Abstract
Enterocytozoon bieneusi is a unicellular enteric fungal pathogen and the most common cause of human microsporidiosis. The frequent detection of this organism in animals, including companion animals, livestock and wildlife, has raised the question of the importance of animal reservoirs in the epidemiology of this pathogen. A partial sequence of the ribosomal internal transcribed spacer (ITS) has been widely used as a genetic marker for studying the molecular epidemiology of E. bieneusi. With the aim of comparing E. bieneusi ITS genotypes originating from different host species, and assess the potential for zoonotic transmission, E. bieneusi ITS sequences retrieved from GenBank were analyzed using two metrics of diversity, rarefaction and phylogenetic distance. In spite of the human ITS sample being geographically more diverse, ITS sequence diversity in animals exceeded that of humans. In both host groups much of the ITS diversity remains to be sampled. Using quantitative phylogenetic tests we found evidence for a partial but significant segregation of E. bieneusi ITS sequences according to host species. Host-specific segregation was confirmed by hierarchical analysis of molecular variation. To improve our understanding of the epidemiology of human microsporidiosis and strengthen the study of E. bieneusi populations, efforts to genotype additional E. bieneusi isolates from wildlife and companion animals should be prioritized and the geographic and species diversify of animal samples should be increased. Due to the possibility of genetic recombination in this species, additional unlinked genetic markers need to be developed and included in future studies.
Keywords: Enterocytozoon bieneusi, microsporidiosis, rarefaction, phylogeny, AMOVA
1. INTRODUCTION
Based on the sequence of a small number of loci (Hirt et al., 1999; Keeling, 2003) the microsporidia are thought to represent an early-diverging branch of the Fungi (James et al., 2006). Several unique characteristics are shared by Microsporidia including the presence of a single ribosomal internal transcribed spacer (ITS), the lack of mitochondria, and the presence of a polar tube serving to propel the spore content into the host cell. Microsporidia are parasitic, and one species, E. bieneusi, is the most common cause of human intestinal microsporidiosis and an opportunistic infection in AIDS.
The epidemiology of E. bieneusi, and in particular the importance of zoonotic transmission, remains unclear. The study of the molecular epidemiology of human microsporidiosis has been driven by surveys of geographically restricted parasite populations collected from humans (Breton et al., 2007; Leelayoova et al., 2006; Rinder et al., 1997; Sulaiman et al., 2003a; ten Hove et al., 2009; Tumwine et al., 2002), livestock (Buckholt et al., 2002; Dengjel et al., 2001; Mathis et al., 1999; Santin et al., 2005; Sulaiman et al., 2004), or wildlife species (Lobo et al., 2006; Sulaiman et al., 2003b). These studies are all based on the sequence of the ribosomal ITS and have contributed to a growing collection of sequences of this locus (Santin and Fayer, 2009). Comparisons of E. bieneusi ITS genotypes from different host species have revealed an apparent lack of host specificity, as ITS genotypes are shared among human and animal hosts. These observation have been interpreted as evidence of zoonotic transmission (Drosten et al., 2005), a view supported by experiments demonstrating transmission between different host species (Feng et al., 2006; Kondova et al., 1998; Tzipori et al., 1997).
As a consequence of several sequencing efforts initiated over 10 years ago (Rinder et al., 1997), a relative large number of ITS sequences have been deposited in GenBank. These sequence data are far from being random in space or with respect to host origin, but pooling data from disparate surveys into a global collection enables testing for the presence of discrete ITS populations, particularly as they relate to host species.
Although a partial sequence of the E. bieneusi genome is available (Akiyoshi et al., 2009), and the sequence of a few other loci has been obtained from multiple isolates (Akiyoshi et al., 2007), the ITS is the only marker which has been used for studying the molecular epidemiology of this species. The reliance on a single locus constrains the interpretation of the data, particularly for an organism such as E. bieneusi for which information on the occurrence of sexual recombination is lacking. In the absence of recombination a single genetic marker would be adequate for studying the epidemiology of this pathogen, but if genetically distinct genotypes recombine in nature, extrapolating from a single locus to the entire genome may lead to the wrong conclusions.
Bearing in mind the limitations of the E. bieneusi ITS database and the constrains of the single-locus typing method, we report here an analysis of the GenBank ITS sequence collection using diversity analysis, phylogenetic tests and analysis of molecular variation (AMOVA). The results indicate host-specific structuring of the E. bieneusi ITS diversity.
