Abstract
High mobility group box 1 (HMGB1) is a nonhistone architectural protein that is involved in many biological processes including chromatin remodeling, transcription, cell signaling of inflammation, DNA damage repair and others. Recent studies have identified the cross-link of HMGB1 with a DNA base excision repair intermediate indicating that this protein is involved in base excision repair (BER) pathway. Further characterization of the roles HMGB1 in BER demonstrates that the protein acts as a cofactor to regulate BER sub-pathways by inhibiting single-nucleotide BER and stimulating long-patch BER through modulating the activities of base excision repair enzymes. Directing of base lesion repair to the long-patch sub-pathway can result in trinucleotide repeat instability suggesting an important role of HMGB1 in modulating genome stability.
Introduction and Background
High mobility group box 1 protein (HMGB1) belongs to the family of the high mobility group (HMG) nuclear proteins. It is an abundant non-histone chromosomal protein with a concentration of 106 per cell [1]. The HMGB1 protein contains an N-terminal Box A domain, a central Box B domain and C-terminal acidic tail (Figure 1) [2, 3]. The HMGB1 Box A and B domains contain 80 to 90 basic amino acids and share ~30% identity in their amino acid sequences. The C-terminal acidic tail contains ~30 consecutive aspartate and glutamate residues [4]. It has been found that the HMGB1 Box A domain is responsible for HMGB1 binding to DNA damage [5], whereas the Box B exhibits pro-inflammatory activities, in addition to DNA binding [6]. The acidic tail of HMGB1 is involved in regulating DNA binding and DNA damage repair [7, 8].
Figure 1.

Structures of HMGB1 protein domains. Tertiary structures of HMGB1 Box A-DNA and Box B were produced according to published data (refs. 5 and 9) using Chimera [58].
Structural studies have revealed that both Box A and Box B domains contain three alpha-helices, named helix I, helix II and helix III and two loops, loop I and loop II [5, 9]. These domains adopt a unique “L” shape structure that has two arms of 31 and 36 Å in length, respectively, in the case of the rat HMGB1 Box B domain (Figure 1)[9]. The short arm contains helix I and helix II. The long arm consists of helix III and an N-terminal unstructured segment in parallel with the helix (Figure 1). The “L” shape structure of the HMGB1 Box B domain is maintained by a cluster of highly conserved amino acid residues that are located in the junction between the two arms (Phe14, Phe17, Trp45, Lys53 and Tyr56) (Figure 1). The conserved basic residues (Lys26, Lys 39 and Arg 22 etc.) are mainly distributed around the concave surface in between the two arms, indicating that they may be involved in DNA binding (Figure 1). These residues may allow the concave surface to contact with the minor groove of double-strand DNA. Interestingly, it has been found that HMGB1 Box A domain also can bind to the DNA minor grove by intercalating the aromatic side chain of Phe37 at helix II into a hydrophobic cleft between two base pairs induced by DNA damage such as cisplatin crosslinking (Figure 1) [5]. This results in the bending of the DNA duplex towards the major groove by 61° indicating that HMGB1 can induce a significant change in DNA structures.
The intrinsic capacity of HMGB1 for altering DNA structures allows it to participate in many biological processes [10, 11][12]. These include regulation of chromatin structure [13], transcription [14], DNA damage repair and recombination [12]. HMGB1 also has been shown to participate in inflammation by acting as an inflammatory signal transducer [15]. The importance of HMGB1 in DNA damage and repair was initially identified in studies that revealed the ability of HMGB1 to bind to a variety of bulky DNA lesions induced by chemotherapeutic agents such as cisplantin [5], psoralen combined with UVC [16, 17], as well as environmental carcinogens such as acetylaminofluorene [18] and UVC [19]. Cellular and genetics studies also demonstrated that mammalian HMGB1 and its yeast homologue were critical for maintaining genome stability [20]. Since DNA damage is one of the mechanisms that cause genome instability, this study further suggests that HMGB1 might participate in DNA repair toward maintaining genome stability. Recent cell biology and biochemical studies indicated that HMGB1 is actively involved in modulating the efficiency of all four major DNA repair pathways, i.e., nucleotide excision repair (NER) [21, 22], base excision repair (BER) [23], mismatch repair [24] and double strand break repair and nonhomologous end-joining [25, 26].
The mechanisms by which HMGB1 modulates DNA repair are still not well understood and are under intensive study. It has been found that HMGB1 binds to DNA lesions based on specific structure recognition and bends DNA [5]. This allows HMGB1 to modulate interactions between repair proteins and lesion-containing DNA [22] thereby affecting damage repair efficiency [8, 22, 27, 28]. Since many DNA damage recognition proteins bind to lesions within distorted DNA much tighter than in normal B-form DNA [29], it is expected that HMGB1-induced DNA bending would facilitate damage recognition. However, the effects of HMGB1 on DNA repair are more complex than the DNA-bending model suggests and also may depend on the type of lesion being processed. For example, in the nucleotide excision repair pathway, it was shown that binding of HMGB1 to DNA cross-link damage induced by (2R, 3R)-diaminobutanedichloroplatinum (II) blocked the accessibility of the damage to NER proteins, resulting in inhibition of repair [28]. In contrast, HMGB1 binding to triplex-directed psoralen DNA interstrand cross-links (ICLs) facilitated the NER of these lesions [16, 22].
HMGB1 also may modulate DNA repair efficiency by directly interacting with repair enzymes and cofactors [12, 17]. Interaction between HMGB1 and replication protein A (RPA) resulted in formation of a HMGB1-RPA-psoralen DNA ternary complex that was proposed to help in the recruitment of NER proteins to damaged sites [16]. Moreover, HMGB1 can form a protein-DNA ternary complex with XPC-RAD23B and RPA simultaneously [17], suggesting that HMGB1 may coordinate damage recognition by these repair proteins thereby accelerating NER. In mismatch repair pathway, HMGB1 was shown to play a role in the initial mismatch recognition and incision steps by interacting with mismatch proteins such as MSH2 and MLH1 [24]. In addition, HMGB1 can substitute for the function of RPA to mediate the excision step of exonuclease 1 [30]. In vitro studies also identified HMGB1 stimulatory effects on DNA-PK and Ku proteins by interacting with and targeting these proteins to the ends of double-strand DNA breaks; these HMGB1 interactions facilitated damage repair [31]. In addition, HMGB1 was found to be involved in modulating nonhomologous end joining and V(D)J recombination by interacting with the key repair proteins in these pathways such as RAG proteins [32, 33] and DNA ligase IV [34]. Recently, we discovered a new role of HMGB1 in that it stimulates DNA base excision repair through its interaction with BER enzymes [23].
