Abstract
Two known recurrent constitutional translocations, t(11;22) and t(17;22), as well as a non-recurrent t(4;22), display derivative chromosomes that have joined to a common site within the low copy repeat B (LCR-B) region of 22q11.2. This breakpoint is located between two AT-rich inverted repeats that form a nearly perfect palindrome. Breakpoints within the 11q23, 17q11 and 4q35 partner chromosomes also fall near the center of palindromic sequences. In the present work the breakpoints of a fourth translocation involving LCR-B, a balanced ependymoma-associated t(1;22), were characterized not only to localize this junction relative to known genes, but also to further understand the mechanism underlying these rearrangements. FISH mapping was used to localize the 22q11.2 breakpoint to LCR-B and the 1p21 breakpoint to single BAC clones. STS mapping narrowed the 1p21.2 breakpoint to a 1990 bp AT-rich region, and junction fragments were amplified by nested PCR. Junction fragment-derived sequence indicates that the 1p21.2 breakpoint splits a 278 nt palindrome capable of forming stem–loop secondary structure. In contrast, the 1p21.2 reference genomic sequence from clones in the database does not exhibit this configuration, suggesting a predisposition for regional genomic instability perhaps etiologic for this rearrangement. Given its similarity to known chromosomal fragile site (FRA) sequences, this polymorphic 1p21.2 sequence may represent one of the FRA1 loci. Comparative analysis of the secondary structure of sequences surrounding translocation breakpoints that involve LCR-B with those not involving this region indicate a unique ability of the former to form stem–loop structures. The relative likelihood of forming these configurations appears to be related to the rate of translocation occurrence. Further analysis suggests that constitutional translocations in general occur between sequences of similar melting temperature and propensity for secondary structure.
INTRODUCTION
The 22q11.2 region is a hotspot for chromosomal rearrangements where deletions, duplications and translocations occur at least once in every 3000–4000 live births (1). Deletions within this region are associated with congenital disorders in development including DiGeorge (DGS), velocardiofacial (VCFS) and conotruncal anomaly face syndromes (2). An inverted interstitial duplication results in the creation of a bisatellited supernumerary chromosome giving rise to cat-eye syndrome (2,3). Additionally, a number of constitutional and neoplastic translocations involving 22q11.2 have been described, including the recurrent constitutional t(11;22) (4–8), and t(17;22) (9,10). Balanced t(11;22) carriers have no phenotype, but 3:1 meiotic malsegregation of the der(22) chromosome can result in progeny with supernumerary-der(22) t(11;22) syndrome (11). The balanced t(17;22) is associated with neurofibromatosis type 1 in which disruption of the NF1 gene on 17q11.2 is thought to be the primary cause of the phenotype (9,10).
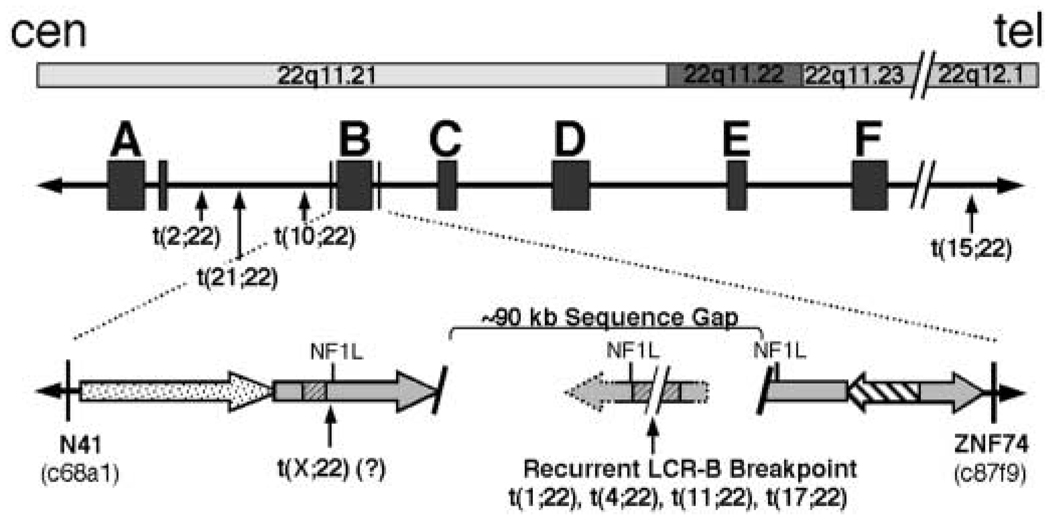
The molecular etiology of these rearrangements is related to the genomic structure of the 22q11.2 region. Long stretches of repeated sequence having greater than 95% identity are clustered together into at least eight regions termed low copy repeats (LCRs). Most interstitial deletions and inverted duplications occurring within 22q11 take place between homologous sequences contained within six of these LCRs (LCR-A to -F; Fig. 1) (12,13). Because these rearrangements appear to occur between sequences having greater than 95% identity, current models for their formation have invoked a homologous recombination mechanism involving unequal inter- and intrachromosomal crossover (2,13). Constitutional translocations, including the recurrent t(11;22) (4–8,11,14), t(17;22) (9,10), and a recently described non-recurrent t(4;22) (15) share the same 22q11.2 breakpoint within LCR-B. Among the repeated sequence modules contained in LCR-B are at least three copies of an NF1-like (NF1L) repeat module (Fig. 1, open gray arrows). The common breakpoint of these three translocations occurs at the center of an AT-rich palindrome located near the center of one of these NF1L repeats. Because the 11q23, 17q11.2 and 4q35.1 breakpoints also fall at the center of inverted repeated sequences, a palindrome-mediated mechanism has been invoked for the formation of these rearrangements (6,10,15).
Figure 1.
Schematic diagram illustrating the location of constitutional translocation breakpoints on chromosome 22 relative to chromosomal bands and LCRs. The relative position of six of eight known LCRs (13) are shown below their respective banding locations (LCRs A–F are shown). The repeated sequence modules comprising LCR-B are shown as large filled arrows in the expanded view. The locations of cosmid clones (c68a1 and ZNF74) used as probes in FISH studies to localize breakpoints to LCR-B of 22q11.2 are shown. The common LCR-B breakpoint is present within an ~90 kb gap in the known 22q11.21 sequence. This breakpoint is flanked by palindromic AT-rich repeats (gray hatched regions) found within one of three NF1L modules (gray filled arrows) present within LCR-B.
The specific sequences required for the mechanisms of translocations in general are poorly understood. Somatic translocations observed in rapidly dividing neoplastic cells occur during mitosis while constitutional rearrangements are meiotic in origin (16). The non-random clustering of somatic translocation breakpoints is probably due to the selection of cells harboring activated oncogenes that provide a growth advantage. However, spatial proximity of breakpoint loci due to higher-order genome organization, also appears to contribute to breakpoint clustering (17). Recurrent constitutional translocations may employ a different mechanism. Breakpoints of these rearrangements tend to localize to specific, potentially unstable, sequences as evidenced by the characterization of t(11;22), t(17;22) and t(4;22) breakpoints. A recent cytogenetic survey by Spiteri et al. (18), suggested that constitutional breakpoints localize to dynamic or potentially unstable regions of the genome such as 22q11.2. Owing to the low resolution of these studies, however, they provide little insight into the specific sequences contributing to the molecular mechanism of constitutional translocations in general or those involving the common LCR-B breakpoint on 22q11.2.
The present work provides definitive information concerning the sequence motifs and DNA secondary structures that predispose chromosomal sequences to translocations with LCR-B of 22q11.2. For this work, we analyze the breakpoint sequences of several constitutional translocations including a previously uncharacterized ependymoma-associated t(1;22) involving the common LCR-B junction (19,20). Characterization of the t(1;22) breakpoint junctions not only provides some information related to the molecular etiology of this patient’s phenotype, but also contributes to our understanding of the mechanisms responsible for the generation of translocations involving the common LCR-B breakpoint. By comparing the breakpoints from these translocations with those that do not involve LCR-B, we are able to demonstrate the unique ability of sequences involved in translocations with the common breakpoint of LCR-B to form secondary structure. Our findings also provide an explanation for the relative frequency of translocations occurring between specific partner chromosome sequences and the common LCR-B breakpoint. The current work also suggests that constitutional translocations in general occur between chromosomal regions having similar sequence characteristics.