2. MATERIALS AND METHODS
2.1. Sequences
E. bieneusi ITS sequences were downloaded from GenBank and aligned with Clustal W accessed through the Accessory Applications menu of BioEdit (Hall, 1999). Sequences were trimmed to 243 nucleotides corresponding to nucleotide position 56–298 in GenBank sequence AB359945 as this region is present in a majority of GenBank entries. Sequences which did not span this region were excluded. A total of 135 sequences were retained including 68 sequences from human infections, 16 sequences of bovine origin, 14 sequences from infected pigs and 15 sequences from cats and dogs combined. Unless stated otherwise, bovine, porcine, feline and canine hosts are referred to as “livestock”. An additional 22 sequences came from E. bieneusi isolated from wildlife, including raccoons, muskrats, beavers, marmosets and birds.
2.2. Sequence analysis
Rarefaction analysis (Gotelli and Colwell, 2001; Sanders, 1968) was used to compare the diversity among E. bieneusi ITS sequences originating from different hosts. Aligned ITS sequences were exported in FASTA format to Microsoft Excel and sorted such that identical sequences were located on adjacent rows. Each unique sequence was then numbered and sequence/sample/abundance combinations saved in plain text format as described in EstimateS User Guide at http://viceroy.eeb.uconn.edu/EstimateS and by Hughes and Hellmann (Hughes and Hellmann, 2005). In the User Guide this format is referred to as “Format 3”. Individual-based analytical rarefaction estimates and their standard deviation were calculated with this program.
A phylogenetic tree was constructed by importing aligned ITS sequences (n=135) into Mega 4.1 (Tamura et al., 2007) downloaded from http://megasoftware.net/. Evolutionary distances were computed with the Maximum composite Likelihood method (Tamura et al., 2004).
The program Unifrac (Lozupone et al., 2006) was used to investigate the extent and statistical significance of any clustering of E. bieneusi ITS sequences according to host species. Unifrac tests whether related sequences cluster according to a user-defined property, such as the environment from which they were obtained. The test measures the phylogenetic distance among populations by comparing the lengths of the branches in a phylogenetic tree which are unique to an environment to the total of the branch lengths in the tree. To determine the statistical significance of the fraction of unique branch length, the environments are randomized and the distribution of the Unifrac Distances (UD) over many replicates determined. The p value represents the number of randomized trees that generate UD values greater than the value obtained from the experimental tree. Here, we extend Unifrac’s application to testing whether E. bieneusi ITS sequences cluster according to host species. The web interface of the program at http://bmf2.colorado.edu/unifrac/index.psp was used. The program requires the input of two files: a tree file in Newick format (http://evolution.genetics.washington.edu/phylip/newick_doc.html) encoding a phylogenetic tree generated from the aligned sequences, and an “environmental file” described below. To create the tree file, a pairwise distance matrix was calculated from ITS sequences aligned with BioEdit using the Dnadist program of PHYLIP (Felsenstein, J. 2005. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle) accessed from the BioEdit Accessory Applications menu. The distance matrix was saved in text format and input into the Neighbor program or Fitch program of the PHYLIP package. Trees (Saitou and Nei, 1987) were drawn with these programs using the default values, except that an outgroup sequence was defined (see below). The file output by these programs was then uploaded into Unifrac. Also uploaded was the “environmental file” which is a tab-delimited text file linking the designation of each sequence (in this case the GenBank accession number) with the “environment”, i.e., the host species or group of host species (i.e. livestock) from which each E. bieneusi sequence originated. The outgroup was excluded. An Unifrac Distance (UD) of 1 indicates complete segregation, whereas a distance of zero is the theoretical lower bound of the UD obtained if sequences and environment (i.e., host) are not associated.
Consistent with the extensive divergence among the Microsporidia (James et al., 2006), no microsporidian ITS from GenBank could be aligned with the E. bieneusi ITS. Even ITS sequences from other Enterocytozoon or Encephalitozoon species were too divergent. To satisfy Unifrac’s need for phylogenetic trees rooted with an outgroup, an artificial outgroup sequence was created by combining 100-bp fragments from three E. bieneusi ITS identified by the following GenBank accession numbers: EF014430, DQ683757, DQ885585. These sequences were chosen because they are located on the most divergent branches of the E. bieneusi ITS phylogeny. In addition, random SNPs were introduced at ~100-bp intervals to ensure that the artificial outgroup would not group with any E. bieneusi ITS branch. To ensure that the artificial outgroup did not affect the results, the analysis was replicated with a different outgroup, which was derived from the Nucleospora salmonis ITS. Because the genetic distance between E. bieneusi ITS sequences and that of N. salmonis exceeded the threshold set by Dnadist, the distance was reduced by manually “mutating” 30% of the N. salmonis positions to that found in E. bieneusi AF242475 of human origin. The divergence of the outgroups with respect to the population of E. bieneusi ITS sequences was individually verified by inspecting a Neighbor Joining tree drawn from all ITS sequences plus the outgroup generated in silico. Finally, UDs were converted into a “heatmap” using a Microsoft Excel macro written by Yukihiro Yabuta downloaded from http://homepage.mac.com/yabyab/my.html. UDs were color coded such that the largest value is represented with the warmest color (red), and smaller distances with colder colors. If present, an UD of 0 would be shown in deep blue. Principal Coordinate Analysis (PCoA) as implemented in the Unifrac program was used to cluster the host-associated ITS sequences. Host-specific segregation of ITS alleles was also investigated with a 2-level hierarchical AMOVA (Excoffier et al., 2005). A model of four groups corresponding to ITS from human, bovine, canine/feline and porcine E. bieneusi isolates was tested.
