathways attack tRNA from 3’ to 5 in the nucleus, from 5’ to 3’ in the nucleus or cytoplasm, or by endonucleolytic cleavage in the cytoplasm upon stress.
tRNA nuclear surveillance by the TRAMP complex and the exosome
The yeast TRAMP complex was the first tRNA turnover pathway described and it was discovered by mutations of yeast genes, RRP44 and TRF4, that stabilized hypomodified tRNAMeti missing m1A58 due to mutations of TRM6 or TRM61 that encode the methyltransferase [76]. Loss of m1A58 in tRNAMeti causes altered interaction between the D and T loops; hence this tRNA turnover pathway serves a nuclear surveillance function to assure that only functional appropriately structured tRNAs are delivered to the protein synthesis machinery in the cytoplasm. The pathway includes the nuclear complex of proteins, Trf4, Air1/Air2, and Mtr4, that recognizes hypomodified tRNAMeti, alters it, and activates the nuclear exososme that degrades the hypomodified tRNA in the 3’ to 5’ direction [77]. Air1/Air2 are RNA binding proteins [77]. Mtr4 is an RNA helicase [78], and Trf4/Trf5 are poly (A) polymerases that add short poly(A) tails to the hypomodified tRNA [76,79,80] targeting tRNA for degradation. Rpr44 is a nuclease and a component of the nuclear exosome ([81] and references therein).
Although discovered via hypomodified substrates, it is clear that the TRAMP complex serves a more general function in quality control of tRNAs and other noncoding RNAs. For example, mutant U6 small nuclear RNA and a 3’-truncated 5S rRNA are Trf4 substrates [79]. Furthermore, the TRAMP complex in collaboration with Rex1, a 3’ to 5’ exonuclease, functions in turnover of tRNAs with unprocessed 3’ extensions and likely also CCA turnover [82,83]. The TRAMP complex and the nuclear exosome are conserved and ortholgues to the TRAMP complex proteins exist in Archaea, S. pombe and humans (Review [75]).
A rapid tRNA decay surveillance pathway in both the nucleus and the cytoplasm
Hypomodified tRNAs in yeast are degraded via a second pathway – the rapid tRNA decay (RTD) pathway, discovered by the very fast turnover of tRNAs lacking certain pairs of tRNA modifications [84]. For example, at elevated temperatures tRNAValAAC lacking m7G, normally catalyzed by Trm8, and m5C, normally catalyzed by Trm4, has a half-life typical of mRNAs (ie., minutes), rather than hours or days typical of fully modified tRNAs [84]. The RTD pathway is independent of the TRAMP complex and the nuclear exosome. Rather, genetic analyses of suppressor mutations that correct the temperature sensitive growth caused by tRNA hypomodification uncovered roles for Rat1 and Xrn1, two 5’ to 3’ exonucleases that function in pre-rRNA processing. Interestingly, Rat1 and Xrn1 primarily reside in different subcellular compartments whereas Rat1 is located in the nucleus and Xrn1 is located in the cytoplasm. Because growth at nonpermissive temperatures and tRNA stability are only partially restored by disruption of either RAT1 or XRN1, but are nearly completely restored by double deletions of both RAT1 and XRN1, the RTD pathway is proposed to occur in both the nucleus and the cytoplasm [85].