RESULTS
Localization of t(1;22) breakpoints to LCR-B of 22q11 and 1p21.2
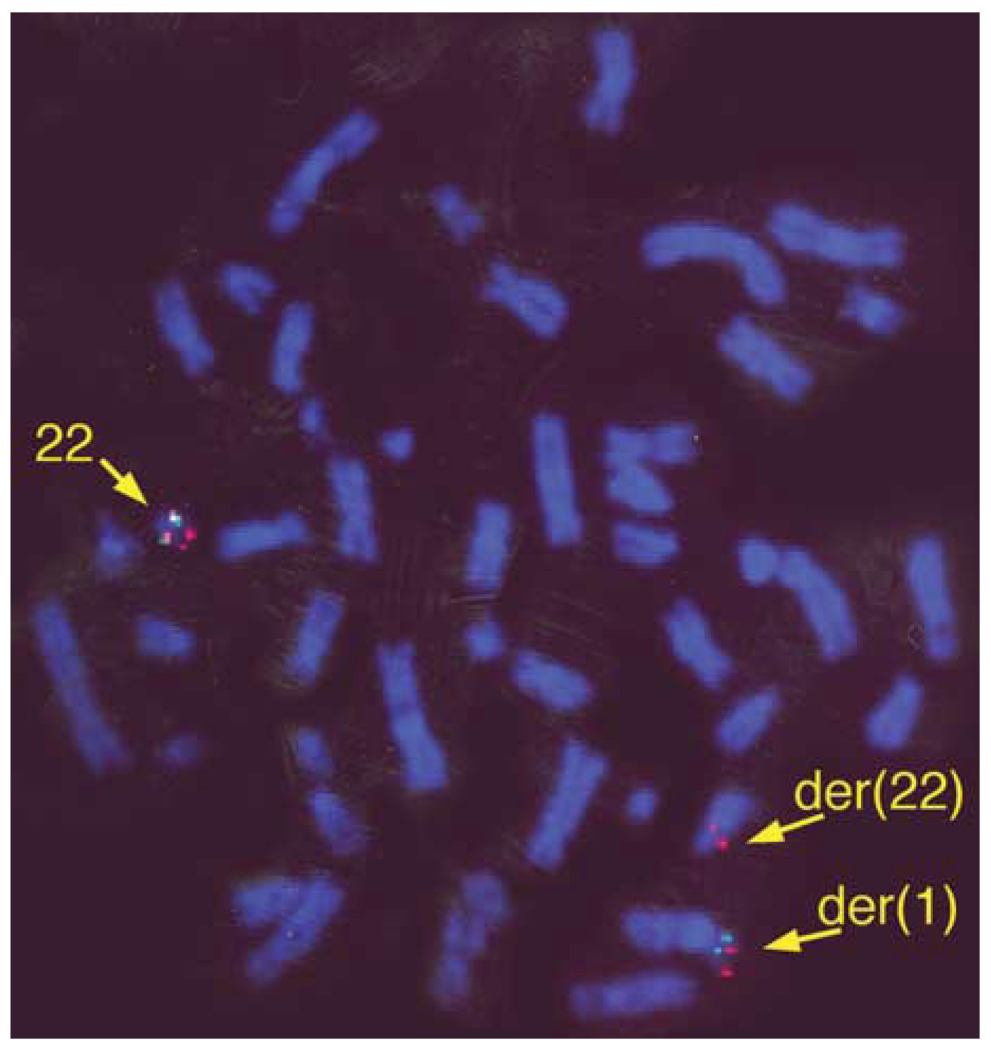
The t(1;22) was initially identified in the prenatal karyotype of a patient who subsequently developed an ependymoma at five years of age (19). Previous STS mapping of somatic cell hybrid DNA identified the t(1;22) breakpoint in a 22q11 region containing LCR-B (19). To further narrow the t(1;22) breakpoint within 22q11.2, fluorescence in situ hybridization (FISH) experiments were done with cosmid probes for N41 and ZNF74 sequences immediately flanking LCR-B (Fig 1 and Fig 2). Metaphase chromosomes derived from t(1;22) patient lymphoblasts exhibit N41 signal on both the normal 22 and der(22) chromosomes (Fig. 2, red), whereas ZNF74 sequence is found on the normal 22 and der(1) (Fig. 2, green). These results indicate that the t(1;22) breakpoint falls between N41 and ZNF74, confining it to LCR-B of 22q11.21. The precise location of the breakpoint within the ~135 kb LCR-B was predicted to lie at the center of the palindromic AT-rich region—the common breakpoint found in recurrent t(11;22)s (4–8,14) and t(17;22)s (9,10), as well as a recently characterized t(4;22) (15) (Fig. 1).
Figure 2.
The t(1;22) breakpoint is located within LCR-B of 22q11.21. FISH analysis was done on metaphase chromosomes of peripheral lymphoblasts derived from the t(1;22) patient. Cosmid clone probes specific for N41 (c68a1) and ZNF74 (c87f9) were labeled with rhodamine (red) and fluorescein (green), respectively. An additional rhodamine-labeled marker probe for D22S39 (22q13.3) used to mark the distal arm of chromosome 22 can be seen near the telomere on both normal and der(1) chromosomes.
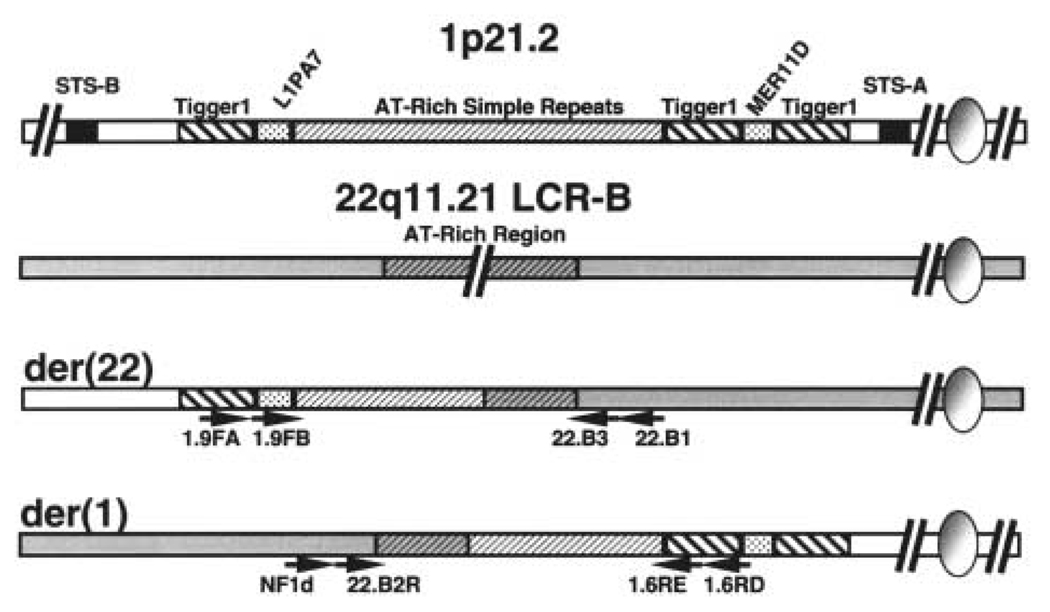
The chromosome 1 breakpoint was initially localized to a region between STS markers D1S2896 and D1S3395 by PCR mapping of the derivative chromosomes isolated in human : hamster somatic hybrid cells (20). Further BAC probe FISH mapping led to the identification of a clone (336L20) that displayed signal on the normal chromosome 1, der(1) and der(22) (Supplementary Fig. S1). This result indicates that the sequence contained within this 179 kb BAC spans the t(1;22) breakpoint at what is currently designated 1p21.2 (November 2002 freeze; www.genome.ucsc.edu/). To fine-map the t(1;22) breakpoint by PCR, primers corresponding to sequences contained in 336L20 were used to amplify STS markers. Using somatic cell hybrid DNA containing either der(1) or der(22) chromosome template (20), this approach successfully localized the t(1;22) breakpoint to a 5.73 kb region flanked by STS-A and STS-B (Fig. 3, data not shown). Southern blots of DNA isolated from t(1;22) patient and somatic hybrid cells probed with sequence immediately distal to STS-B substantiated this localization (Supplementary Fig. S1). Further PCR mapping using the hybrid cell DNA narrowed the breakpoint to a 1990 bp sequence containing several AT-rich simple repeats (Fig. 3, data not shown).
Figure 3.
Strategy for amplification of t(1;22) junction fragments. The 1p22 breakpoint was localized between STS-A and STS-B markers by PCR mapping of somatic cell hybrid DNA. Repetitive elements present in this 6.7 kb breakpoint region of 1p22 are depicted. Chromosomes 1 and 22 are shown as white and shaded bars, respectively. AT-rich repeats are shown as hatched bars on both 1p22 and at the common breakpoint region of the 22q11.21 LCR-B. Sequence-specific nested primers used for junction fragment amplification are shown as arrows.
Amplification of der(1) and der(22) junction fragments
Junction fragments spanning the t(1;22) breakpoint on both the der(1) and der(22) chromosomes were amplified by nested primer PCR. Nested primers were designed against sequences flanking the narrowed breakpoint regions in both 1p21.2 and LCR-B of 22q11.2. In LCR-B, primer sequences were located just outside the palindromic AT-rich region surrounding the recurrent 22q11.2 breakpoint. On 1p21.2, specific primers were designed against sequences flanking the 1990 bp AT-rich region containing the breakpoint as delineated by PCR mapping experiments above (Fig. 3). Junction fragments were then amplified from t(1;22) patient-derived lymphoblast DNA in two successive rounds of PCR with oppositely directed primers from each chromosome. The der(22) fragment, amplified first with 1.9FA and 22.B1 primers and subsequently with nested 1.9FB and 22.B3 primers, contained 982 bp of AT-rich 1p21.2 sequence joined to 300 bp of 22q11.2 sequence (Fig. 4A). The der(1) junction, amplified with oppositely directed primers, begins with 337 bp of 22q11.2 sequence distal to the breakpoint and gives way to 1148 bp of 1p21.2 sequence. Strikingly, the breakpoints within LCR-B of 22q11.2 fall within 15 bp of those found previously for the t(4;22) (15), two different t(17;22)s (9,10) and most of the recurrent t(11;22)s (6,7,14). The 1p21.2 breakpoint, on the other hand, falls near the center of the 1990 bp AT-rich repeat region.
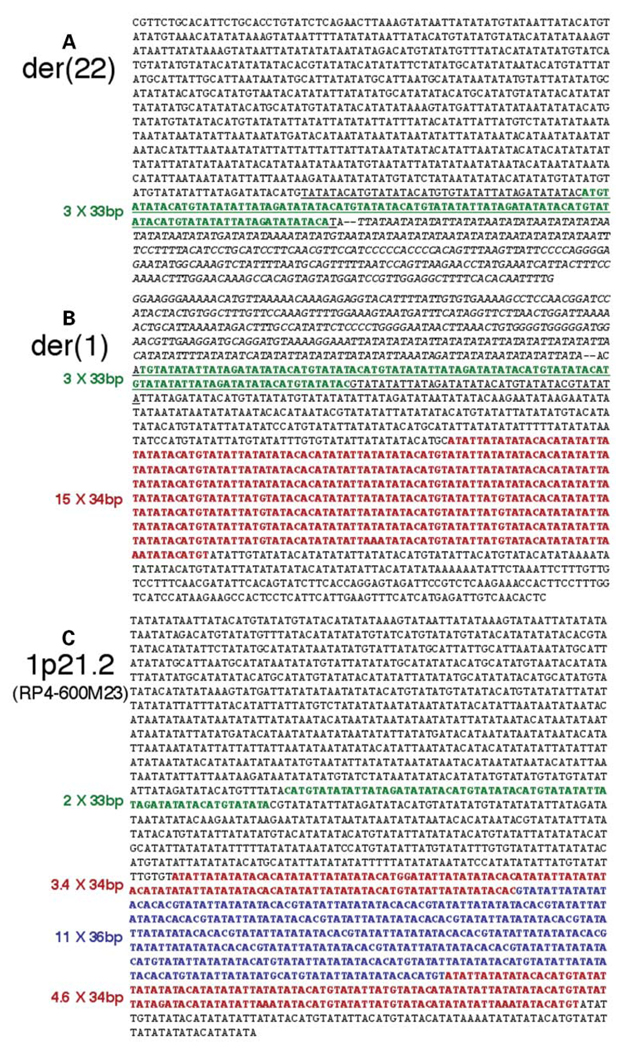
Figure 4.