In addition to interacting with damaged DNA and DNA repair proteins, HMGB1 can regulate DNA damage repair through modulating chromatin dynamics and structure. As one of chromatin architectural proteins, HMGB1 can stimulate chromatin remodeling by presenting bended DNA to chromatin remodeling factors, e.g., ACF and CHRAC thereby facilitating nucleosome sliding [35]. In addition, it has been found that HMGB1 is critical for UV damage-induced histone acetylation [22]. These HMGB1-mediated processes can facilitate unpacking of chromatin and increase the accessibility of DNA lesions to repair proteins. Furthermore, HMGB1 may help to restore normal chromatin structure after completion of DNA repair [22]. Thus, HMGB1 appears to accelerate the whole process of repair that includes lesion removal and repair as well as unpacking and restoring chromatin structure.
A most recent study revealed some aspects of the molecular basis on how HMGB1 may interact with cisplatin-damaged nucleosomes [7, 36]. The results indicated that HMGB1 interacted with damaged sites using its C-terminal acidic tail, and this interaction inhibited repair of the lesion. Posttranslational modifications of HMGB1, such as acetylation and phosphorylation, were found to play an important role in mediating binding to cisplatin-induced damage and in the inhibitory effects on the damage repair [7].
We have discovered that HMGB1 can modulate base excision repair (BER) by interacting with the major players in BER pathway [23]. This was the first evidence indicating that HMGB1 can participate in repair of small base lesions. Since DNA base damage is an abundant form of DNA lesions in cells, this could cause the genome instability that is associated with common human diseases such as cancer and neurodegeneration. This linkage between HMGB1, maintenance of genome stability, and prevention of human diseases further emphasizes the importance of understanding the roles of HMGB1.
In this review, we will discuss the roles of HMGB1 during cellular repair of DNA base lesions and single-strand DNA breaks and their impact on base damage-induced genome instability.
Discovery of HMGB1 in association with a BER intermediate
Earlier, an unknown protein that interacted with a BER intermediate, the single-nucleotide gapped DNA with 5′-deoxyribose group (dRP), was identified in mouse embryonic fibroblasts (MEF) cell extract by making use of a photoaffinity labeling approach [37] (Figure 2, lane 2, band I) This protein was not detected in our laboratory standard bovine testis nuclear extract (BTNE) (Figure 2, lane 3). Initial efforts to identify this unknown protein were not successful, but subsequent efforts involving recovery of the protein-DNA complex and mass spectrometery revealed the protein to be HMGB1 with a high degree of certainty [23]. We then found that HMGB1 had been removed from the BTNE during a preparative step, explaining the failure to observe its cross-linking in this extract (Figure 2, lane 3).
Figure 2.

Photoaffinity labeling of HMGB1 in Cell extracts (from Figure S2 of ref. 23). (A) Diagram of BER intermediate with radiolabeled uracil and the scheme of enzymatic reactions. (B) NaBH4 crosslinking of proteins in various cell extracts with the substrate treated by UDG.
In other studies conducted in parallel, we used a different approach to further examined proteins in cells extracts that are capable of interacting with the BER intermediate containing the 5′-dRP group. In this approach, the C1′ aldehyde of the dRP group sugar forms a Schiff base complex with a lysine residue (or other nucleophile) in a protein and the complex is reduced by sodium borohydride treatment. Schiff base formation is an early step in the dRP lyase reaction that removes the dRP group from the BER intermediate. A DNA probe that mimics the BER intermediate with a single-nucleotide gapped DNA and a downstream natural 5′-dRP group was used, as shown in Figure 3A [23]. This DNA probe was constructed by annealing a synthesized oligonucleotide with a 32P-labeled 5′-deoxyuridine and 3′-biotin to a template oligonucleotide along with an upstream oligonucleotide (Figure 3A). The 5′-deoxyuridine then was treated with uracil-DNA glycosylase (UDG) and to remove uracil and generate a single-nucleotide gapped molecule with 32P-labeled 5′-dRP flap. This BER intermediate probe then was incubated with extracts from Pol β null cells and null cells complemented with Flag epitope-tagged Pol β. The mixtures were treated with sodium borohydride (NaBH4) allowing covalent cross-linking of the proteins capable of binding and Schiff base formation with the dRP group of the DNA probe [23]. Protein-DNA complexes were then captured by streptavidin-coated magnetic beads, eluted and resolved by SDS-PAGE. With this approach, we identified protein-DNA complexes that migrated at around 50- and 37-kDa, in the extracts that expressed Flag-tagged Pol β (Figure 3B lane 2). The mass of the 50-kDa protein-DNA complex corresponded to the mass of Pol β cross-linked with the 32P-labeled DNA probe, and this cross-linked product was not detected in Pol β null cell extracts (Figure 3B, lane 3). The mobility of the 50-kDa complex also corresponded to that of purified Pol β cross-linked with the DNA probe (Figure 3B, lane 5). These results indicated that the 50-kDa protein-DNA complex was the Pol β-DNA cross-linking product. The 37-kDa protein-DNA complex appeared to be an unknown protein cross-linked to the dRP group. As in the results from the photoaffinity labeling experiments described above (Figure 2, lane 2, Band I), the cross-linked 37-kDa protein-DNA complex was not detected in the bovine testis nuclear extract (Figure 3B, lane 5). In a control experiment where the DNA probe was not pretreated with UDG, only weak background level cross-linking signals were detected (Figure 3C), indicating that the unknown protein specifically bound and cross-linked to a native dRP group. To reveal the identity of the unknown protein, we performed in-gel trypsin digestion and mass spectrometry. The protein was identified as HMGB1 [23]. Thus, in the MEF cell extract, HMGB1 was capable of binding to the BER intermediate (Figure 2) and forming a Schiff base lyase reaction intermediate with the dRP group.
Figure 3.