Endonucleolytic cleavage to tRNA halves upon stress
In addition to the 3’ to 5’ and 5’ to 3’ tRNA turnover pathways that rid cells of faulty tRNAs, cells respond to a variety of stresses by endonucleolytic cleavage of tRNAs in the anticodon loop into halves. First described in nutrient starved Tetrahymena [86], similar responses to oxidative stress have been documented in yeast [87], in stressed mammalian cells and tissues [88,89], and upon nutrient deprivation of bacteria and other protozoa [90,91]. The enzymes that catalyze the endonucleolytic cleavage have been identified in yeast and mammalian cells. In yeast, it is Rny1 [92], a member of the RNase T2 family that normally resides in the vacuole (yeast lysosome equivalent). Upon oxidative stress Rny1 is released from the vacuole to the cytoplasm to act on mature tRNAs residing in the cytoplasm [74,92]. In human cells a similar process is catalyzed by angiogenin, a member of the RNase A family. Angiogenin is a secreted endonuclease that is internalized via endocytosis and then kept in an inactive state by interaction with an inhibitor, RNH1 [88]. Upon stress angiogenin is released to act on tRNAs in the cytoplasm. Since levels of mature tRNAs do not decrease upon stress (summarized in Thompson and Parker, [74]), the function of this endonucleolytic process is not to decrease the levels of cellular tRNAs. Rather, it appears, at least for mammalian cells, that the 5’ tRNA half-molecules inhibit protein synthesis [88]. Other possible roles for the tRNA halves have been described [74]. In sum, the process of cutting tRNAs in halves is catalyzed by secreted or endocytosed endonucleases in the cytoplasm. Discover of the process of cutting tRNAs into half-molecules upon various cellular stresses has likely uncovered new cellular functions for tRNAs.
Summary – tRNA biosynthesis is organized for unique necessities
The tRNA biosynthetic pathways need to satisfy a large combination of constraints not faced by any other RNA family. Foremost, the entire family has to conform to a very narrowly defined structural window to allow proper recognition by the translation machinery. In order to get sufficient sequence diversity to also allow the diversity of amino acid charging needed for the genetic code, an unusually complex series of structural “tweaks” have been evolved in the form of covalent modifications and nucleolytic processing. Early steps in biosynthesis are held in common among most tRNAs, while later ones are often individualized for specific tRNAs. This has apparently led to the spatial organization of the highly diversified biosynthetic pathways as well as multiple turnover routes found in separate subcellular compartments. Dynamic movement of tRNA and tRNA genes within the nucleus and tRNAs between the nucleus and the cytoplasm renders tRNA biosynthesis, function, and turnover unique among eukaryotic functional cellular RNAs. Most of the recent understandings of the cell biology of eukaryotic tRNA biogenesis and response to physiological events have been gained by studies employing yeast. Future studies are required to learn the biological function(s) for separating tRNA biosynthesis, distribution, and turnover among various subcellular locations and whether the tRNA cell biology evidenced for yeast is conserved among other eukaryotes.
Acknowledgements
The authors acknowledge support by NIH grants GM27930 to A.K.H. and GM34869 to D.R.E. and by the NIH University of Michigan Genetics Predoctoral Training Grant (T32 GM07544) to D.A.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pikaard CS, Haag JR, Ream T, Wierzbicki AT. Roles of RNA polymerase IV in gene silencing. Trends Plant Sci. 2008;13:390–397. doi: 10.1016/j.tplants.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paule MR, White RJ. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haeusler RA, Engelke DR. Spatial organization of transcription by RNA polymerase III. Nucleic Acids Res. 2006;34:4826–4836. doi: 10.1093/nar/gkl656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briand JF, Navarro F, Gadal O, Thuriaux P. Cross talk between tRNA and rRNA synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:189–195. doi: 10.1128/MCB.21.1.189-195.