Junction fragment sequences amplified from t(1;22) patient DNA by nested primer PCR contain distinct numbers of repeated elements relative to control sequence found in genomic assembly clone RP4-600m23. (A) Sequence of the der(22) junction fragment shown in the telomere to centromere orientation. Three copies of a 33 bp VNTR element immediately distal to the breakpoint (two dashes) within the chromosome 1 sequence are shown in green. Chromosome 22 sequence is in italics. (B) The der(1) junction fragment sequence shown telomere to centromere. VNTRs 33 and 34 bp long within 1p21 sequence are green and red, respectively. Chromosome 22 sequence is in italics. (C) Normal genomic sequence surrounding the 1p22 breakpoint from BAC clone RP4-600m23. VNTRs 33, 34 and 36 bp long are green, red and blue, respectively.
Examination of the AT-rich 1p21.2 breakpoint sequence compiled from PCR-amplified der(22) and der(1) junction fragments reveals the presence of at least two different variable number tandem repeats (VNTRs). One of these is a 34 bp repeat that is duplicated 15 times in a region proximal to the breakpoint (Fig. 4B, red bases). Immediately adjacent to the t(1;22) breakpoint on each junction fragment are three copies of a another VNTR that is 33 bp long (Fig. 4A and B, green bases). When the experimentally determined der(1) and der(22) junction fragments are compiled to construct the 1p21.2 breakpoint sequence, these six 33 bp VNTRs are situated symmetrically around the breakpoint site. The sequence of each of these AT-rich 33 bp VNTRs is almost completely palindromic in that the forward sequence nearly matches its reverse compliment with the exception of two bases. Together with adjacent sequence, six copies of this 33 bp VNTR make up a 278 bp palindrome with the t(1;22) breakpoint falling near the center of these inverted repeats (Fig. 4A and B, underlined bases).
The polymorphic nature of this AT-rich repeat region is illustrated by comparing the experimentally determined breakpoint sequence compiled from der(1) and der(22) junction fragments with that of control sequences. Although the size of the AT-rich region surrounding the 1p21.2 breakpoint prohibited PCR amplification of sequences from control DNA templates, normal 1p21.2 sequence was obtainable from two different genomic sequence databases (GenBank and Celera). Junction fragment sequence is identical to that of controls at the distal and proximal ends of the 1990 bp AT-rich region (compare junction fragment sequences in Fig. 4A and B to control sequences in Fig. 4C and Supplementary Fig. S2). Within this region, however, small blocks of divergence are observed that appear to be the result of differential VNTR duplication. Control sequence from the genomic assembly clone, RP4-600m23 (accession no. AC114483.2), contains only eight copies of the 34 bp VNTR (Fig 4C, red bases), and one of these is interrupted by 11 copies of a different, 36 bp VNTR (Fig. 4C, blue bases). The control sequence from a second source, the Celera database, does include 15 copies of the 34 bp VNTR, but diverges from junction fragment sequence at other locations due to the presumed expansion and contraction of other repeat sequences (Supplementary Fig. S2), further demonstrating the apparent polymorphic nature of this AT-rich region.
Interestingly, the 33 bp VNTRs identified near the t(1;22) breakpoint are repeated only twice in the control sequence of RP4-600m23 relative to the six copies seen in the experimental sequence derived from t(1;22) junction fragments. The control sequence from the Celera database exhibits only one copy of this 33 bp VNTR with a sequence gap replacing the remaining 33 bp VNTRs observed in the experimentally-determined 1p21.2 breakpoint sequence (Supplementary Fig. S2). Because palindromic sequences are inefficiently cloned into yeast and bacterial vectors used in the determination of human genomic sequence (21,22), the gap in this control sequence suggests that the source of DNA used to generate the Celera sequence also contained similar palindromic sequence. These palindromic 33 bp VNTRs have the potential to influence secondary structure at the 1p21.2 breakpoint.
The 1p21.2 breakpoint from the t(1;22) patient is predisposed to forming stem–loop structures
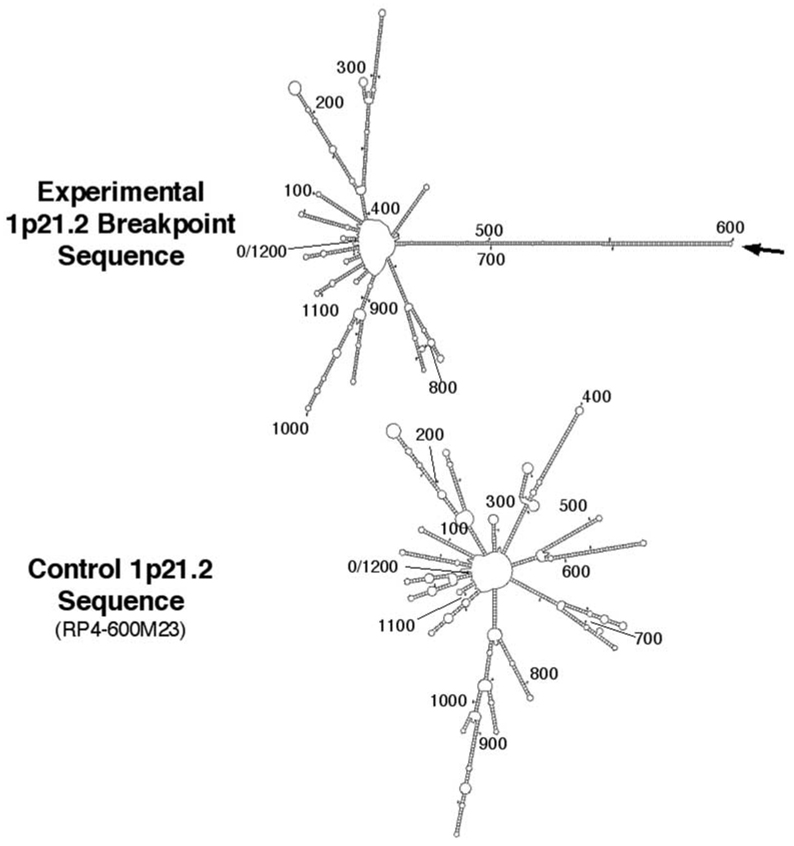
Palindromic and inverted repeat sequences in genomic DNA are predicted to form stem–loop structures. To examine the potential secondary structures formed at the 1p21.2 breakpoint, 1200 nucleotides of sequence surrounding the experimentally determined 1p21.2 breakpoint (600 nucleotides on either side) were analyzed using the M-Fold sequence analysis package (http://bioweb.pasteur.fr/seqanal/interfaces/mfold-simple.html). As shown in Figure 5 (top), the AT-rich region surrounding the experimentally determined sequence surrounding the 1p21.2 breakpoint is capable of forming numerous stem–loops, the most prominent of which is a 141 nucleotide (nt) stem having only 12 mismatches (including four asymmetrical bases). The stem-like structure is predicted to form from the 278 nt palindrome described above and is capped by a small three-nucleotide loop (Supplementary Fig. S3). Interestingly, the t(1;22) breakpoint occurs at tip of the predicted stem-loop (Fig. 5, Supplementary Fig. S3).
Figure 5.
Junction fragment sequence surrounding the 1p21.2 breakpoint is capable of forming a prominent stem–loop structure, while control 1p21.2 AT-rich sequence does not. 1200 nt of single-strand sequence surrounding the 1p21.2 breakpoint compiled from junction fragments amplified from the t(1;22) patient (top) or normal sequence from the genomic assembly clone RP4-600m23 (bottom) were analyzed by M-Fold to determine potential secondary structures. Graphic outputs depicting the two-dimensional stem–loop base pairings are shown. See Supplementary Figure S3 for the sequence alignment of the 281 bp stem–loop at the experimentally determined 1p21.2 breakpoint.
The importance of the six 33 bp VNTRs in the formation of potential secondary structure at the t(1;22) breakpoint is evident in comparisons between experimentally derived 1p21.2 sequence from the t(1;22) patient and that contained in the genomic assembly clone, RP4-600m23. When analyzed by M-Fold, this sequence gives rise to numerous stem-loop structures, owing to the AT-rich nature of this sequence, but a prominent stem–loop structure formed from a single long palindrome is not seen (Fig. 5, bottom). These results indicate that the presence of six 33 bp VNTRs contribute to the potential secondary structure at the t(1;22) breakpoint, and raise the possibility that this sequence could have conferred a predisposition to the formation of the of the t(1;22).