NaBH4 Crosslinking of HMGB1 in Cell Extracts (from Figure 1 of ref. 23). NaBH4 crosslinking of proteins in various cell extracts with the UDG-untreated substrate.
Biological function of HMGB1 in BER
To explore if HMGB1 could play a role in DNA base damage repair, we initially studied accumulation of GFP-tagged human HMGB1 on the damaged sites induced by 405 nm laser light in HeLa cells [23]. We found that GFP-HMGB1 accumulated at damaged sites after cells were irradiated by 500 scans (50–75 μJ/mm2) of laser light generating base damage and single-strand DNA breaks [38](Figure 4, upper panel). Human DNA glycosylases, i.e., GFP-OGG1 and GFP-NTH1 as well as double-strand break repair proteins, GFP-Ku70 and GFP-RAD52, were examined for accumulation at these laser-irradiated sites (Figure 4, upper panel). Using a photosensitizer, 8-MOP (methoxypsoralen) that can produce 8-hydroxy-2-deoxyguanosine by absorbing and emitting laser energy [39], we observed a significant increase in accumulation of HMGB1, OGG1 and NTH1 at the damaged sites induced by 10 scans of 405 nm laser light (Figure 4, bottom panel). However, the double-strand break factors Ku70 and RAD52 did not accumulate at these damaged sites (Figure 4, bottom panel), suggesting that base lesions without double-strand breaks were introduced under this irradiation condition. Importantly, these results indicated that HMGB1 is concentrated at sites of base damage.
Figure 4.

Accumulation of GFP-tagged Proteins at Sites of DNA damage in Living Cells (from Figure 3 of ref. 15). The recruitment of GFP-tagged human OGG1, NITH1, HMGB1, Ku70 and Rad52 on DNA damage induced by 405 nm scanning laser in the absence and presence of photosensitizer (8-MOP).
To explore the molecular basis underlying a possible function of HMGB1 in BER, we initially characterized the dRP lyase activity of HMGB1, since its Schiff base cross-linking to the dRP group had been detected (Figure 3B). To measure the dRP lyase activity, i.e., the release of the 5′-dRP group from a BER intermediate, a duplex oligonucleotide with a U:G mismatch was pretreated with UDG and AP endonuclease 1 (APE1) as shown in Figure 5A. This resulted in a single-nucleotide gapped DNA with a 5′-dRP group, the substrate for measuring dRP lyase activity. Release of the dRP group was then monitored as the production of a DNA fragment that is one-nucleotide shorter than the substrate (Figure 5A). With a substrate labeled with 32P at the 3′-end of the dRP group-containing oligonucleotide, the product and substrate were detected by phosphor-Imager after separation in a urea-denaturing sequencing gel (Figure 5B). HMGB1 exhibited some lyase activity of releasing the dRP group, but with ~600-fold less efficiency than Pol β (Figure 5, compare lanes 3 and 4 with lanes 5 and 6). This suggested that the weak dRP lyase activity of HMGB1 may not be biologically significant [23]. Unexpectedly, the Pol β dRP lyase was inhibited in the presence of purified HMGB1 protein (Figure 5B, lanes 7 and 8), suggesting that these two proteins may compete to bind to the dRP group. To further confirm the activities of dRP cross-linking and the dRP lyase of HMGB1 and exclude any possibilities that these activities could be due to contamination from other proteins, we re-examined the activities in samples of renatured HMGB1 protein isolated from SDS-PAGE gel (Figure 5C and 5D) [23]. The results demonstrated that the renatured HMGB1 protein cross-linked well with the dRP group (Figure 5D) and exhibited ~10% as much dRP lyase activity as the native purified HMGB1 protein (Figure 5E and 5F). This indicates that HMGB1 can bind to the dRP group and function as a dRP lyase. Furthermore, we found that HMGB1 inhibited single-nucleotide BER (Figure 5G), suggesting HMGB1 may bind to the dRP group and block the access of Pol β to perform dRP lyase activity. To estimate the binding ability of HMGB1 for BER substrates containing the dRP group, we performed gel mobility shift assays (Figure 6A). These assays measured the apparent binding constant of the protein for various BER intermediates (Figure 6B and 6C). The results showed that HMGB1 can bind to various BER intermediates including DNA containing the abasic site, nicked DNA, and gapped DNA, as well as nicked and single-nucleotide gapped DNA with the 5′-dRP group. Interestingly, HMGB1 bound to the intermediates containing the dRP group with highest affinity (Figure 6C). These results indicated that the dRP group is important for HMGB1 binding to BER intermediates, and this may explain why HMGB1 was specifically cross-linked with the dRP group-containing probe in the experiments with crude extracts. The suppression of Pol β dRP lyase activity suggests that HMGB1 could have a regulatory role in BER, as discussed below.
Figure 5.
Characterization of HMGB1 Isolated from HeLa Cells (from Figure 4 of ref. 23). (A) Schematic representation of the dRP lyase substrate generated by treatment of the radiolabeled oligonucleotide substrates with UDG and APE1. (B) The dRP lyase activities of Pol β and HMGB1. (C) The purified HMGB1 (lane 2 and 3, 0.45 and 0.9 μg, respectively) was renatured (lane 4, ~0.2 μg) and analyzed by NuPAGE Bis-Tris gel electrophoresis. (D) NaBH4 crosslinking of renatured HMGB1 at the concentrations of 500 nM (lane 2), 140 nM (lane 3) and 280 nM (lane 4). (E) The dRP lyase activity of renatured HMGB1 at concentration of 420 nM. (F) Quantification of the dRP lyase activities of purified (open circles) and renatured HMGB1 (filled circles). (G) The effects of HMGB1 on single-nucleotide BER.
Figure 6.