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302:1399–1401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 2008;22:2204–2214. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22:2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coughlin DJ, Babak T, Nihranz C, Hughes TR, Engelke DR. Prediction and verification of mouse tRNA gene families. RNA Biol. 2009;6:195–202. doi: 10.4161/rna.6.2.8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pombo A, Jackson DA, Hollinshead M, Wang Z, Roeder RG, Cook PR. Regional specialization in human nuclei: visualization of discrete sites of transcription by RNA polymerase III. Embo J. 1999;18:2241–2253. doi: 10.1093/emboj/18.8.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker SC, Engelke DR. Ribonuclease P: the evolution of an ancient RNA enzyme. Crit Rev Biochem Mol Biol. 2006;41:77–102. doi: 10.1080/10409230600602634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao S, Scott F, Fierke CA, Engelke DR. Eukaryotic ribonuclease P: a plurality of ribonucleoprotein enzymes. Annu Rev Biochem. 2002;71:165–189. doi: 10.1146/annurev.biochem.71.110601.135352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertrand E, Houser-Scott F, Kendall A, Singer RH, Engelke DR. Nucleolar localization of early tRNA processing. Genes Dev. 1998;12:2463–2468. doi: 10.1101/gad.12.16.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishiguro A, Kassavetis GA, Geiduschek EP. Essential roles of Bdp1, a subunit of RNA polymerase III initiation factor TFIIIB, in transcription and tRNA processing. Mol Cell Biol. 2002;22:3264–3275. doi: 10.1128/MCB.22.10.3264-3275.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matera AG, Frey MR, Margelot K, Wolin SL. A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract-binding protein, hnRNP I. J Cell Biol. 1995;129:1181–1193. doi: 10.1083/jcb.129.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobson MR, Cao LG, Taneja K, Singer RH, Wang YL, Pederson T. Nuclear domains of the RNA subunit of RNase P. J Cell Sci. 1997;110(Pt 7):829–837. doi: 10.1242/jcs.110.7.829. [DOI] [PubMed] [Google Scholar]
- 17.Kufel J, Tollervey D. 3'-processing of yeast tRNATrp precedes 5'-processing. RNA. 2003;9:202–208. doi: 10.1261/rna.2145103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuo Y, Deutscher MP. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubrovsky EB, Dubrovskaya VA, Levinger L, Schiffer S, Marchfelder A. Drosophila RNase Z processes mitochondrial and nuclear pre-tRNA 3' ends in vivo. Nucleic Acids Res. 2004;32:255–262. doi: 10.1093/nar/gkh182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hazbun TR, et al. Assigning function to yeast proteins by integration of technologies. Mol Cell. 2003;12:1353–1365. doi: 10.1016/s1097-2765(03)00476-3. [DOI] [PubMed] [Google Scholar]
- 21.Czerwoniec A, Dunin-Horkawicz S, Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Grosjean H, Rother K. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res. 2009;37:D118–D121. doi: 10.1093/nar/gkn710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin NC, Hopper AK. How single genes provide tRNA processing enzymes to mitochondria, nuclei and the cytosol. Biochimie. 1994;76:1161–1167. doi: 10.1016/0300-9084(94)90045-0. [DOI] [PubMed] [Google Scholar]
- 23.Hopper AK, Phizicky EM. tRNA transfers to the limelight. Genes Dev. 2003;17:162–180. doi: 10.1101/gad.1049103. [DOI] [PubMed] [Google Scholar]
- 24.Xing F, Hiley SL, Hughes TR, Phizicky EM. The specificities of four yeast dihydrouridine synthases for cytoplasmic tRNAs. J Biol Chem. 2004;279:17850–17860. doi: 10.1074/jbc.M401221200. [DOI] [PubMed] [Google Scholar]
- 25.De Robertis EM, Black P, Nishikura K. Intranuclear location of the tRNA splicing enzymes. Cell. 1981;23:89–93. doi: 10.1016/0092-8674(81)90273-7. [DOI] [PubMed] [Google Scholar]