3. RESULTS
3.1. Analysis of E. bieneusi ITS diversity
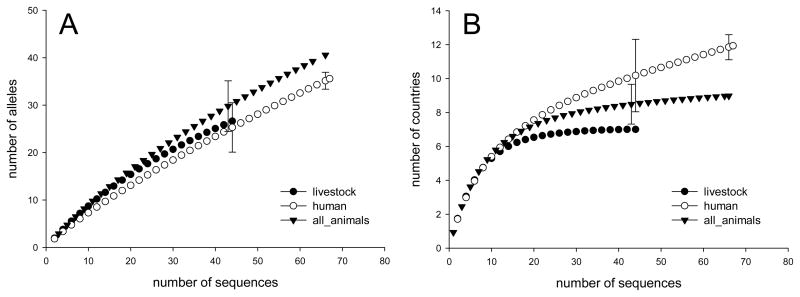
Rarefaction is commonly used to compare taxonomic diversity among samples of different size. To assess whether ITS diversity differs among host species, rarefaction curves were plotted for sequences originating from humans, livestock and all the other animal hosts combined (Fig. 1A). The graph shows that many of the ITS sequences collected to date are unique. The steep slopes and the lack of a plateau indicate that much of ITS diversity remains to be sampled.
Fig. 1.
Rarefaction analysis comparing the diversity of E. bieneusi ITS isolated various host species. A) Rarefaction analysis indicates that ITS diversity in E. bieneusi isolated from animals exceeds that of E. bieneusi from humans. B) Comparison of geographical diversity for ITS originating from different host species demonstrates that lower ITS diversity in humans is not a result of a smaller geographical diversity, as human geographical diversity exceeds that of livestock and animals.
Using rarefaction analysis we compared the ITS diversity among parasites originating from different host species. A total of 41 ITS genotypes were found in the 13 animal species (livestock and wildlife) represented in the database. This sample displays greater taxonomic richness than sequences from humans (n=67, 35.1 genotypes; 95% confidence interval (C.I.) 33.7–36.9), but did not differ significantly from the diversity found in each of the four livestock species (cattle, pig, cat and dog). Rarefied to the livestock sample size of 45, the estimated ITS richness for all animal species combined was 30.3. The 95% C.I. of 24.7–35.6 genotypes includes the livestock ITS richness of 27, indicating a lack of significant difference. Similarly, the rarefied human E. bieneusi ITS diversity of 25.3 (95% C.I. 20.1–30.6) is not significantly different from that of the livestock samples. The main conclusion we are drawing from this analysis is that E. bieneusi ITS sequences in humans are less diverse than in all animals combined. Among the three groups of host species, non-livestock animal host (wildlife) are the smallest groups (n=22) and were therefore not analyzed separately.
Because the geographical diversity of the ITS host-specific groups could impact the analyses, the geographical diversity of human, animals, and livestock ITS collections were compared (Fig. 1B) using the same approach. Rarefied to the animal sample size of 67, the geographic diversity of human ITS (11.8 countries, 95% C.I. 11.1–12.6) was significantly higher that that of the ITS sequences originating from all animals combined (9 countries). If the human and animal groups are rarefied to 45, the livestock sample size, the human and the animal samples were geographically more diverse than the former group which originated from 7 countries only. We infer that lack of geographic diversity is unlikely to be the cause of the lower ITS diversity found in human infections. To the contrary, the higher taxonomic diversity in animals was observed in spite of this group being geographically significantly less diverse than the human sample. We also note that the country rarefaction curves have reached, or are close to a plateau, particularly in the case of the animal collections. This indicates a lack of geographic diversity in these two groups, which is particularly pronounced in the collection from livestock and animals in general.