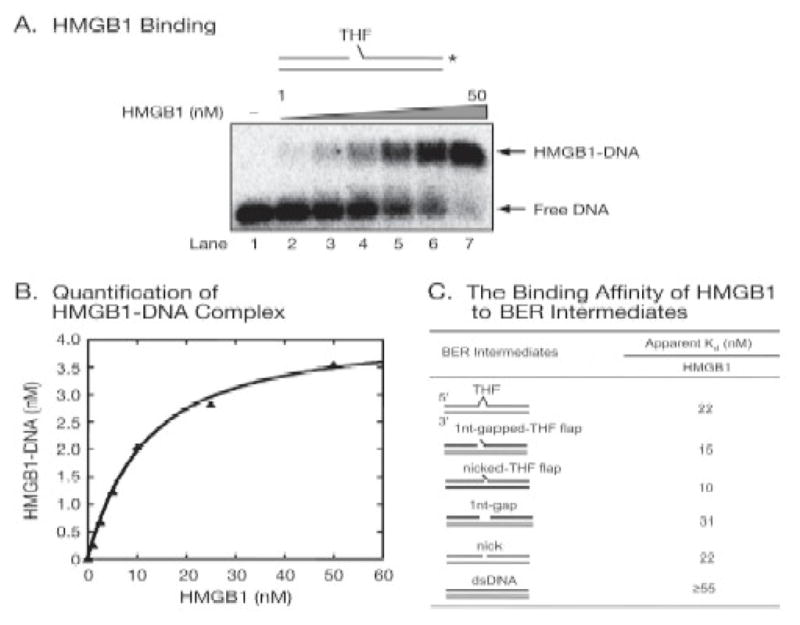
HMGB1 Binding to BER Intermediates (from Figure S4 of ref. 23) (A) HMGB1 binding to the BER intermediate with a single-nucleotide gap and a sugar phosphate measured by gel mobility shift assay. (B) Quantification of the HMGB1-DNA complex was plotted as a function of HMGB1 concentration. (C) The binding affinity (apparent Kd) of HMGB1 on BER intermediate DNA substrates
HMGB1 stimulates production of single-strand DNA breaks during BER
To further understand the effects of HMGB1 on BER in cells, we evaluated base lesion repair using a mouse embryonic fibroblast cell line with HMGB1 gene knockout (HMGB1−/−) and compared the capacity of the repair with that of an isogenic wild type cell line (HMGB1+/+) [23]. Repair was initially studied by determining the cell sensitivity to the methylating agent MMS. Surprisingly, a modest decrease and increase, respectively, was detected in MMS sensitivity for the HMGB1−/− and the HMGB1+/+ cells, (Figure 7A). This indicated that the absence of HMGB1 did not significantly impact MMS-induced damage repair. To verify the identity of these cell lines, cellular expression of HMGB1 was measured by immunoblot using HMGB1-specific monoclonal antibody (inset i) of Figure 7A). In both cell lines, Pol β expression was found to be the same (inset ii) of Figure 7A). Since increased cellular MMS sensitivity often reflects the accumulation of BER intermediates [40], the slight increase in MMS sensitivity of the HMGB1+/+ cell line suggested to us that more BER intermediates accumulated in these cells than the HMGB1−/− cells. If this were true, then we would expect HMGB1 to stimulate the production of strand break intermediates and that this effect may be more prominent under conditions of limiting APE1 incision of abasic site. Such an APE1 limiting condition can be created by treating cells with MMS in combination with methoxyamine (MX), a chemical known to react with AP sites in cells to generate MX-adducted AP sites that are more resistant to APE1 incision than native AP sites [41–43]. Indeed, under these conditions (the presence of MX) it was found that the HMGB1+/+ cells were much more sensitive to MMS than the HMGB1−/− cells in (Figure 7B). Consistent with this finding, more strand break products were detected by comet assay in the HMGB1+/+ cells than in the HMGB1−/− cells when the cells were treated with MMS and MX simultaneously (Figure 7C bottom panel and 7D). These results indicated that HMGB1 can stimulate APE1 incision activity on MX-adducted AP site. To confirm this stimulatory effect of HMGB1, we measured APE1 5′-incision activity on MX-adducted AP site in the presence and absence of purified HMGB1. APE1 incision on this substrate was significantly stimulated with increasing concentrations of HMGB1 (Figure 7E and 7F). This result further predicts that HMGB1 may also stimulate APE1 incision on native AP site when the amount of the enzyme is limited. In fact, the incision of a native AP site by limiting amount of APE1 (0.1 nM) was stimulated about 10-fold by HMGB1 under steady-state conditions (Figure 8A and 8B). Thus, HMGB1 can stimulate APE1 incision activity leading to accumulation of cellular DNA strand breaks. The stimulatory effects could be mediated through its binding to the AP site (Figure 6C) as well as its physical interaction with APE1 [23]. The mechanisms underlying the effect need to be further elucidated, but HMGB1 bending of the substrate/product may increase the enzyme-product complex off rate under steady-state conditions [44].
Figure 7.
Biological and Biochemical Analysis of HMGB1+/+ and HMGB1−/− MEFs after Treatment with MMS (from Figure 7 of ref. 23). (A) MMS sensitivity of HMGB1+/+ (red open circles) and HMGB1−/− (blue open circles) cells. (B) MMS sensitivity of HMGB1+/+ (red filled symbols) and HMGB1−/− (blue open symbols) cells in the absence (circles) and presence of MX for 1 (squares) or 4 hr (diamonds). (C) Comet assay of MMS-induced single-strand DNA breaks in HMGB1+/+ and HMGB1−/− cells in the absence and presence of MX. (D) Quantification of Olive tail moment (OTM) of comet from (C) in HMGB1+/+ (red bars) and HMGB1−/− cells (blue bar). (E) HMGB1 stimulates APE1 incision on MX-adducted DNA substrate. (F) Quantification of HMGB1 stimulatory effects on APE1 incision activity.
Figure 8.