- 26.Jiang HQ, Motorin Y, Jin YX, Grosjean H. Pleiotropic effects of intron removal on base modification pattern of yeast tRNAPhe: an in vitro study. Nucleic Acids Res. 1997;25:2694–2701. doi: 10.1093/nar/25.14.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purushothaman SK, Bujnicki JM, Grosjean H, Lapeyre B. Trm11p and Trm112p are both required for the formation of 2-methylguanosine at position 10 in yeast tRNA. Mol Cell Biol. 2005;25:4359–4370. doi: 10.1128/MCB.25.11.4359-4370.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rose AM, Belford HG, Shen WC, Greer CL, Hopper AK, Martin NC. Location of N2,N2-dimethylguanosine-specific tRNA methyltransferase. Biochimie. 1995;77:45–53. doi: 10.1016/0300-9084(96)88103-x. [DOI] [PubMed] [Google Scholar]
- 29.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinemann IU, Soil D, Randau L. Transfer RNA processing in archaea: Unusual pathways and enzymes. FEBS Lett. 2009 doi: 10.1016/j.febslet.2009.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trotta CR, Miao F, Arn EA, Stevens SW, Ho CK, Rauhut R, Abelson JN. The yeast tRNA splicing endonuclease: a tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell. 1997;89:849–858. doi: 10.1016/s0092-8674(00)80270-6. [DOI] [PubMed] [Google Scholar]
- 32.Abelson J, Trotta CR, Li H. tRNA splicing. J Biol Chem. 1998;273:12685–12688. doi: 10.1074/jbc.273.21.12685. [DOI] [PubMed] [Google Scholar]
- 33.Phizicky EM, Consaul SA, Nehrke KW, Abelson J. Yeast tRNA ligase mutants are nonviable and accumulate tRNA splicing intermediates. J Biol Chem. 1992;267:4577–4582. [PubMed] [Google Scholar]
- 34.Culver GM, McCraith SM, Zillmann M, Kierzek R, Michaud N, LaReau RD, Turner DH, Phizicky EM. An NAD derivative produced during transfer RNA splicing: ADP-ribose 1"–2" cyclic phosphate. Science. 1993;261:206–208. doi: 10.1126/science.8392224. [DOI] [PubMed] [Google Scholar]
- 35.Mitchell M, Xue S, Erdman R, Randau L, Soll D, Li H. Crystal structure and assembly of the functional Nanoarchaeum equitans tRNA splicing endonuclease. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harding HP, Lackey JG, Hsu HC, Zhang Y, Deng J, Xu RM, Damha MJ, Ron D. An intact unfolded protein response in Trpt1 knockout mice reveals phylogenic divergence in pathways for RNA ligation. RNA. 2008;14:225–232. doi: 10.1261/rna.859908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paushkin SV, Patel M, Furia BS, Peltz SW, Trotta CR. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3' end formation. Cell. 2004;117:311–321. doi: 10.1016/s0092-8674(04)00342-3. [DOI] [PubMed] [Google Scholar]
- 38.Lund E, Dahlberg JE. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science. 1998;282:2082–2085. doi: 10.1126/science.282.5396.2082. [DOI] [PubMed] [Google Scholar]
- 39.Clark MW, Abelson J. The subnuclear localization of tRNA ligase in yeast. J Cell Biol. 1987;105:1515–1526. doi: 10.1083/jcb.105.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshihisa T, Yunoki-Esaki K, Ohshima C, Tanaka N, Endo T. Possibility of cytoplasmic pre-tRNA splicing: the yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol Biol Cell. 2003;14:3266–3279. doi: 10.1091/mbc.E02-11-0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takano A, Endo T, Yoshihisa T. tRNA actively shuttles between the nucleus and cytosol in yeast. Science. 2005;309:140–142. doi: 10.1126/science.1113346. [DOI] [PubMed] [Google Scholar]
- 42.Shaheen HH, Hopper AK. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2005;102:11290–11295. doi: 10.1073/pnas.0503836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 44.Englert M, Latz A, Becker D, Gimple O, Beier H, Akama K. Plant pre-tRNA splicing enzymes are targeted to multiple cellular compartments. Biochimie. 2007;89:1351–1365. doi: 10.1016/j.biochi.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 45.Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS. Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci U S A. 2005;102:3691–3696. doi: 10.1073/pnas.0405570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hopper AK, Shaheen HH. A decade of surprises for tRNA nuclear-cytoplasmic dynamics. Trends Cell Biol. 2008;18:98–104. doi: 10.1016/j.tcb.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 47.Duchene AM, Pujol C, Marechal-Drouard L. Import of tRNAs and aminoacyl-tRNA synthetases into mitochondria. Curr Genet. 2009;55:1–18. doi: 10.1007/s00294-008-0223-9. [DOI] [PubMed] [Google Scholar]