3.2. Comparison of E. bieneusi ITS originating from different host species
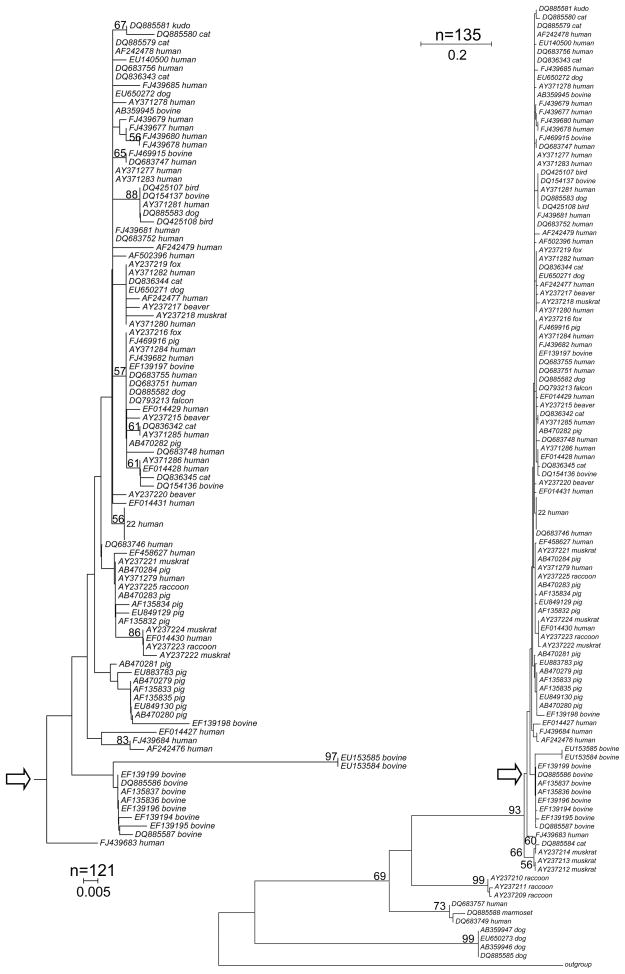
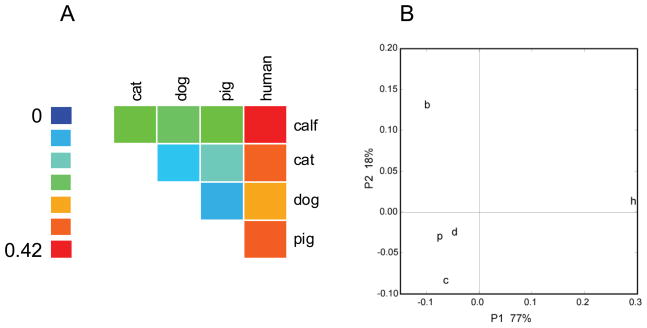
Phylogenetic trees (Fig. 2) are commonly drawn to visualize the relationship among sequences. Several studies have used such methods to visualize the relations among E. bieneusi ITS sequences from different collections. A limitation of this approach is that evolutionary trees are typically interpreted based on visual inspection. To test whether there is a statistically significant difference among E. bieneusi ITS sequences isolated from different hosts, we applied the Unifrac method (Lozupone et al., 2006) and the “Phylogenetic Test” (P test) (Martin, 2002) to the entire collection of 135 GenBank entries. To rule out bias due to ITS sequences from rare or highly divergent host species, these analyses were performed with different samples; i.e., the complete sample of 135 ITS sequences, the human and livestock subsample of 113 sequences, a human and lifestock subsample of 109 sequences which excludes the four most divergent dog samples, and a subsample of 99 sequences originating from the most abundant hosts species, human, bovine, canine and feline. To facilitate the interpretation, the UDs are represented as a heat map and a cluster diagram (Fig. 3). In a comparison of sequences from humans and each animal subgroup (wildlife excluded) UD values exceeded those among animal groups. UD between human and bovine sequences was the largest (0.42), followed by human-cat (0.38), human-pig (0.38) and human-dog (0.35). In contrast, comparisons among the four animal subgroups yielded smaller UD ranging from 0.08 for cat-dog and pig-dog, to 0.22 for cat-calf. Although none of the human-animal UD approaches the maximum value of 1 which would indicate complete separation, the higher distance in human-animal comparisons is apparent (Fig. 3A). A similar picture emerged from a Principal Coordinate Analysis (PCoA), which shows a pig/dog/cat cluster which is well separated from the bovine group and even more distant from the human group (Fig. 3B). When all host species were tested together, both Unifrac and P test revealed a significant clustering by host (p<0.01), regardless whether all 135 sequences were included, or only human and livestock (n=113), or human and livestock minus pigs (n=99) were analyzed. In addition we also tested whether removal of four E. bieneusi ITS sequences from dogs (AB359947, AB359946, EU650273, DQ885585) would affect the results. Because these sequences are divergent from the remaining sequences (Fig. 2), we wanted to ensure that Unifrac significance was not a result of the long single-species branch formed by these four sequences. The results of the analysis of the remaining 109 sequences (113−4=109) again showed significant clustering by host (p=0.01). PCoA with n=109 generated a similar clustering as observed when 113 sequences were included (not shown). The choice of outgroup (see Materials and Methods) did not affect these results, nor did the algorithm used to draw the phylogenetic tree. Using a Fitch-Margoliash tree (as opposed to Neighbor-Joining) led to same conclusion. UD values and P-test thus are indicative of partial host-specificity among E. bieneusi ITS sequences, particularly with respect to sequences originating from humans and animals.