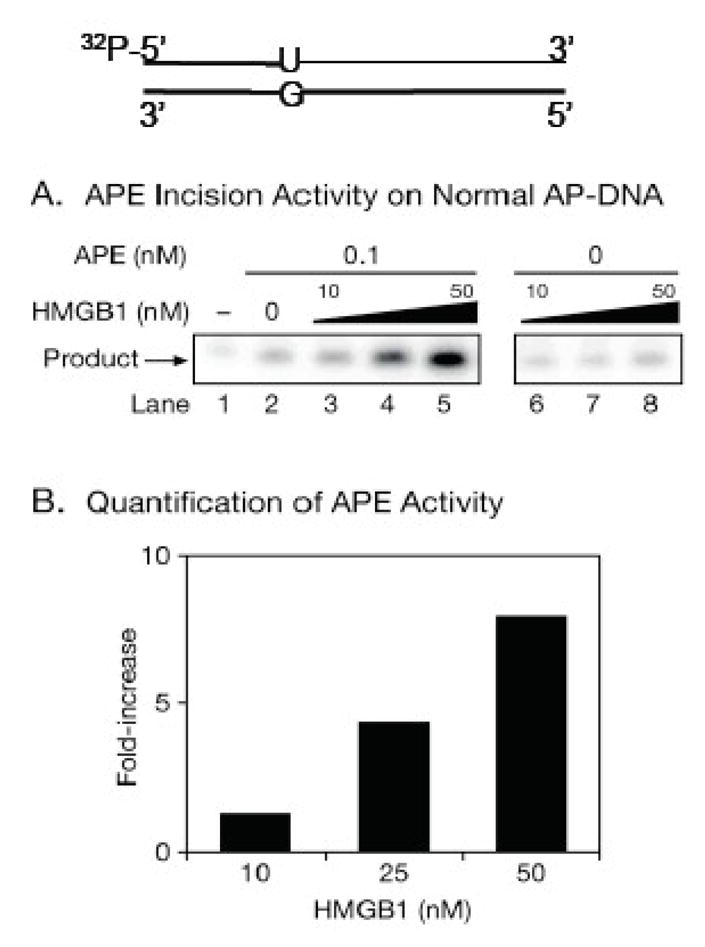
Stimulation of APE1 Activity by HMGB1 (from Figure 5 of ref. 23) (A) HMGB1 stimulates APE1 incision activity on normal AP site. (B) Quantification of the HMGB1 stimulatory effects on APE1.
HMGB1 stimulates Pol β dependent long-patch BER
The binding of HMGB1 to nicked DNA with the 5′-dRP group (Figure 6C) and its inhibition of single-nucleotide BER (Figure 5G) suggests that HMGB1 also could be involved in long-patch BER. Pol β-dependent long-patch BER can be accomplished through two different modes. In one mode, Pol β coordinates its single-nucleotide gap-filling synthesis with flap endonuclease 1 (FEN1) to remove the dRP flap on a nicked DNA. This results in replacement of two nucleotides involving alternate gap-filling synthesis by Pol β and cleavage of the dRP flap by FEN1, i.e., the “Hit and Run” mechanism for Pol β and FEN1 cooperation in long-patch BER [45]. In a less efficient repair mode, Pol β has to perform strand-displacement synthesis generating a 5–6 nt flap containing the 5′-dRP group, and the flap is subsequently cleaved by FEN1 [46]. To identify the roles of HMGB1 in long-patch BER, we examined the HMGB1 effect on FEN1 cleavage of a nicked-3 nt flap containing a tetrahydrofuran (THF) residue, a mimic of modified sugar subjected to long-patch BER (Figure 9A and 9B), as well as its impact on FEN1 cleavage of a nicked-THF flap (Figure 9C and 9D). The results showed that FEN1 cleavage on these flaps was significantly stimulated at increasing concentrations of HMGB1. This indicated that HMGB1 can enhance the efficiency of long-patch BER by stimulating FEN1 flap cleavage activity. It appeared that the stimulatory effect may be accomplished through physical interaction between HMGB1 and FEN1 [23] during long-patch BER. However, the molecular mechanisms underlying its effect on FEN1 remains to be elucidated.
Figure 9.
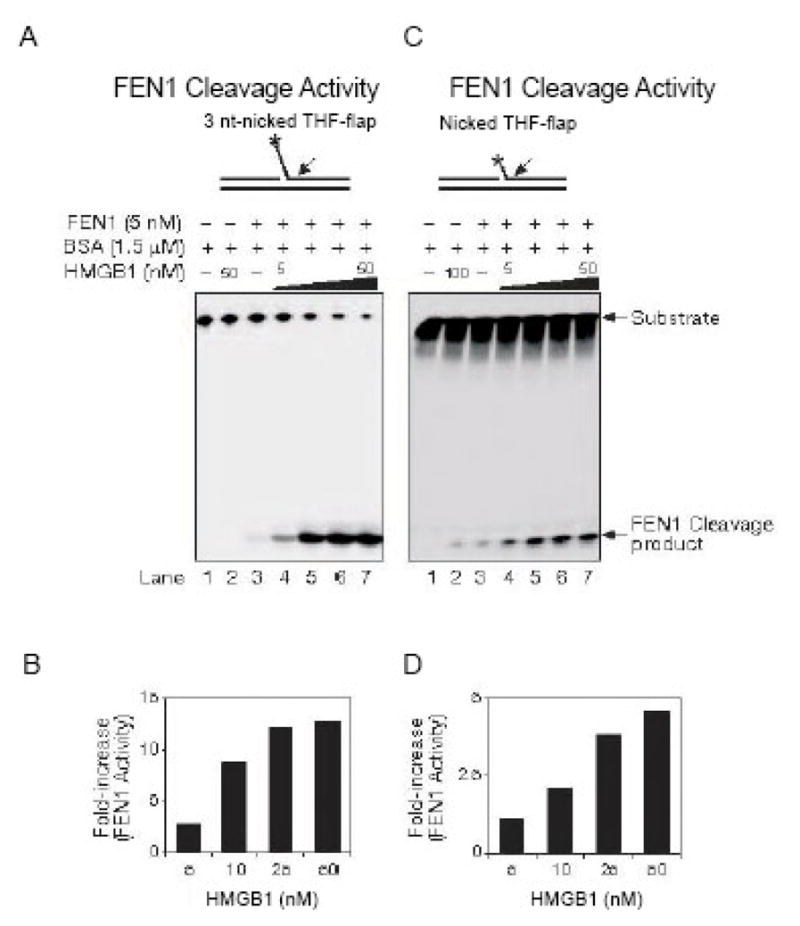
Stimulation of FEN1 by HMGB1 (from Figure 5 of ref. 23). (A) The effect of HMGB1 on FEN1 cleavage activity on a nicked-3nt flap substrate. (B) Quantification of FEN1 activity in (A). (C) HMGB1 stimulates FEN1 cleavage on a nicked-THF flap substrate. (D) The quantification of FEN1 activity in (C).