Translocation breakpoints involving LCR-B of 22q11.2 occur at potential stem–loop structures
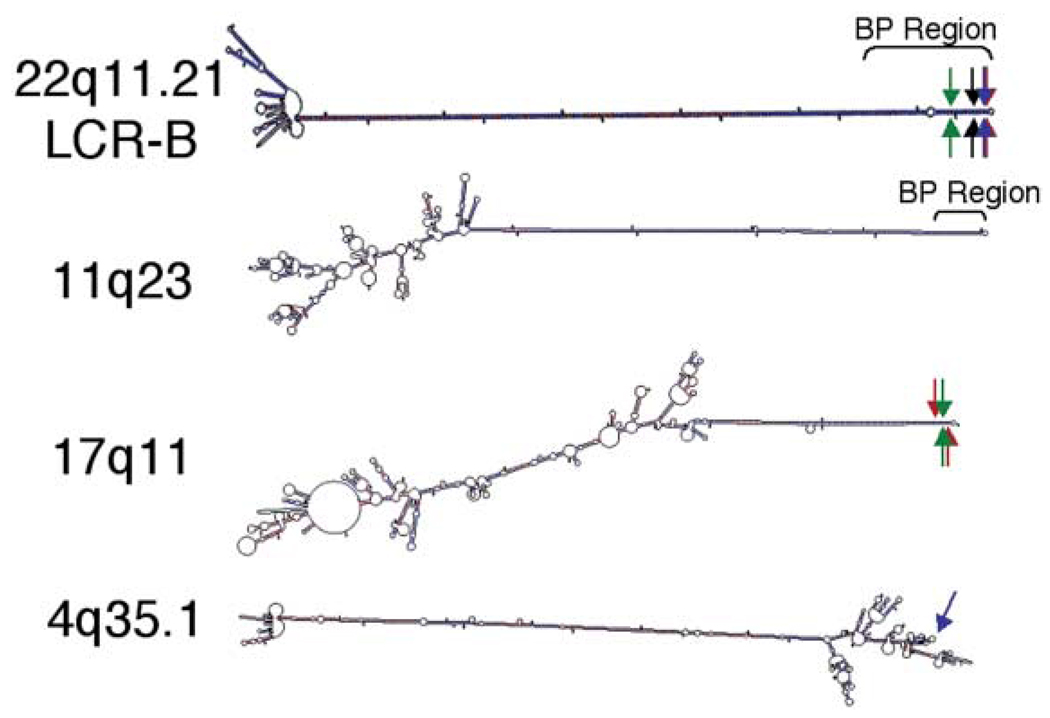
The t(1;22) is formed by a junction between the AT-rich region of 1p21.2 and a common breakpoint localized to LCR-B of 22q11.2. This site, which is also involved in the recurrent t(11;22), the recurrent t(17;22) and the recently described t(4;22), occurs at the center of a palindromic sequence containing an AT-rich repeat. Likewise, palindromic sequences have now been described at the breakpoints of all four partner chromosomes involved in translocations with LCR-B, suggesting that stem–loop structures form at each of these junctions. Indeed, when single-strand sequence surrounding each of these breakpoints is analyzed by M-Fold, stem–loops greater than 95 nt are seen, and in every case, the location of the breakpoint is near the terminal end of these structures (Fig. 6, Supplementary Fig. S4). In the known sequence surrounding the common 22q11.2 breakpoint, a 589 nt palindrome can form a 294 bp stem with only four mismatched bases (one asymmetrical base). In the formation of any particular translocation with LCR-B, a symmetrical number of 22q11.2 bases are usually lost such that the ‘stem’ is truncated equally on both strands (6) (Fig. 6, Supplementary Fig. S4). The potential structures formed around the breakpoints of the three other partner chromosomes (11q23, 17q11, 4q35.1) are just as striking. The 11q23 breakpoint contains a 441 nt palindrome capable of forming a 219 bp stem with five mismatches (three asymmetrical bases), and the junction at 17q11 lies in the center of a 202 nt palindrome capable of forming a 97 bp stem with nine mismatches (eight asymmetrical bases; Fig. 6, Supplementary Fig. S4). Of particular interest is the sequence surrounding the 4q35.1 breakpoint. In this case, a 277 bp stem is capped by an unusually long 547 nt loop, wherein the breakpoint lies (Fig. 6, Supplementary Fig. S4). Together, these results suggest that chromosomal sequences capable of forming stem–loop structures are predisposed to forming translocations with LCR-B of 22q11.2.
Figure 6.
Sequences surrounding translocation breakpoints involving the LCR-B of 22q11.2 contain palindromic sequences capable of forming prominent stem-loop structures. 1200 nt of single-strand sequence surrounding each breakpoint are shown. Breakpoint regions on 22q11.2 and 11q23 refer to sites at which numerous symmetrical breakpoints have been documented in t(11;22) recurrent translocations (6). Red and green arrows, t(17;22) breakpoints (9, 10, respectively); blue arrows, t(4;22) breakpoint (15); black arrow, t(1;22) breakpoint described herein.
For these secondary structures to form, double-stranded chromosomal DNA must first denature, and complimentary sequence contained in a single strand must then re-anneal into stem–loops. The stability of a particular strand of DNA in its double-stranded configuration or in secondary structure formed within a single strand can be described by separate Gibbs free energy (G) values. Because a maximum amount of base pairing occurs in double-stranded DNA, it is inherently more stable and always has a free energy value (GDS) that is more negative than that of a single strand folding within itself (GSTRUC). The free energy of a single strand of sequence forming stem–loops depends on the position and number of complementary base pairs as well as the GC content of these nucleotides. A nearly perfect palindrome, similar to that seen in LCR-B of 22q11.2, will be attributed a more negative GSTRUC that approaches that of the same sequence annealed to its complimentary strand (GDS). The propensity for a given strand of DNA sequence to form a stem–loop structure, therefore, is inversely related to the difference between these free energies such that a given strand of DNA sequence exhibiting a small difference between GDS and GSTRUC (ΔG) would be more likely to form a stem–loop than those sequences having a greater ΔG.
As shown in Table 1, breakpoint sequences involved in translocations with LCR-B of 22q11.2 show a propensity for secondary structure formation that is related to the observed recurrence of these rearrangements. The common LCR-B breakpoint exhibits the smallest ΔG values, as expected given the prolific tendency of this site to be involved in translocations. Sequences surrounding the 11q23 and 17q11 breakpoints have slightly higher free energy differences when analyzing only the stem–loop region or 1200 nt of sequence surrounding these junctions. When these ΔG values are normalized for the number of nucleotides contained in the analyzed sequence (ΔG/nt), free energy differences of 0.025, 0.034 and 0.075 kcal/mol are found for sequences surrounding the 22q, 11q and 17q breakpoints, respectively. These values are substantially lower than those found for breakpoints involved in the non-recurrent t(4;22) and t(1;22) rearrangements (Table 1). Although the inverted repeat sequence surrounding the 4q35.1 breakpoint is larger than that found at either of the 11q and 17q breakpoints, a higher ΔG/nt value of 0.400 is seen, due to the large intervening loop region that gives rise to a GSTRUC that is less negative relative to the GDS for this sequence (Fig. 6, Supplementary Fig. S4). This indicates that inverted repeats contained within a single strand must be near one another for secondary structure to occur and for a particular chromosomal sequence to be a good substrate for translocations with LCR-B.
Table 1.
Secondary structure characteristics of breakpoint sequences involved in constitutional 22q11.2 LCR-B translocations
| Translocation BP | Stem–loop region | 1200 nt | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stem–loop regiona |
Palindrome/ mismatches |
Stem– loop |
Tm (°C) |
GDS (kcal/mol) |
GSTRUC (kcal/mol) |
ΔG (kcal/mol) |
ΔG/ntb (kcal/mol) |
Tm (°C) |
GDS (kcal/mol) |
GSTRUC (kcal/mol) |
ΔG (kcal/mol) |
ΔG/nt (kcal/mol) |
|
| Ch22q11.2 (LCR-B) | 593 nt | 590/4 | 294/4 | 72.1 | −331.2 | −316.6 | 14.6 | 0.025 | 71.4d | −476.6d | −355.3d | 121.3d | 0.138d |
| Ch11q23 @ t(11;22) | 445 nt | 441/5 | 219/4 | 64.0 | −197.0 | −181.7 | 15.3 | 0.034 | 70.8 | −647.3 | −246.1 | 401.2 | 0.334 |
| Ch17q11 @ t(17;22) | 205 nt | 202/9 | 97/3 | 67.9 | −104.0 | −88.6 | 15.4 | 0.075 | 76.2 | −737.2 | −203.9 | 533.3 | 0.444 |
| Ch4q35.1 @ t(4;22) | 1072 nt | 554/38 | 277/547 | 79.4 | −709.4 | −280.3 | 429.1 | 0.400 | 79.4 | −792.4 | −292.4 | 500.0 | 0.417 |
| Ch1p21.1 @ t(1;22)c (junction fragment) |
281 nt | 278/12 | 141/3 | 66.9 | −134.3 | −86.8 | 47.5 | 0.169 | 66.8 | −558.8 | −266.7 | 292.1 | 0.243 |
| Average | 0.141 | 369.6e | 0.360e | ||||||||||
| SEM | 0.078 | 83.9e | 0.052e | ||||||||||
| Ch1p21.1 @ t(1;22) (RP4-600m23) |
— | — | — | — | — | — | — | — | 66.6 | −558.8 | −230.8 | 328.0 | 0.274 |
Sequences surrounding breakpoints involved in 22q11.2 LCR-B translocations were analyzed by M-Fold. Nucleotide sequence forming the primary stem loop or 1200 nt surrounding the breakpoint (600 nt on each side) were analyzed to determine melting temperature, and the free energy of this single strand of sequence annealed to its complimentary strand to form a double-stranded conformation (GDS) or that contained within the single-strand secondary structure (GSTRUC).
References for characterized breakpoint sequences: 22q11.2 (6), 11q23 (4–7), 17q11 (9,10), 4q35.1 (15).
Number of nucleotides contained in a the stem–loop region surrounding the indicated translocation breakpoint. This includes palindromic and loop sequence, as well as mismatched bases.
ΔG/nt, for a given sequence, the difference in free energy between that contained in the double-stranded and single-strand secondary structure configurations per nucleotide [(GDS − GSTRUC)/(number of base pairs analyzed)].
1p21.2 sequence surrounding the t(1;22) breakpoint in the current work was deduced from junction fragment sequence.
Only 878 bp of sequence surrounding the LCR-B breakpoint are currently known.
Statistical analysis does not include 22q11.2 LCR-B breakpoint data as 1200 bp of sequence surrounding this breakpoint are not known.
Sequence surrounding the 1p21.2 breakpoint obtained from junction fragments of the t(1;22) patient display ΔG values lower than that of normal sequence, indicating that the affected chromosome is more likely to form single-strand secondary structures. 1200 nt of sequence surrounding the breakpoint on the derivative chromosomes has a ΔG of 292.1 kcal/mol (0.243 kcal/mol per nucleotide), while the control sequence has a value of 328.0 kcal/mol (0.274 kcal/mol per nucleotide), thus providing a relative quantitative assessment of the predisposition of the progenitor chromosomes of the t(1;22) to form this translocation. Owing to its AT-rich repeat nature, the normal 1p21.2 sequence is capable of forming a multitude of small stem–loops that contribute to a stable structure having a ΔG value that is actually quite low relative to free energy differences of translocation breakpoints not involving LCR-B of 22q11.2.