Fig. 2.
Neigbor-Joining tree of 135 E. bieneusi ITS sequences. The percentage of trees displaying a specific branch in 500 replicates is shown if >50%. A total of 220 nucleotide positions were included in the analysis. The scales show the number of substitutions per site. The inset (left) shows a subtree of 121 sequences. The point of attachment of the subtree to the main tree is marked with arrows. Twenty-two identical sequences of human origin were collapsed to inprove clarity of the image.
Fig. 3.
Unifrac distances among 113 E. bieneusi ITS sequences originating from humans and four livestock species. A) Heatmap of UD: larger UD (more divergent populations) are represented in warm colors. The largest UD is 0.42 (human - bovine) and is indicated in red. The smallest distance (0.08) separates dog - cat and dog – pig populations and is shown in light blue. The color scale is shown left. B) Principal Coordinate Analysis of the UD matrix of 5 host species indicates that E. bieneusi ITS from pigs (p), dogs (d) and cats (c) form a cluster which is distinct from bovine (b) and human (h) sequences. The X and Y axis show the first and second coordinate, respectively, and the associated variation in percent.
To verify the results obtained with Unifrac, AMOVA was used to test the model of four (human, bovine, feline, canine) host-associate E. bieneusi ITS groups. Consistent with the above analyses, variation among groups was highly significant (p≈0), regardless whether the four divergent sequences from dogs were included or omitted (Table 1). In the AMOVA of 113 human and livestock sequences, variation among groups accounted for 23.8% of total variation. As expected based on their significant sequence divergence, removal of the four sequences from dogs lowered the percentage of variation among populations, which now accounted for 16.7% of the total.
Table 1.
Test of genetic structure of E. bieneusi ITS based on a model of four host-associated groups (humans, calves, pigs, cats/dogs).
| n=113 | n=109b | |||||||
|---|---|---|---|---|---|---|---|---|
| source of variation | d.f.a | sum of squares | variance components | % variation | d.f. | sum of squares | variance components | % variation |
| among populations | 3 | 161.2 | 2.1 | 23.8 | 3 | 58.5 | 0.8 | 16.7 |
| within population | 109 | 744.1 | 6.8 | 76.2 | 105 | 401.4 | 3.8 | 83.3 |
| total | 112 | 905.3 | 8.9 | 108 | 459.9 | 4.6 | ||
d.f., degrees of freedom.
excludes a cluster of four divergent ITS sequences from infected dogs (see text).
4. DISCUSSION
Our understanding of the epidemiology of human microsporidiosis is still superficial and will evolve as more samples, particularly sequences originating from currently under-sampled host species and geographical locations, are analyzed. The present study represents a preliminary analysis of global E. bieneusi ITS diversity in different host species based on statistical methods to test for host-specific segregation. The main limitations of these analyses is that only one locus is available and that some host species, which may potentially be important for understanding transmission, are missing or are under-sampled. The lack of geographical diversity, particularly for the animal sequences, was already mentioned above in the context of the rarefaction analysis.