HMGB1 and trinucleotide repeat expansion
Recently, we found that oxidative DNA base lesions and BER play a role in aging-dependent CAG repeat expansion in the Huntington’s disease mouse model [47]. The single-strand DNA break resulting from removal of the oxidative base lesion 8-oxo-guanine (8-oxoG) by 8-oxoG DNA glycosylase (OGG1) was required for age-dependent repeat expansion. It also was demonstrated that a single-strand DNA break in the context of the CAG repeat sequence can initiate limited CAG repeat expansion in vitro by promoting DNA slippage and formation of hairpin structures [48]. This process resulted in production of DNA repair intermediates with multi-nucleotide gaps and hairpins. Subsequent long-patch BER gap-filling on these intermediates lead to repeat amplification and hairpin ligation, ultimately resulting in stable repeat expansion [47]. Overall, these results provided evidence that a base lesion, accompanied by a glycosylase-mediated strand break, and BER may be involved in DNA repeat sequence instability associated with several human diseases. Since HMGB1 can facilitate the production of single-strand DNA breaks by stimulating APE1 incision of AP sites, it is proposed that HMGB1 could promote CAG repeat expansion by inducing more strand breaks during BER in the repeat sequence context. In addition, an earlier study suggested that HMGB1 might affect the stability of dinucleotid repeats by binding and stabilizing loops formed within the repeat sequence context [49]. Thus, HMGB1 also may employ a similar mechanism in facilitating CAG repeat expansion.
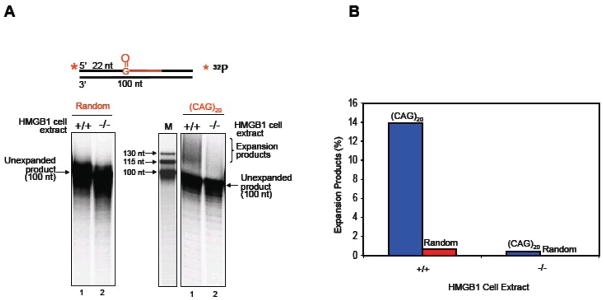
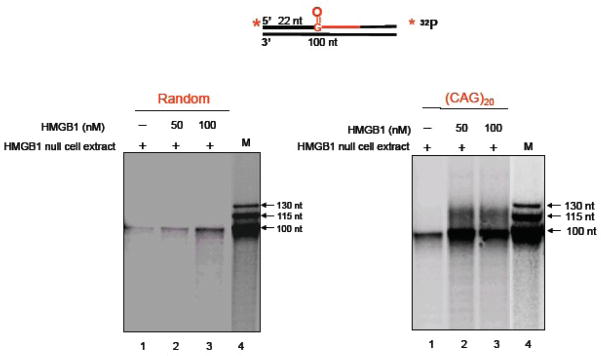
Using MEF extracts isolated from the HMGB1−/− and HMGB1+/+ cells and oligonucleotide substrates with an 8-oxoG imbedded in either a random sequence or (CAG)20 repeats, we initially studied the importance of HMGB1 in trinucleotide repeat expansion in vitro (Figure 10). When the (CAG)20-containing repeat substrate was incubated with the HMGB1+/+ cell extract, limited expansion products were detected (Figure 10A, right panel, lane 1). However, these products were not detected with the HMGB1−/− cell extract (Figure 10A, right panel, lane 2). The expansion products were not detected when the random sequence substrate was incubated with either of these cell extracts (Figure 10A, left panel). Quantification of these results demonstrated that the presence of HMGB1 increased the production of expansion products by ~20-fold (Figure 10B). Addition of purified HMGB1 to the HMGB1−/− cell extract also resulted in production of the expansion products from (CAG)20 -containing substrate, but not from the random sequence substrate (Figure 11). These results indicated that HMGB1 can promote CAG repeat expansion. This effect appeared to be mediated through HMGB1 stimulation of APE1’s incision activity producing more single-strand break intermediates during the BER process. This notion was further supported by in vitro experiments that demonstrated HMGB1’s stimulation of APE1-mediated CAG repeat expansion, when the APE1 concentration was limiting (Figure 12). In this case, increasing concentrations of HMGB1 stimulated APE1 incision of an AP site in the context of (CAG)20 repeats by ~10-fold (Figure 13).
Figure 10.
HMGB1 Facilitates CAG Repeat Expansion (from Figure 6 of ref. 48). (A) Repair products from BER reactions mediated by HMGB1+/+ and HMGB1−/− cell extracts in the context of CAG repeats. (B) Quantification of expansion products from HMGB1+/+ and HMGB1−/− cell extracts.
Figure 11.
Recombinant HMGB1 Stimulates CAG Repeat Expansion (From Figure 6 of ref. 48). The effect of HMGB1 on CAG repeat expansion was determined by addition of purified HMGB1 to the HMGB1 null cell extract. Size markers (M) are shown.
Figure 12.
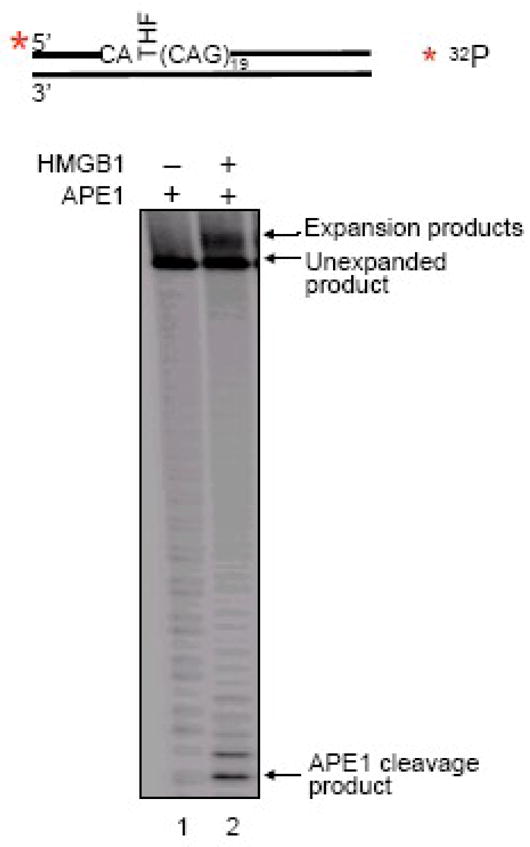
HMGB1 Stimulates CAG Repeat Expansion mediated by APE1 (from Figure 7 of ref. 48). The effect of HMGB1 on the CAG repeat expansion mediated by APE1 was examined under limiting concentration of APE1 (0.1 nM).