Stem–loop structures are not typical of constitutional translocations
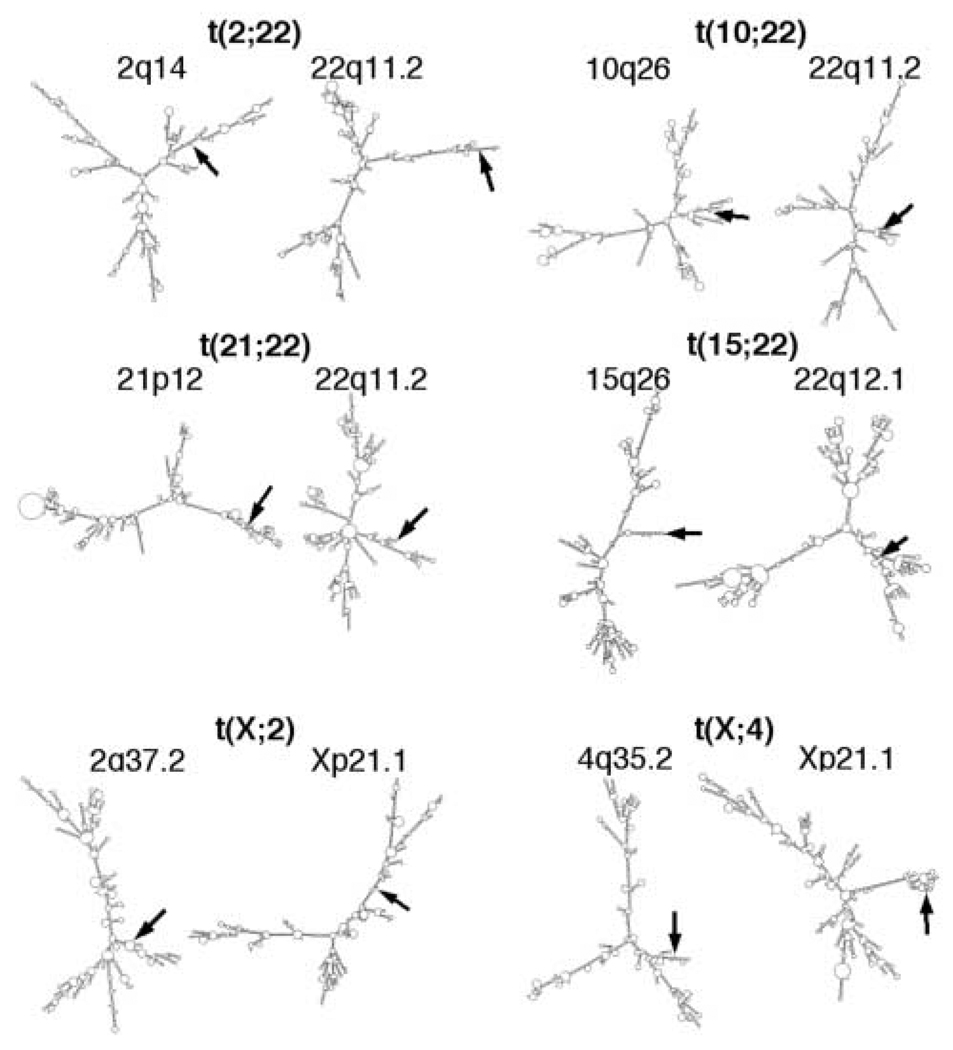
To investigate the potential for single-strand secondary structure to form at breakpoints of translocations not involving the 22q11.2 LCR-B, M-Fold was used to analyze 1200 nt of sequence surrounding translocation sites from rearrangements involving other regions of chromosome 22 [t(2;22) (23); t(10;22) (24); t(15;22) (18); t(21;22) (25), see Fig. 1] as well as two different translocations involving the × chromosome [t(X;2), t(X;4) (26)]. In no case was a palindrome greater than 40 nucleotides observed, and breakpoints were never found to reside at the end of a prominent stem–loop structure (Fig. 7). In two cases, individual breakpoints were found on small stem–loops. These were the breakpoints within 22q11.2 of the t(2;22) and 15q26 of the t(15;22). These breakpoints, however, were not found at the center of inverted repeat sequences like those seen in translocations involving the common LCR-B breakpoint. Free energy analysis supports these findings, since the average ΔG and ΔG/nt values for the non-LCR-B translocation breakpoints were 627.2 and 0.523 kcal/mol, respectively (Table 2). These free energy values are significantly higher than those for breakpoints that do involve LCR-B (P < 0.01; compare the results in Table 2 with those of the 1200 bp values found in Table 1). This analysis indicates that sequences participating in translocations with the common LCR-B breakpoint are unique in their propensity to form stem–loops and are statistically more likely to form secondary structure than sequences surrounding other constitutional breakpoints.
Figure 7.
Breakpoints of constitutional translocations not involving LCR-B of 22q11.2 do not contain palindromic sequences larger than 30 bp capable of forming prominent stem–loop structures. 1200 bp of single-strand sequence surrounding each of the indicated breakpoints were analyzed by M-Fold and are illustrated in two-dimensional graphical form. Breakpoint bases are marked by arrows.
Table 2.
Secondary structure and sequence characteristics of regions surrounding breakpoints of constitutional translocations not involving LCR-B of 22q11.2
| Translocation BP | 1200 nt | ||||
|---|---|---|---|---|---|
| Tm (°C) | GDS (kcal/mol) | GSTRUC (kcal/mol) | ΔG (kcal/mol) | ΔG/nt (kcal/mol) | |
| Ch2q14 @ t(2;22) | 86.8 | −903.2 | −254.4 | 648.8 | 0.541 |
| Ch22q11.21 @ t(2;22) | 86.0 | −885.1 | −232.4 | 652.7 | 0.544 |
| Ch10q26 @ t(10;22) | 86.8 | −903.0 | −256.2 | 646.8 | 0.539 |
| Ch22q11.2 @ t(10;22) | 88.3 | −920.1 | −277.8 | 642.1 | 0.535 |
| Ch15q26.2 @ t(15;22) | 76.8 | −751.3 | −136.6 | 614.7 | 0.512 |
| Ch22q12.1 @ t(15;22) | 77.6 | −759.2 | −146.9 | 612.3 | 0.510 |
| Ch21p12 @ t(21;22) | 78.9 | −788.2 | −160.6 | 627.6 | 0.523 |
| Ch22q11.2 @ t(21;22) | 82.9 | −848.0 | −206.4 | 641.6 | 0.535 |
| Ch2q37.2 @ t(X;2) | 79.9 | −801.1 | −166.5 | 634.6 | 0.529 |
| ChXp21.2 @ t(X;2) | 78.9 | −781.2 | −160.1 | 621.1 | 0.518 |
| Ch4q35.2 @ t(X;4) | 74.5 | −716.3 | −128.6 | 587.7 | 0.490 |
| ChXq21.1 @ t(X;4) | 75.2 | −723.3 | −127.2 | 596.1 | 0.497 |
| Average | 627.2 | 0.523 | |||
| Standard error | 6.1 | 0.005 | |||
| P-value (t-test)a | <0.001a | <0.001a | |||
| ChXq27 @ t(X;22) (inferred)b | 79.5 | −788.6 | −141.4 | 647.2c | 0.539c |
| Ch22q11.21 @ t(X;22) (inferred)b | 75.8 | −736.5 | −125.2 | 611.3c | 0.509c |
M-Fold analyses was performed on 1200 nt of sequence surrounding breakpoints of constitutional translocations not involving the common 22q11.2 LCR-B breakpoint.
References for characterized breakpoint sequences involved in the indicated translocations: t(2;22) (23); t(10;22) (24); t(15;22) (18); t(21;22) (25); t(X;2) and t(X;4) (26); t(X;22) (37).
Statistical analyses were done by comparing ΔGs and ΔG/nts for breakpoints involved in (2;22), (10;22), (15;22), (21;22) (X;2) and (X;4) translocations shown in this table with 1200 nt of sequence surrounding the breakpoints involved in the 22q11.2 LCR-B translocations [t(11;22), t(17;22), t(4;22), t(1;22)] listed in Table 1.
Breakpoint sequences used in these analyses were inferred from der(X) junction fragment sequence (34), and assumed no loss of sequence between derivative and normal chromosomes. Owing to greater sequence identity, the location of the 22q11.2 breakpoint in this t(X;22) was taken to be located within a second NF1-like repeat found in the LCR-B (see Fig. 1 for breakpoint location). 1200 nt of sequence surrounding the inferred Xq27 and 22q11.2 breakpoints were analyzed.
ΔGs and ΔG/nts found for t(X;22) breakpoints were not included in statistical analyses since these breakpoint junctions have not been fully characterized.
Although non-LCR-B translocation breakpoints do not form large stem–loops, partner chromosome junction sequences do appear to share similar melting temperatures and ΔG values for the formation of secondary structure. As Table 2 shows, the 1200 nt of sequence surrounding breakpoints of a particular translocation have Tms that are similar to one another, but not necessarily to that of the breakpoints involved in other translocations. For example, the 2q14 and 22q11.2 breakpoint sequences involved in the t(2;22) are 86.8 and 86.0°C, respectively—only a 0.8°C difference (Table 2). On the other hand, breakpoint sequences involved in the t(X;4) have Tms that are over 10°C lower than those involved in the t(2;22), while the Xq21.1 and 4q35.2 partner breakpoints are only 0.7°C different from one another. The greatest difference between Tms of partner chromosomes is just 4°C [breakpoints involved in the t(21;22)], whereas Tms of these translocation breakpoints in general range from 88.3 to 74.5°C—almost 14°C. The same trend is also observed in the free energies for the formation of single-strand secondary structure in that ΔG and ΔG/nt values for breakpoints of a particular translocation appear more similar to one another than they are to breakpoint sequences taking part in other translocations (Table 2). These analyses suggest that constitutional translocations, in general, occur between chromosomal regions having similar sequence characteristics.
DISCUSSION
In addition to providing valuable insight into sequence characteristics underlying the mechanism of constitutional translocations, the present work localizes the breakpoint of an ependymoma-associated t(1;22) and offers a starting point from which the molecular etiology of this patient’s disorder might be better understood. BLAT algorithm searches of the UCSC database, however, suggest that this rearrangement does not exert its effect by disrupting the intron–exon structure of any known gene or EST. The two nearest genes are located 250.4 kb proximal and 311.6 kb distal to the 1p21.2 breakpoint, and encode Optimedin (OLFM3, accession no. AF397397) (27,28) and endothelial differentiation protein 1 (EDG-1, accession no. NM_001400) (29), respectively. Future work may shed more light on the effect of this translocation on the expression of these and other 1p21.2 genes.