- 48.Shaheen HH, Horetsky RL, Kimball SR, Murthi A, Jefferson LS, Hopper AK. Retrograde nuclear accumulation of cytoplasmic tRNA in rat hepatoma cells in response to amino acid deprivation. Proc Natl Acad Sci U S A. 2007;104:8845–8850. doi: 10.1073/pnas.0700765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zaitseva L, Myers R, Fassati A. tRNAs promote nuclear import of HIV-1 intracellular reverse transcription complexes. PLoS Biol. 2006;4:e332. doi: 10.1371/journal.pbio.0040332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terry LJ, Shows EB, Wente SR. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318:1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- 51.Arts GJ, Kuersten S, Romby P, Ehresmann B, Mattaj IW. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J. 1998;17:7430–7441. doi: 10.1093/emboj/17.24.7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E, Gorlich D. Identification of a tRNA-specific nuclear export receptor. Mol Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- 53.Hunter CA, Aukerman MJ, Sun H, Fokina M, Poethig RS. PAUSED encodes the Arabidopsis exportin-t ortholog. Plant Physiol. 2003;132:2135–2143. doi: 10.1104/pp.103.023309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarkar S, Hopper AK. tRNA nuclear export in saccharomyces cerevisiae: in situ hybridization analysis. Mol Biol Cell. 1998;9:3041–3055. doi: 10.1091/mbc.9.11.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hellmuth K, Lau DM, Bischoff FR, Kunzler M, Hurt E, Simos G. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol Cell Biol. 1998;18:6374–6386. doi: 10.1128/mcb.18.11.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cook AG, Fukuhara N, Jinek M, Conti E. Structures of the tRNA export factor in the nuclear and cytosolic states. Nature. 2009;461:60–65. doi: 10.1038/nature08394. [DOI] [PubMed] [Google Scholar]
- 57.Lipowsky G, Bischoff FR, Izaurralde E, Kutay U, Schafer S, Gross HJ, Beier H, Gorlich D. Coordination of tRNA nuclear export with processing of tRNA. RNA. 1999;5:539–549. doi: 10.1017/s1355838299982134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghavidel A, Kislinger T, Pogoutse O, Sopko R, Jurisica I, Emili A. Impaired tRNA nuclear export links DNA damage and cell-cycle checkpoint. Cell. 2007;131:915–926. doi: 10.1016/j.cell.2007.09.042. [DOI] [PubMed] [Google Scholar]
- 59.Qiu H, Hu C, Anderson J, Bjork GR, Sarkar S, Hopper AK, Hinnebusch AG. Defects in tRNA processing and nuclear export induce GCN4 translation independently of phosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol Cell Biol. 2000;20:2505–2516. doi: 10.1128/mcb.20.7.2505-2516.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaffman A, Rank NM, O'Neill EM, Huang LS, O'Shea EK. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature. 1998;396:482–486. doi: 10.1038/24898. [DOI] [PubMed] [Google Scholar]
- 61.Shibata S, Sasaki M, Miki T, Shimamoto A, Furuichi Y, Katahira J, Yoneda Y. Exportin-5 orthologues are functionally divergent among species. Nucleic Acids Res. 2006;34:4711–4721. doi: 10.1093/nar/gkl663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bohnsack MT, Regener K, Schwappach B, Saffrich R, Paraskeva E, Hartmann E, Gorlich D. Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J. 2002;21:6205–6215. doi: 10.1093/emboj/cdf613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Calado A, Treichel N, Muller EC, Otto A, Kutay U. Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J. 2002;21:6216–6224. doi: 10.1093/emboj/cdf620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 65.O'Connor JP, Peebles CL. PTA1, an essential gene of Saccharomyces cerevisiae affecting pre-tRNA processing. Mol Cell Biol. 1992;12:3843–3856. doi: 10.1128/mcb.12.9.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feng W, Hopper AK. A Los1p-independent pathway for nuclear export of intronless tRNAs in Saccharomycescerevisiae. Proc Natl Acad Sci U S A. 2002;99:5412–5417. doi: 10.1073/pnas.082682699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grosshans H, Hurt E, Simos G. An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes Dev. 2000;14:830–840. [PMC free article] [PubMed] [Google Scholar]