The diversity of E. bieneusi is expected to increase as additional loci are included, particularly if recombinant genotypes are found. If the life cycle of E. bieneusi includes a sexual phase, the ITS genotypes by themselves would not be sufficient as isolates carrying identical ITS alleles may differ at other loci. In the absence of genetic recombination the global E. bieneusi population could be viewed as a collection of clonally reproducing genotypes which can be adequately characterized with a single marker. However, evidence of meiotic recombination in other microsporidia, such as Kneallhazia solenopsae, a parasite of fire ants (Sokolova and Fuxa, 2008), Andreanna caspii, a mosquito parasite (Simakova et al., 2008), and Vairimorpha, parasites of Lepidoptera (Jouvenaz and Ellis, 1986), indicates that sexual recombination in E. bieneusi should be investigated. The fact that sex is widespread among fungi supports this argument. Examining the possibility of genetic recombination will require the identification of unlinked polymorphic markers and the development of multilocus genotyping tools. Multilocus genotypes would reveal the extent of linkage disequilibrium in E. bieneusi populations, and enable other population biology analyses. On the other hand, a purely asexual mode of reproduction would imply that genetic diversity can be captured by typing a single locus. Moving to a multilocus genotyping method would be particularly productive if all participating laboratories were to adopt the same markers. The recent evidence of a unified approach to E. bieneusi ITS nomenclature (Santin and Fayer, 2009) indicates that a coordinated effort may also be possible for expanding from the current single-locus method to a multi-locus system. Country rarefaction curves indicate that sampling efforts aiming at diversifying the geography of animal collection should be prioritized.
It is unknown to what extent the ITS sequence is under selection and whether any of the polymorphisms included in the analyses are the result of positive selection. In addition, since it is unknown if E. bieneusi genomes recombine, speculating on the effect that such polymorphisms may have on the parasite’s phenotype would be premature.
The detection of significant clustering of ITS in phylogenetic tests and AMOVA, and the difference in taxonomic richness between humans and animal hosts indicates that ITS alleles are not distributed randomly among host species. Consistent with previously published ITS phylogenies (Drosten et al., 2005), UD values smaller than 1 indicate partial segregation by host. This conclusion was also inferred by others on the basis of sequence comparisons (Cama et al., 2007; Dengjel et al., 2001; Leelayoova et al., 2008; Mathis et al., 2005; Santin et al., 2005; Sulaiman et al., 2004). Recently, Ten Hove and co-workers found that in a group of 57 isolates from the Netherlands and Malawi ITS sequences isolated from pigs and cattle formed separate clusters, and that E. bieneusi from immunosuppressed individuals shared the ITS genotype C (ten Hove et al., 2009). Together with the present analysis of a global ITS collection, these studies are consistent with partial segregation of ITS genotypes according to host and with the occurrence of zoonotic transmission. If future multilocus genotypes confirm this model, it is conceivable that E. bieneusi may comprise genotypes with different host range, some infecting only certain species and other circulating among a wider range of host species, including humans. Improved genotyping methods, in particular multilocus genotyping protocols, and the expansion of genotype information to other hosts and countries will be needed to further dissect the epidemiology of human microsporidiosis.
Supplementary Material
Acknowledgments
Financial support from the NIAID (grants R01AI052781 and R21AI064118) is gratefully acknowledged. Our thanks to Zijin Wu for assistance with data analysis and Alejandro Greenberg for critical comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akiyoshi DE, Weiss LM, Feng X, Williams BA, Keeling PJ, Zhang Q, Tzipori S. Analysis of the beta-tubulin genes from Enterocytozoon bieneusi isolates from a human and rhesus macaque. J Eukaryot Microbiol. 2007;54:38–41. doi: 10.1111/j.1550-7408.2006.00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi DE, Morrison HG, Lei S, Feng X, Zhang Q, Corradi N, Mayanja H, Tumwine JK, Keeling PJ, Weiss LM, Tzipori S. Genomic survey of the non-cultivatable opportunistic human pathogen, Enterocytozoon bieneusi. PLoS Pathog. 