The present work contributes substantially to our understanding of the sequence motifs underlying the formation of constitutional translocations, particularly those involving the common 22q11.2 LCR-B breakpoint. These analyses found the most recurrent rearrangements involving LCR-B to occur at breakpoint sequences having a greater relative propensity to form secondary structure. This was determined by comparing the free energy of these sequences in the double-stranded configuration with that of the same sequence folded into secondary structure. Stable secondary structures, such as those arising from palindromic sequences, have free energy values approaching that of double-stranded DNA, and are therefore more likely to occur. In theory, a perfect inverted repeat sequence should be sufficient to predispose a chromosomal site to forming a translocation with the common LCR-B breakpoint regardless of the A/T content. The 4q35.1 breakpoint involved in the t(4;22) approaches this condition with 554 nt of inverted repeat sequence that is relatively G/C-rich (15). However, the A/T content of a breakpoint sequence is also likely to be a contributing factor; low melting temperatures would be expected to allow double-stranded DNA to denature more readily, allowing the subsequent formation of stem–loop structures. A combination of low melting temperature and palindromic sequences are found at the t(17;22) and t(11;22) breakpoints, and these two factors presumably underlie the more recurrent nature of these rearrangements. In this regard, the 22q11.2 LCR-B breakpoint, with its ~72°C Tm and 590 nt palindrome, satisfies both of these criteria. The 1p21.2 breakpoint is also extremely AT-rich, and, at least in the t(1;22) patient’s breakpoint sequence, contains a palindrome capable of forming a prominent stem–loop structure. If palindromic sequences at this polymorphic region of 1p21.2 persist in the population, the t(1;22) may actually be expected to recur. Although analysis of other ependymoma cases may uncover additional similar rearrangements, abnormalities of chromosome 1p are rarely reported in these tumors based on data available by searching the Cancer Genome Anatomy Project (CGAP) web site (http://cgap.nci.nih.gov).
These analyses predict that palindromic sequences are susceptible to forming translocations with the common 22q11.2 LCR-B breakpoint. It does appear that the progenitor of the t(1;22) contained a palindrome that could have predisposed this individual to this rearrangement. Unfortunately, the presence of this sequence configuration in control DNA samples could not be successfully amplified by PCR. Nevertheless, control sequences present in both the NCBI and Celera databases demonstrate the apparent polymorphic nature of this AT-rich 1p21.2 breakpoint region. Furthermore, the presence of a gap at the 1p21.2 breakpoint in the control Celera sequence suggests the presence of a palindromic sequence, since inverted repeat sequences are not stably cloned into bacterial and yeast vectors (21,22). In this regard, other gaps in the current human genome assembly may represent sites with which the common LCR-B breakpoint is predisposed to forming translocations.
The 33 to 36 bp VNTRs in the region surrounding the 1p21.2 breakpoint closely resemble the AT-rich minisatellite repeats (VNTRs) found at chromosomal fragile sites FRA10B and FRA16B at 10q25.2 and 16q22.1, respectively (30,31). The repeated sequences of both FRA10B and FRA16B have partial palindromic character and have been hypothesized to form hairpin structures (31), much like the AT-rich region of 1p21.2. Fragile sites are induced in cultured cells grown under particular conditions and/or in the presence of mutagenic agents. They are thought to represent unstable chromosomal regions involved in the rearrangements that lead to genetic disorders and cancer. In the case of FRA10B and FRA16B, induction by bromodeoxyuridine and distamycin A, respectively, is associated with the expansion of VNTRs contained at these sites (30,31). Several fragile sites have been identified cytogenetically on 1p21–22, including FRA1D, FRA1E, FRA1F and FRA1M (32,33), but none of these has been characterized at the sequence level. Given its similarity to the VNTRs of known fragile site sequence, the t(1;22) breakpoint on 1p21.2 may actually represent one of these FRA1 sites. Further, it has been shown that inverted repeat sequences introduced transgenically in mice confer genomic instability (34–36). Perhaps the sequences present at the AT-rich palindromes of the 1p21.2 breakpoint, FRA10B and FRA16B, underlie the apparent genomic instability at these loci.
The insertion of mobile repetitive elements, such as Alus, may also create palindromic sites susceptible to translocations with the common LCR-B breakpoint. For such a site to be generated, however, an element would need to insert itself sufficiently close to an identical sequence oriented in the opposite direction. This appears to be the case for the recently described t(4;22) translocation in which a 554 nt inverted repeat sequence, partially comprised of oppositely oriented Alu elements, are separated by 547 nt of sequence (15). This prompts the question of what defines repetitive elements that are sufficiently close? The existing dataset of characterized 22q11.2 LCR-B translocations is not yet sufficient to place an upper limit on the distance between mobile repeat elements permissive to the formation of stem–loop structures. It is also worth emphasizing that this secondary structure motif only holds for LCR-B translocations, since constitutional translocations in general do not appear to involve palindromic sequences capable of forming stem–loop structures.
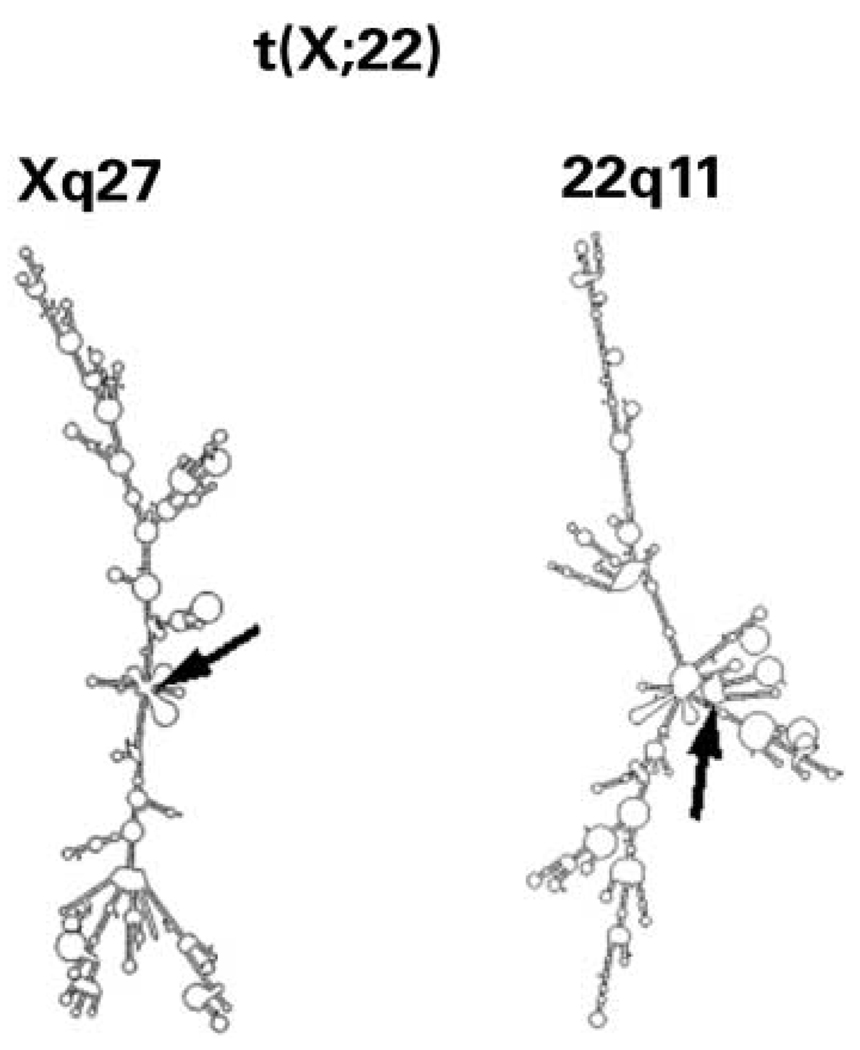
One translocation, the t(X;22), has been suggested to occur at the common LCR-B breakpoint, a reasonable conclusion based on FISH results localizing the breakpoint to this region of 22q11.2 and sequence data from a single junction fragment derived from the der(X) chromosome (37). Examination of the chromosome 22 sequence contained in this junction fragment, however, indicates that this breakpoint is likely to reside within an NF1L repeat module that is distinct from the one containing the common LCR-B breakpoint (see Fig. 1 for putative breakpoint location). The fact that repeated attempts to amplify the der(22) sequence by PCR had failed (37) supports this conclusion, since the alternative NF1L sequence does not contain primer annealing sequences needed for these amplifications. M-Fold analysis shows that neither of the sequences surrounding the Xq27 or 22q11.2 breakpoints is capable of forming a stem–loop structure (Fig. 8). Furthermore, the free energy differences for these junctions [647.2 kcal/mol (0.539 kcal/mol per nucleotide) and 611.3 kcal/mol (0.509 kcal/mol per nucleotide), respectively; Table 2], lie outside that typically seen for LCR-B translocations [369.6 ± 83.9 kcal/mol (0.360±0.052 kcal/mol per nucleotide), Table 1]. These results suggest that the 22q11.2 breakpoint involved in the t(X;22) is located at a different site within the LCR-B distinct from the common breakpoint. Amplification of the der(22) junction fragment with primers designed against the alternative NF1L sequence would confirm this hypothesis.
Figure 8.
Single-strand sequences surrounding t(X;22) breakpoints do not give rise to prominent stem–loop secondary structures. Sequences on both sides of the t(X;22) breakpoints were inferred from the single der(X) junction fragment sequence (34), and were assumed to have no loss of sequence during the formation of this rearrangement. The 22q11.2 sequence found in the chromosome 22 portion of the der(X) junction fragment directly matched that contained in a second NF1L sequence found in genomic cosmid clone HK89 (accession no. AC024070) [see Fig. 1 for t(X;22) breakpoint location within the LCR-B]. 1200 nt of each breakpoint sequence were analyzed by M-Fold and the two-dimensional graphical output presented. Arrows indicate breakpoint location.
The present work provides important findings regarding the sequences required for the formation of constitutional translocations. In general, these rearrangements appear to take place between chromosomal regions having similar characteristics, but not necessarily sequence homology. For example, translocations involving LCR-B occur between sequences containing palindromes capable of forming stem–loop structures that do not share extensive sequence homology. Other translocations appear to form between partner chromosome sequences of both similar G/C content and propensity to form secondary structure. However, sequences surrounding translocation breakpoints, in general, do not appear to share common sequence characteristics. This suggests that the mechanism responsible for constitutional translocations does not require a specific sequence motif, but only that breakpoint sequences somehow interact such that double-strand break and repair processes join two juxtaposed chromosomes. Because partner chromosomes seem to share the same propensity to form secondary structure, chromosomal sequences may interact with one another through some higher-order structure that is not detectable in the simple analysis performed here. Single-strand structures in vivo are undoubtedly quite complex with DNA binding proteins stabilizing the formation of single-strand structures interspersed with regions annealed to the complimentary strand. M-Fold cannot predict such structures, nor can it predict the conditions required for these structures to form in vivo. Nevertheless these analyses do provide a meaningful measure of the relative likelihood of a given strand of DNA to adopt a secondary structure.