Figure 13.
HMGB1 Stimulates APE1 Incision Activity in the Context of CAG Repeat. APE1 (0.1 nM) incision activity on a synthesized abasic site (THF) imbedded in CAG repeats is significantly increased by increasing concentrations of HMGB1 (Lane 2–5). Lane 1 represents APE1 (0.1 nM) incision in the absence of HMGB1. Lane 2–5 represents APE1 incision in the presence of 10, 20, 50 and 100 nM HMGB1, respectively. Substrate and product are indicated by arrows. The quantification of APE1 incision product is illustrated in lower panel.
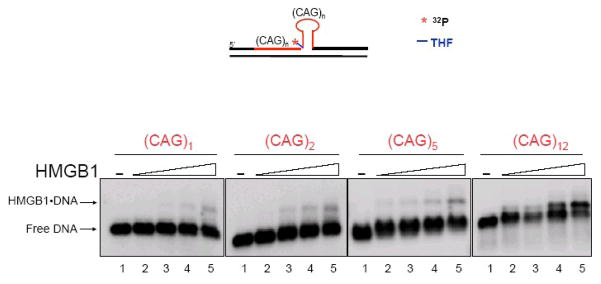
We explored the possibility that HMGB1 could be involved in stabilizing hairpin structures formed by CAG repeats, as noted above for dinucleotide repeats. HMGB1 binding to pre-formed hairpin structures that contained one, two, five and twelve CAG repeats, i.e. (CAG)1, (CAG)2, (CAG)5 and (CAG)12 was determined (Figure 14). The results demonstrated that HMGB1 bound to the small hairpin containing (CAG)1 only weakly (Figure 14). However, the HMGB1 binding ability was significantly enhanced with increasing sizes of hairpins, with highest binding affinity for the hairpin containing (CAG)12. Thus, HMGB1 bound the hairpin structure, suggesting that HMGB1 may facilitate repeat expansion by stabilizing hairpin intermediates formed during BER. In summary, the results have suggested that HMGB1 may promote CAG repeat expansion by stimulating production of single-strand break BER intermediates and stabilizing hairpin structures in BER intermediates.
Figure 14.
HMGB1 Binds to CAG Repeat Hairpins. The binding ability of HMGB1 to various size of CAG repeat hairpin was examined by gel mobility shift assay. HMGB1 binding to (CAG)1, (CAG)2 and (CAG)5 was poor. However, its binding to (CAG)12 was significantly enhanced with its increasing concentrations (Lane 2–4) suggesting that HMGB1 prefers to bind to larger CAG repeat hairpin. Lane 1 represents binding mixture without HMGB1. Lane 2–4 represents binding mixture containing 20, 50, 100 and 200 nM HMGB1, respectively. DNA substrate and HMGB1-DNA complex are indicated by arrows.
Models for HMGB1 coordination of BER and genome stability
New roles of HMGB1 in the BER pathway have been identified: HMGB1 can act as a cofactor to modulate BER enzymatic activities, and HMGB1 can recognize and bind to DNA base lesions (Figure 6). These effects are in addition to HMGB1’s well-known ability to recognize bulky DNA lesions induced by chemotherapeutic drugs and environmental carcinogens [12, 17, 22]. Regarding BER intermediates, HMGB1 exhibits tight binding to the AP site intermediate and a strong stimulation of APE1 incision for producing the single-strand break intermediate. This could become significant in the cell when the APE1 enzymatic activity is limited either by its sub-cellular compartmentalization/low expression [50] or by a chemical modification of AP sites [41]. Under these conditions, APE1 incision becomes a rate-limiting step of BER, and HMGB1 appears to enhance the overall efficiency of BER. However, the high affinity binding of HMGB1 to the dRP group-containing BER intermediate (Figure 6) and its cellular abundance (estimated to be much higher than that of Pol β) [23] renders HMGB1 capable of competing with Pol β. This could block Pol β binding to the sugar phosphate group-containing BER intermediate, resulting in inhibition of single-nucleotide BER mediated by the Pol β dRP lyase (Figure 5G). On the other hand, HMGB1 can stimulate the activity for FEN1, a long-patch BER enzyme, to remove the dRP flap (Figure 9). Furthermore, HMGB1 also can moderately stimulate the Pol β strand-displacement synthesis activity that is fundamental to long-patch BER (data shown). These effects may enhance the efficiency of long-patch BER. Thus, HMGB1 may play distinct roles in modulating the sequential BER steps and sub-pathways of BER. It is possible that HMGB1 binds to the dRP group or a longer flap with the 5′-dRP group on a nicked-DNA, inducing bending of the DNA and allowing a favorable configuration for Pol β strand-displacement synthesis and FEN1 cleavage on the dRP-containing flap. However, such a bent DNA-HMGB1 complex may not be favorable for Pol β binding to the dRP group to perform its dRP lyase activity for single-nucleotide BER. The protein-protein interactions between HMGB1 and Pol β, as well as FEN1, also could play a role in modulating Pol β and FEN1 activities determining which of the two BER sub-pathways is pursued. In summary, the results to date indicate that HMGB1 can act as modulator to regulate the steps of BER and determine BER sub-pathway choice. The molecular mechanisms underlying the effects of HMGB1 in modulating BER still remain to be elucidated.