2009;5:e1000261. doi: 10.1371/journal.ppat.1000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton J, Bart-Delabesse E, Biligui S, Carbone A, Seiller X, Okome-Nkoumou M, Nzamba C, Kombila M, Accoceberry I, Thellier M. New highly divergent rRNA sequence among biodiverse genotypes of Enterocytozoon bieneusi strains isolated from humans in Gabon and Cameroon. J Clin Microbiol. 2007;45:2580–2589. doi: 10.1128/JCM.02554-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckholt MA, Lee JH, Tzipori S. Prevalence of Enterocytozoon bieneusi in swine: an 18-month survey at a slaughterhouse in Massachusetts. Appl Environ Microbiol. 2002;68:2595–2599. doi: 10.1128/AEM.68.5.2595-2599.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cama VA, Pearson J, Cabrera L, Pacheco L, Gilman R, Meyer S, Ortega Y, Xiao L. Transmission of Enterocytozoon bieneusi between a child and guinea pigs. J Clin Microbiol. 2007;45:2708–2710. doi: 10.1128/JCM.00725-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengjel B, Zahler M, Hermanns W, Heinritzi K, Spillmann T, Thomschke A, Loscher T, Gothe R, Rinder H. Zoonotic potential of Enterocytozoon bieneusi. J Clin Microbiol. 2001;39:4495–4499. doi: 10.1128/JCM.39.12.4495-4499.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C, Laabs J, Kuhn EM, Schottelius J. Interspecies transmission of Enterozytozoon bieneusi supported by observations in laboratory animals and phylogeny. Med Microbiol Immunol. 2005;194:207–209. doi: 10.1007/s00430-005-0240-y. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol Bioinf Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Feng X, Akiyoshi DE, Sheoran A, Singh I, Hanawalt J, Zhang Q, Widmer G, Tzipori S. Serial propagation of the microsporidian Enterocytozoon bieneusi of human origin in immunocompromised rodents. Infect Immun. 2006;74:4424–4429. doi: 10.1128/IAI.00456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotelli NJ, Colwell RK. Quantifying biodiversity: procedures and pitfalls in the measurement and comparison of species richness. Ecology Letters. 2001;4:379–391. [Google Scholar]
- Hall T. BioEdit, a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Res Ser. 1999;41:95–98. [Google Scholar]
- Hirt RP, Logsdon JM, Jr, Healy B, Dorey MW, Doolittle WF, Embley TM. Microsporidia are related to Fungi: evidence from the largest subunit of RNA polymerase II and other proteins. Proc Natl Acad Sci U S A. 1999;96:580–585. doi: 10.1073/pnas.96.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JB, Hellmann JJ. The application of rarefaction techniques to molecular inventories of microbial diversity. Methods Enzymol. 2005;397:292–308. doi: 10.1016/S0076-6879(05)97017-1. [DOI] [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, Lumbsch HT, Rauhut A, Reeb V, Arnold AE, Amtoft A, Stajich JE, Hosaka K, Sung GH, Johnson D, O’Rourke B, Crockett M, Binder M, Curtis JM, Slot JC, Wang Z, Wilson AW, Schussler A, Longcore JE, O’Donnell K, Mozley-Standridge S, Porter D, Letcher PM, Powell MJ, Taylor JW, White MM, Griffith GW, Davies DR, Humber RA, Morton JB, Sugiyama J, Rossman AY, Rogers JD, Pfister DH, Hewitt D, Hansen K, Hambleton S, Shoemaker RA, Kohlmeyer J, Volkmann-Kohlmeyer B, Spotts RA, Serdani M, Crous PW, Hughes KW, Matsuura K, Langer E, Langer G, Untereiner WA, Lucking R, Budel B, Geiser DM, Aptroot A, Diederich P, Schmitt I, Schultz M, Yahr R, Hibbett DS, Lutzoni F, McLaughlin DJ, Spatafora JW, Vilgalys R. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- Jouvenaz DP, Ellis EA. Vairimorpha invictae n. sp (Microspora, Microsporida), a parasite of the red imported fire ant, Solenopsis invicta Buren (Hymenoptera, Formicidae) J Protozool. 1986;33:457–461. [Google Scholar]
- Keeling PJ. Congruent evidence from alpha-tubulin and beta-tubulin gene phylogenies for a zygomycete origin of microsporidia. Fungal Genet Biol. 2003;38:298–309. doi: 10.1016/s1087-1845(02)00537-6. [DOI] [PubMed] [Google Scholar]
- Kondova I, Mansfield K, Buckholt MA, Stein B, Widmer G, Carville A, Lackner A, Tzipori S. Transmission and serial propagation of Enterocytozoon bieneusi from humans and Rhesus macaques in gnotobiotic piglets. Infect Immun. 1998;66:5515–5519. doi: 10.1128/iai.66.11.5515-5519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leelayoova S, Subrungruang I, Suputtamongkol Y, Worapong J, Petmitr PC, Mungthin M. Identification of genotypes of Enterocytozoon bieneusi from stool samples from human immunodeficiency virus-infected patients in Thailand. J Clin Microbiol. 2006;44:3001–3004. doi: 10.1128/JCM.00945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leelayoova S, Suputtamongkol Y, Subrungruang I, Worapong J, Chavalitshewinkoon-Petmitr P, Mungthin M. Evidence supporting the zoonotic and non-zoonotic transmission of Enterocytozoon bieneusi. Ann Trop Med Parasitol. 2008;102:459–461. doi: 10.1179/136485908X300823. [DOI] [PubMed] [Google Scholar]