- 68.Whitney ML, Hurto RL, Shaheen HH, Hopper AK. Rapid and reversible nuclear accumulation of cytoplasmic tRNA in response to nutrient availability. Mol Biol Cell. 2007;18:2678–2686. doi: 10.1091/mbc.E07-01-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hurto RL, Tong AH, Boone C, Hopper AK. Inorganic phosphate deprivation causes tRNA nuclear accumulation via retrograde transport in Saccharomyces cerevisiae. Genetics. 2007;176:841–852. doi: 10.1534/genetics.106.069732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sarkar S, Azad AK, Hopper AK. Nuclear tRNA aminoacylation and its role in nuclear export of endogenous tRNAs in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1999;96:14366–14371. doi: 10.1073/pnas.96.25.14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gu W, Hurto RL, Hopper AK, Grayhack EJ, Phizicky EM. Depletion of Saccharomyces cerevisiae tRNA(His) guanylyltransferase Thg1p leads to uncharged tRNAHis with additional m(5)C. Mol Cell Biol. 2005;25:8191–8201. doi: 10.1128/MCB.25.18.8191-8201.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Azad AK, Stanford DR, Sarkar S, Hopper AK. Role of nuclear pools of aminoacyl-tRNA synthetases in tRNA nuclear export. Mol Biol Cell. 2001;12:1381–1392. doi: 10.1091/mbc.12.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 74.Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009;138:215–219. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 75.Reinisch KM, Wolin SL. Emerging themes in non-coding RNA quality control. Curr Opin Struct Biol. 2007;17:209–214. doi: 10.1016/j.sbi.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 76.Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 78.Wang X, Jia H, Jankowsky E, Anderson JT. Degradation of hypomodified tRNA(iMet) in vivo involves RNA-dependent ATPase activity of the DExH helicase Mtr4p. RNA. 2008;14:107–116. doi: 10.1261/rna.808608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kadaba S, Wang X, Anderson JT. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA. 2006;12:508–521. doi: 10.1261/rna.2305406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vanacova S, et al. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schneider C, Leung E, Brown J, Tollervey D. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res. 2009;37:1127–1140. doi: 10.1093/nar/gkn1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Copela LA, Fernandez CF, Sherrer RL, Wolin SL. Competition between the Rex1 exonuclease and the La protein affects both Trf4p-mediated RNA quality control and pre-tRNA maturation. RNA. 2008;14:1214–1227. doi: 10.1261/rna.1050408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ozanick SG, Wang X, Costanzo M, Brost RL, Boone C, Anderson JT. Rex1p deficiency leads to accumulation of precursor initiator tRNAMet and polyadenylation of substrate RNAs in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:298–308. doi: 10.1093/nar/gkn925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell. 2006;21:87–96. doi: 10.1016/j.molcel.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 85.Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5'-3' exonucleases Rat1 and Xrn1. Genes Dev. 2008;22:1369–1380. doi: 10.1101/gad.1654308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee SR, Collins K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem. 2005;280:42744–42749. doi: 10.1074/jbc.M510356200. [DOI] [PubMed] [Google Scholar]
- 87.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fu H, et al. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583:437–442. doi: 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 90.Haiser HJ, Karginov FV, Hannon GJ, Elliot MA. Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor. Nucleic Acids Res. 2008;36:732–741. doi: 10.1093/nar/gkm1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li Y, Luo J, Zhou H, Liao JY, Ma LM, Chen YQ, Qu LH. Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 2008;36:6048–6055. doi: 10.1093/nar/gkn596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thompson DM, Parker R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol. 2009;185:43–50. doi: 10.1083/jcb.200811119. [DOI] [PMC free article] [PubMed] [Google Scholar]