To fully understand the genesis of constitutional translocations, the biochemical mechanisms that require these sequence motifs must be better understood. Proteins involved in the translocation mechanism must recognize and bring these DNA secondary structures together, nick both strands of DNA and ligate the partner chromosomes together. Although the breakpoints of partner chromosomes do not appear to share extensive sequence identity, the present results suggest that genomic regions of similar melting temperature and propensity to form secondary structure interact to form these rearrangements. In the case of translocations involving the LCR-B, these structures take the form of stem-loops or cruciforms. Constitutional translocations appear to occur during meiosis (16), and potentially concurrent with homologous recombination. Given a lack of sequence identity between breakpoint sequences, constitutional translocations are not likely to involve a simple homologous recombination mechanism, but may employ similar protein factors. A further understanding of these mechanisms as well as those involved in DNA repair, chromosome cohesion and segregation may ultimately lend insight into constitutional translocation processes.
MATERIALS AND METHODS
Fluorescence in situ hybridization
The de novo t(1;22) patient was originally identified by prenatal diagnosis. A lymphoblastoid cell line was established subsequently and used for cytogenetic and molecular analysis (19,20). Metaphase FISH to localize the 22q11.2 breakpoint to LCR-B was performed using standard methodology (25). Probes were labeled by nick translation (38) and included cosmid clones c68a1 (N41) and c87f9 (ZNF74) which flank LCR-B and a cosmid containing D22S39 to mark the distal region of 22q. Using similar methods, BAC 336L20 (see below) was verified by FISH as the 1p21.2 breakpoint-spanning BAC.
Narrowing the 1p21.2 breakpoint and STS mapping using somatic hybrid cell DNA
A human genomic pBeloBAC library (Research Genetics) was screened by PCR using several STS markers adjacent to the 1p21.2 breakpoint to identify relevant clones (20). One of these clones, BAC 336L20, was found to span the 1p21.2 breakpoint by STS analysis and FISH (see above). Fragments of 336L20 were subcloned, sequenced and STS markers were generated from subclone end sequences. PCR mapping of somatic cell hybrid DNA (19) identified two sequence tagged sites, ‘STS-A’ and ‘STS-B’, contained separately in the der(1) and der(22) hybrids, respectively. Primers to amplify these markers were the following: STS-A.u, TGAGGTTAAGGTTGAAAGC; STS-A.l, GGGAAGAGTCCAGAATGTAT; STS-B.u, AAGTAGTTTGTTCTGTGGCT; STS-B.l, GTTTGATTTTGATGATAGCA. Southern blots of SpeI and BstXI/SpeI-digested DNA from the t(1;22) patient and somatic cell hybrid DNA probed with a 1.8 kb probe immediately telomeric to STS-B confirmed the breakpoint localization. Together these results placed the t(1;22) breakpoint within a 5.73 kb region on 1p21.2 found within genomic assembly clone, RP4-600m23 (accession no. AC114483.2). To further position the breakpoint within this 5.73 kb sequence, it was analyzed by RepeatMasker (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker) to identify repetitive sequence, and several unique primer pairs were used in PCR mapping experiments to narrow the breakpoint to the 1990 bp AT-rich region depicted in Figure 3 and Figure 4C.
Nested PCR amplification of der(1) and der(22) junction fragments
der(1) and der(22) junction fragments were amplified by sequential nested primer PCR using the GeneAmp XL PCR kit (Applied Biosystems) with modifications to the manufacturer’s instructions as previously described (15). ‘Hot-start’ protocols were run with the following parameters: 1 min at 85°C, after which 1 µl of rTth polymerase was added to the reaction, 94°C for 1 min; 12 cycles of 94°C for 30 s (denaturation), 72–61°C (~1°C/cycle) for 45 s (annealing), 74°C for 3 min (extension); followed by 30 cycles of 94°C for 30 s (denaturation), 60°C for 45 s (annealing), 74°C for 3 min (extension). For all primary junction fragment amplifications, template was DNA purified from t(1;22) patient-derived lymphoblasts. Subsequent amplifications with nested primers included 1 µl of product from the primary reaction as template in 50 µl reactions. Sequence-specific primers corresponding to 1p21.2 and 22q11.2 LCR-B sequences were oriented as depicted in Figure 3 and were designed with the following sequences: 1.9FA, GGTACAAATCACCTTTATACTGGCATACCT;1.9FB, CGTTCTGCACATTCTGCACCTGTATCT; 1.6RD, GCCTGTGCTCTACAAATGGATCATTGAAGT; 1.6RE, GAGTGTTGACAATCTCATGATGAAACTTCAATGA; NF1d, AGTGACTTACTACAAATATATTAA; 22.B2R, GGAAGGGAAAAACATGTTAAAAACAAAGAGAGGTAC; 22.B1, CAAAATTGTGTGAAAAGCCTCCAACGG; 22.B3, GGGGGTGGGGGATGGAACGTTGAAGGATC (in some cases, sequence-specific primers were designed against non-conserved base pairs existing within repeat element sequences). der(1) junction fragments were amplified with NF1d and 1.6RD primers followed by nested PCR with 22.B2R and 1.6RE primers. der(22) junction fragments were amplified with 1.9FA and 22.B1 primers, and 1.9FB and 22.B3 subsequently. Products from these reactions were cloned into the pCRII-TopoTA vector (Invitrogen) and sequenced using vector-specific primers (Children’s Hospital of Philadelphia Nucleic Acids Core Facility). der(1) and der(22) junction fragment sequences were compiled from complete sequencing of three and two clones, respectively. The 1p21.2 breakpoint sequence in the t(1;22) patient was constructed from chromosome 1 sequence contained in junction fragments and compared to normal sequence from the genome assembly clone, RP4-600m23, as well as that contained in the Celera database (www.celeradiscoverysystem.com/index.cfm). Variable number tandem repeats were determined by visual inspection of sequences plotted at a period length equal to the length of the particular VNTR.
Sequence and single-strand secondary structure analysis
The location of all gene loci, BAC clones, polymorphic markers and breakpoint sequences relative to G-banding patterns were determined according to the UCSC Genome Bioinformatics website (November 2002 freeze; www.genome.ucsc.edu/). Differences between this current database and older ones have prompted us to update the t(1;22) breakpoint from the original 1p22 designation (19,20) to 1p21.2 in the current paper.
Breakpoint sequences from constitutional translocations in which both derivative chromosome junctions had been sequenced were obtained from published reports [t(11;22) (q23;q11.2) (4–7), t(17;22)(q11.2;q11.2) (9,10), t(4;22)(q35.1; q11.2) (15), t(2;22)(q14;q11.2) (23); t(15;22)(q26;q11.2) (18); t(21;22)(p12;q11.2) (25); t(X;2)(p21.2;q37.2) and t(X;4) (p21.1;q35.1) (26)], a doctoral thesis, [t(10;22)(q26;q11.2) (24)] or junction fragment amplification [t(1;22) (determined in present work)]. These translocations were chosen on the basis of their full characterization at the sequence level. For the (X;22)(q27;q11.2) translocation, only the der(X) junction fragment sequence has been reported (37). Sequence surrounding the putative X and 22 breakpoints was identified under the assumption that no sequence was lost during the translocation. For this reason, these data were not included in further statistical analyses. For each translocation, 2500 bp of sequence surrounding each breakpoint was obtained through BLASTN searches of genomic sequence (htgs) within the National Center for Biotechnology’s web site (www.ncbi.nlm.nih.gov/BLAST/), or BLAT searches of genome assembly within the UCSC Genome Bioinformatics web site. With the exception of the common LCR-B breakpoint for which normal genomic sequence is not available across the breakpoint, published breakpoint sequences were used only if less than 20 bp were lost due to the translocation as compared to genomic control sequence contained in the NCBI database.
Potential secondary structure formed within a single strand of DNA sequence was determined by entering 2500 (not shown) and 1200 bp of sequence surrounding indicated breakpoints (1250 and 600 bp, respectively, on either side) into the European M-Fold server (http://BiBiServ.TechFak.Uni-Bielefeld.DE/mfold/). Default parameters were used except for the ‘DNA’ setting. Plots of nucleotide sequence folding (Supplementary Material) and two-dimensional figures were obtained as output and free energy values were recorded (GSTRUC). To remain consistent in determining free energy values for the same sequence annealed to its complimentary strand (GDS), the 1200 nt sequence along with its reverse compliment was analyzed and the resulting output halved. Free energy for the formation of secondary structure (ΔG) is the GDS − GSTRUC difference, and ΔG/nt is this value normalized to the amount of sequence analyzed. Because numerous structures (>50) are typically obtained for any particular sequence entry, the most stable structure, or that with the most negative free energy value, was used in subsequent analyses. To evaluate only the stem–loop region of translocation breakpoints involving LCR-B (Table 1, first eight columns), sequences from the first to last nucleotide of palindromes identified in larger 2500 and 1200 nt structures were reanalyzed by M-Fold.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Drs Hiroki Kurahashi and Manjunath Nimmakayalu for helpful discussions. Access to preliminary t(1;22) breakpoint mapping performed by Katherine M. Call, Randall Little and Karen Braunschweiger, and t(10;22) breakpoint cloning and sequencing achieved by Carla Chieffo are gratefully acknowledged. These studies were supported by funds from the American Cancer Society (Dartmouth-Hitchcock Medical Center institutional grant), the Charles E. H. Upham chair in Pediatrics (B.S.E.) and grants R43-CA69996-01 and CA29926 (B.S.E.) from the National Cancer Institute of the NIH.
Footnotes
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG Online.