- Lobo ML, Xiao L, Cama V, Magalhaes N, Antunes F, Matos O. Identification of potentially human-pathogenic Enterocytozoon bieneusi genotypes in various birds. Appl Environ Microbiol. 2006;72:7380–7382. doi: 10.1128/AEM.01394-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AP. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl Environ Microbiol. 2002;68:3673–3682. doi: 10.1128/AEM.68.8.3673-3682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis A, Breitenmoser AC, Deplazes P. Detection of new Enterocytozoon genotypes in faecal samples of farm dogs and a cat. Parasite. 1999;6:189–193. doi: 10.1051/parasite/1999062189. [DOI] [PubMed] [Google Scholar]
- Mathis A, Weber R, Deplazes P. Zoonotic potential of the microsporidia. Clin Microbiol Rev. 2005;18:423–445. doi: 10.1128/CMR.18.3.423-445.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinder H, Katzwinkel-Wladarsch S, Loscher T. Evidence for the existence of genetically distinct strains of Enterocytozoon bieneusi. Parasitol Res. 1997;83:670–672. doi: 10.1007/s004360050317. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sanders HL. Marine benthic diversity - a comparative study. American Naturalist. 1968;102:243–282. [Google Scholar]
- Santin M, Trout JM, Fayer R. Enterocytozoon bieneusi genotypes in dairy cattle in the eastern United States. Parasitol Res. 2005;97:535–538. doi: 10.1007/s00436-005-1482-8. [DOI] [PubMed] [Google Scholar]
- Santin M, Fayer R. Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: a consensus. J Eukaryot Microbiol. 2009;56:34–38. doi: 10.1111/j.1550-7408.2008.00380.x. [DOI] [PubMed] [Google Scholar]
- Simakova AV, Vossbrinck CR, Andreadis TG. Molecular and ultrastructural characterization of Andreanna caspii n. gen., n sp (Microsporida: Amblyosporidae), a parasite of Ochlerotatus caspius (Diptera: Culicidae) J Invertebr Pathol. 2008;99:302–311. doi: 10.1016/j.jip.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Sokolova YY, Fuxa JR. Biology and life-cycle of the microsporidium Kneallhazia solenopsaeKnell Allan Hazard 1977 gen n., comb n., from the fire ant Solenopsis invicta. Parasitology. 2008;135:903–929. doi: 10.1017/S003118200800440X. [DOI] [PubMed] [Google Scholar]
- Sulaiman IM, Bern C, Gilman R, Cama V, Kawai V, Vargas D, Ticona E, Vivar A, Xiao L. A molecular biologic study of Enterocytozoon bieneusi in HIV-infected patients in Lima, Peru. J Eukaryot Microbiol. 2003a;50(Suppl):591–596. doi: 10.1111/j.1550-7408.2003.tb00642.x. [DOI] [PubMed] [Google Scholar]
- Sulaiman IM, Fayer R, Lal AA, Trout JM, Schaefer FW, 3rd, Xiao L. Molecular characterization of microsporidia indicates that wild mammals harbor host-adapted Enterocytozoon spp. as well as human-pathogenic Enterocytozoon bieneusi. Appl Environ Microbiol. 2003b;69:4495–4501. doi: 10.1128/AEM.69.8.4495-4501.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulaiman IM, Fayer R, Yang C, Santin M, Matos O, Xiao L. Molecular characterization of Enterocytozoon bieneusi in cattle indicates that only some isolates have zoonotic potential. Parasitol Res. 2004;92:328–334. doi: 10.1007/s00436-003-1049-5. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- ten Hove RJ, Van Lieshout L, Beadsworth MB, Perez MA, Spee K, Claas EC, Verweij JJ. Characterization of genotypes of Enterocytozoon bieneusi in immunosuppressed and immunocompetent patient groups. J Eukaryot Microbiol. 2009;56:388–393. doi: 10.1111/j.1550-7408.2009.00393.x. [DOI] [PubMed] [Google Scholar]
- Tumwine JK, Kekitiinwa A, Nabukeera N, Akiyoshi DE, Buckholt MA, Tzipori S. Enterocytozoon bieneusi among children with diarrhea attending Mulago Hospital in Uganda. Am J Trop Med Hyg. 2002;67:299–303. doi: 10.4269/ajtmh.2002.67.299. [DOI] [PubMed] [Google Scholar]
- Tzipori S, Carville A, Widmer G, Kotler D, Mansfield K, Lackner A. Transmission and establishment of a persistent infection of Enterocytozoon bieneusi, derived from a human with AIDS, in simian immunodeficiency virus-infected rhesus monkeys. J Infect Dis. 1997;175:1016–1020. doi: 10.1086/513962. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.