REFERENCES
- 1.Burn J, Goodship J. Congenital heart disease. In: Rimoin DL, Connor JM, Peyritz RE, Emery AEH, editors. Principles and Practice of Medical Genetics. Vol. 1. New York: Churchill Livingstone; 1996. pp. 767–803. [Google Scholar]
- 2.Emanuel BS, Shaikh TH. Segmental duplications: an ‘expanding’ role in genomic instability and disease. Nat. Rev. Genet. 2001;2:791–800. doi: 10.1038/35093500. [DOI] [PubMed] [Google Scholar]
- 3.McTaggart KE, Budarf ML, Driscoll DA, Emanuel BS, Ferreira P, McDermid HE. Cat eye syndrome chromosome breakpoint clustering: identification of two intervals also associated with 22q11 deletion syndrome breakpoints. Cytogenet. Cell Genet. 1998;81:222–228. doi: 10.1159/000015035. [DOI] [PubMed] [Google Scholar]
- 4.Kurahashi H, Shaikh TH, Hu P, Roe BA, Emanuel BS, Budarf ML. Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22) Hum. Mol. Genet. 2000;9:1665–1670. doi: 10.1093/hmg/9.11.1665. [DOI] [PubMed] [Google Scholar]
- 5.Kurahashi H, Shaikh TH, Zackai EH, Celle L, Driscoll DA, Budarf ML, Emanuel BS. Tightly clustered 11q23 and 22q11 breakpoints permit PCR-based detection of the recurrent constitutional t(11;22) Am. J. Hum. Genet. 2000;67:763–768. doi: 10.1086/303054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurahashi H, Emanuel BS. Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum. Mol. Genet. 2001;10:2605–2617. doi: 10.1093/hmg/10.23.2605. [DOI] [PubMed] [Google Scholar]
- 7.Edelmann L, Spiteri E, Koren K, Pulijaal V, Bialer MG, Shanske A, Goldberg R, Morrow BE. AT-rich palindromes mediate the constitutional t(11;22) translocation. Am. J. Hum. Genet. 2001;68:1–13. doi: 10.1086/316952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurahashi H, Emanuel BS. Unexpectedly high rate of de novo constitutional t(11;22) translocations in sperm from normal males. Nat. Genet. 2001;29:139–140. doi: 10.1038/ng1001-139. [DOI] [PubMed] [Google Scholar]
- 9.Kehrer-Sawatzki H, Assum G, Hameister H. Molecular characterization of t(17;22)(q11.2;q11.2) is not consistent with NF1 gene duplication. Hum. Genet. 1997;111:465–467. doi: 10.1007/s00439-002-0794-3. [DOI] [PubMed] [Google Scholar]
- 10.Kurahashi H, Shaikh TH, Takata M, Toda T, Emanuel BS. The constitutional t(17;22): another translocation mediated by palindromic AT-rich repeats. Am. J. Hum. Genet. 2003;72:733–738. doi: 10.1086/368062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaikh TH, Budarf ML, Celle L, Zackai EH, Emanuel BS. Clustered 11q23 and 22q11 breakpoints and 3;1 meiotic malsegregation in multiple unrelated t(11;22) families. Am. J. Hum. Genet. 1999;65:1595–1607. doi: 10.1086/302666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaikh TH, Kurahashi H, Saitta SC, O’Hare AM, Hu P, Roe BA, Driscoll DA, McDonald-McGinn DM, Zackai EH, Budarf ML, Emanuel BS. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum. Mol. Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- 13.Shaikh TH, Kurahashi H, Emanuel BS. Evolutionarily conserved low copy repeats (LCRs) in 22q11 mediate deletions, duplications, translocations, and genomic instability: an update and literature review. Genet. Med. 2001;3:6–13. doi: 10.1097/00125817-200101000-00003. [DOI] [PubMed] [Google Scholar]
- 14.Tapia-Paez I, Kost-Alimova M, Hu P, Roe BA, Blennow E, Fedorova L, Imreh S, Dumanski JP. The position of t(11;22)(q23;q11) constitutional translocation breakpoint is conserved among its carriers. Hum. Genet. 2001;109:167–177. doi: 10.1007/s004390100560. [DOI] [PubMed] [Google Scholar]
- 15.Nimmakayalu MA, Gotter AL, Shaikh TM, Emanuel BS. A novel sequence-based approach to localize translocation breakpoints identifies the molecular basis of a t(4;22) Hum. Mol. Genet. 2003;12:2817–2825. doi: 10.1093/hmg/ddg301. [DOI] [PubMed] [Google Scholar]
- 16.Miller OJ, Therman E. The causes of structural aberrations. In: Miller OJ, Therman E, editors. Human Chromosomes, Fourth Edition. New York: Springer; 2001. pp. 207–221. [Google Scholar]
- 17.Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. Spatial proximity of translocation-prome gene loci in human lymphomas. Nat. Genet. 2003;34:287–291. doi: 10.1038/ng1177. [DOI] [PubMed] [Google Scholar]
- 18.Spiteri E, Babcock M, Kashork CD, Wakul K, Gogineni S, Lewis DA, Williams KM, Minoshima S, Sasaki T, Shimizu N, et al. Frequent translocations occur between low copy repeats on chromosome 22q11.2 (LCR22s) and telomeric bands of partner chromosomes. Hum. Mol. Genet. 2003;12:1823–1837. doi: 10.1093/hmg/ddg203. [DOI] [PubMed] [Google Scholar]
- 19.Park JP, Chaffeed S, Noll WW, Rhodes CH. Constitutional de novo t(1;22)(p22;q11.2) and ependymoma. Cancer Genet. Cytogenet. 1996;86:150–152. doi: 10.1016/0165-4608(95)00182-4. [DOI] [PubMed] [Google Scholar]
- 20.Rhodes CH, Call KM, Budarf ML, Barnoski BL, Bell CJ, Emanuel BS, Bigner SH, Park JP, Mohandas TK. Molecular studies of an ependymoma-associated constitutional t(1;22)(p22;q11.2) Cytogenet. Cell Genet. 1997;78:247–252. doi: 10.1159/000134667. [DOI] [PubMed] [Google Scholar]
- 21.Leach DRF. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays. 1994;16:893–900. doi: 10.1002/bies.950161207. [DOI] [PubMed] [Google Scholar]
- 22.Gordenin DA, Lobachev KS, Degtyareva NP, Malkova AL, Perkins E, Resnick MA. Inverted DNA repeats: a source of eukaryotic genomic instability. Mol. Cell. Biol. 1993;13:5315–5322. doi: 10.1128/mcb.13.9.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Budarf ML, Collins J, Gong W, Roe B, Wang Z, Bailey LC, Sellinger B, Michaud D, Driscoll DA, Emanuel BS. Cloning a balanced translocation associated with DiGeorge syndrome and identification of a disrupted candidate gene. Nat. Genet. 1995;10:269–278. doi: 10.1038/ng0795-269. [DOI] [PubMed] [Google Scholar]
- 24.Chieffo C. Department of Pathology Doctoral Thesis. University of Pennsylvania; 1998. The isolation and characterization of the TBX1 gene and the t(10;22) breakpoint of GM5878 from the DiGeorge chromosomal region; pp. 67–91. [Google Scholar]
- 25.Holmes SE, Riazi MA, Gong W, McDermid HE, Sellinger BT, Hua A, Chen F, Wang Z, Zhang G, Roe B, et al. Disruption of the clathrin heavy chain-like gene (CLTCL) associated with features of DGS/VCFS: a balanced (21;22)(p12;q11) translocation. Hum. Mol. Genet. 1997;6:357–367. doi: 10.1093/hmg/6.3.357. [DOI] [PubMed] [Google Scholar]
- 26.Bodrug SE, Ray PN, Gonzalez IL, Schmickel RD, Sylvester JE, Worton RG. Molecular analysis of a constitutional X-autosome translocation in a female with muscular dystrophy. Science. 1987;237:1620–1624. doi: 10.1126/science.3629260. [DOI] [PubMed] [Google Scholar]
- 27.Rhodes CH. NOE3, a novel Olfactomedin/Noelin/Pancortin homolog identified near an ependymoma-associated translocation breakpoint. Neuro-Oncology. 2001;3:306. (abstract) [Google Scholar]
- 28.Torrado M, Trivedi R, Zinovieva R, Karavanova I, Tomarev SI. Optimedin: a novel olfactomedin-related protein that interacts with myocilin. Hum. Mol. Genet. 2002;11:1291–1301. doi: 10.1093/hmg/11.11.1291. [DOI] [PubMed] [Google Scholar]
- 29.Hla T, Maciag T. An abundant transcript induced in differentiating human endothelial cells encodes a polypeptide with structural similarities to G-protein-coupled receptors. J. Biol. Chem. 1990;265:9308–9313. [PubMed] [Google Scholar]
- 30.Hewett DR, Handt O, Hobson L, Mangelsdorf M, Eyre HJ, Baker E, Sutherland GR, Schuffenhauer S, Mao J, Richards RI. FRA10B structure reveals common elements in repeat expansion and chromosomal fragile site genesis. Mol. Cell. 1998;1:773–781. doi: 10.1016/s1097-2765(00)80077-5. [DOI] [PubMed] [Google Scholar]
- 31.Yu S, Mangelsdorf M, Hewett D, Hobson L, Baker E, Eyre HJ, Lapsys N, Le Paslier D, Dogget NA, Sutherland GR, Richards RI. Human chromosomal fragile site FRA16B is an amplified AT-rich minisatellite repeat. Cell. 1997;88:367–374. doi: 10.1016/s0092-8674(00)81875-9. [DOI] [PubMed] [Google Scholar]
- 32.Yunis JJ, Soreng AL. Constitutive fragile sites and cancer. Science. 1984;226:1199–1204. doi: 10.1126/science.6239375. [DOI] [PubMed] [Google Scholar]
- 33.Le Beau MM. Chromosomal fragile sites and cancer-specific rearrangements. Blood. 1986;67:849–858. [PubMed] [Google Scholar]
- 34.Collick A, Drew J, Penberth J, Bois P, Luckett J, Scaerou F, Jeffreys A, Reik W. Instability of long inverted repeats within mouse transgenes. EMBO J. 1996;15:1163–1171. [PMC free article] [PubMed] [Google Scholar]
- 35.Akgun E, Zahn J, Baumes S, Brown G, Liang F, Romanienko PJ, Lewis S, Jasin M. Palindrome resolution and recombination in the mammalian germ line. Mol. Cell. Biol. 1997;17:5559–5570. doi: 10.1128/mcb.17.9.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Z-H, Akgun E, Jasin M. repeat expansion by homologous recombination in the mouse germ line at palindromic sequences. Proc. Natl Acad. Sci. USA. 2001;98:8326–8333. doi: 10.1073/pnas.151008498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Debeer P, Mols R, Huysmans C, Devriendt K, Van de Ven WJ, Fryns J-P. Involvement of a palindromic chromosome 22-specific low-copy repeat in a constitutional t(X;22)(q27;q11) Clin. Genet. 2002;62:410–414. doi: 10.1034/j.1399-0004.2002.620510.x. [DOI] [PubMed] [Google Scholar]
- 38.Lichter P, Tang CJ, Call K, Hermanson G, Evans GA, Housman D, Ward DC. High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science. 1990;247:64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.