Our studies demonstrated that HMGB1 may facilitate DNA base lesion-induced trinucleotide (CAG) repeat expansion (Figure 10–12). This effect could be mediated by HMGB1’s stimulation of APE1 incision of the AP site in the sequence context of CAG repeats (Figure 13). This stimulatory effect could promote production of single-strand breaks in the repeats facilitating strand slippage and formation of BER intermediates with hairpin structures and multi-nucleotide gaps for repeat amplification and expansion [48]. Interestingly, we also identified binding of HMGB1 to CAG repeat hairpin structures (Figure 14). In fact, it has been known that HMGB1 preferentially binds to a variety of noncanonical DNA structures that include supercoiled DNA, Z-DNA [51], looped structures [49], triplex DNA [16] and others [12]. Yet, the biological consequences of HMGB1 binding to loop structures and other noncanonical DNA structures formed by repeat sequences remained unknown. As noted above, it was proposed that HMGB1 might influence the stability of dinucleotide repeats by transiently binding and stabilizing loop structures [49]. In a similar fashion, HMGB1 binding to CAG repeat hairpin structures could enhance the stability of these structures. This may prevent removal of the hairpins by nucleases such as FEN1 [52, 53] and facilitate hairpin ligation resulting in repeat expansion. Moreover, it has been found that HMGB1 can interact with the mismatch repair protein MSH2 [24]. Since MSH2 can form a complex with MSH3 to bind and stabilize CAG repeat hairpin structures [54], it is possible that HMGB1 cooperates with MSH2/MSH3 to stabilize hairpins and stimulate FEN1 “alternate cleavage” [48] facilitating hairpin ligation and repeat expansion. In addition, since long-patch BER is the sub-pathway that mediates CAG repeat expansion [48], HMGB1 in directing BER to this sub-pathway, could also promote mediated by Pol β multi-nucleotide gap-filling synthesis ultimately resulting in repeat expansion. These models regarding the biological significance of HMGB1 [44] in DNA damage-induced genomic instability remain to be further explored Studies on stabilization of hairpin structures by HMGB1 and its cooperation with the MSH2/MSH3 complex in modulating stability of tri-and dinucleotide repeats during BER should be promising.
HMGB1 and chromatin structure
As an architectural protein, HMGB1 preferentially binds to distorted DNA structures such as DNA bulges, including certain DNA damages [55]. Such structure-specific recognition suggests that HMGB1 may bind to damaged DNA and then induce a stably bent structure within the DNA. This function may facilitate recruitment of chromatin remodeling factors by accelerating nucleosome sliding [56] allowing access of DNA damage sites for the repair machinery. On the other hand, HMGB1-induced DNA bending may help the recruitment of repair proteins to DNA damage sites, once they become accessible after chromatin remodeling. HMGB1 may affect chromatin remodeling by directly participating in histone deacetylation [22] and helping restore normal chromatin structure after repair [22]. It is conceivable that binding of HMGB1 to damaged sites and hairpin structures can induce further DNA bending that facilitates the recruitment of chromatin remodeling factors to dislodge nucleosomes, allowing the DNA damage sites or hairpin structures to become accessible. HMGB1 binding to hairpin structures could stabilize these structures thereby preventing removal of hairpins and facilitating their ligation, resulting in repeat sequence expansion. The complex roles of HMGB1 in coordinating DNA repair, chromatin structure, and genomic stability remain to be further elucidated.
Conclusions
Recent studies have substantially advanced our understanding of important roles of HMGB1 in chromatin structure, transcription, DNA damage repair, cell signaling of inflammation and potentially in human diseases such as cancer. However, many questions remain about the biological roles of HMGB1 in cellular response and defense to genotoxic stress. For example, it is still unclear how HMGB1 is involved in regulating DNA damage repair and maintaining genome stability in the context of chromatin and cell system. Since HMGB1 has dual roles in DNA damage repair and cellular signaling of inflammation, an important question is how cells can balance HMGB1 function in DNA damage repair and the inflammatory response. HMGB1 release or secretion can trigger a series of inflammation responses and reactions. This, in turn, can induce production of oxidative stress chemicals leading to a variety of DNA base lesions that are subject to the BER pathway. Release of HMGB1 into the extra-cellular matrix could result in depletion of the cellular level of HMGB1. Furthermore, DNA base damage can trigger HMGB1 translocation from nucleus to cytoplasm by inducing poly(ADP)ribosylation of the protein [57]. This could significantly decrease BER capacity and accumulation of DNA base lesions and aberrant strand breaks leading to cell death. Thus, understanding of the molecular mechanisms by which cells maintain a balanced level of HMGB1 to sustain efficient DNA damage repair may help to understand etiology, progression and treatment of human diseases that are related to inflammation.
Acknowledgments
We thank William A. Beard for discussion and assistance with structural analysis of HMGB1. This work is supported by National Institutes of Health Research Projects Z01-ES010158 and Z01-ES050159 from the NIEHS Intramural Research Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol. 1999;19:5237–5246. doi: 10.1128/mcb.19.8.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reeck GR, Isackson PJ, Teller DC. Domain structure in high molecular weight high mobility group nonhistone chromatin proteins. Nature. 1982;300:76–78. doi: 10.1038/300076a0. [DOI] [PubMed] [Google Scholar]
- 3.Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5:331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 4.Shirakawa H, Tsuda K, Yoshida M. Primary structure of non-histone chromosomal protein HMG2 revealed by the nucleotide sequence. Biochemistry. 1990;29:4419–4423. doi: 10.1021/bi00470a022. [DOI] [PubMed] [Google Scholar]
- 5.Ohndorf UM, Rould MA, He Q, Pabo CO, Lippard SJ. Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature. 1999;399:708–712. doi: 10.1038/21460. [DOI] [PubMed] [Google Scholar]
- 6.Li J, Kokkola R, Tabibzadeh S, Yang R, Ochani M, Qiang X, Harris HE, Czura CJ, Wang H, Ulloa L, Warren HS, Moldawer LL, Fink MP, Andersson U, Tracey KJ, Yang H. Structural basis for the proinflammatory cytokine activity of high mobility group box 1. Mol Med. 2003;9:37–45. [PMC free article] [PubMed] [Google Scholar]
- 7.Ugrinova I, Zlateva S, Pashev IG, Pasheva EA. Native HMGB1 protein inhibits repair of cisplatin-damaged nucleosomes in vitro. Int J Biochem Cell Biol. 2009;41:1556–1562. doi: 10.1016/j.biocel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 8.Mitkova E, Ugrinova I, Pashev IG, Pasheva EA. The inhibitory effect of HMGB-1 protein on the repair of cisplatin-damaged DNA is accomplished through the acidic domain. Biochemistry. 2005;44:5893–5898. doi: 10.1021/bi047712c. [DOI] [PubMed] [Google Scholar]
- 9.Weir HM, Kraulis PJ, Hill CS, Raine AR, Laue ED, Thomas JO. Structure of the HMG box motif in the B-domain of HMG1. EMBO J. 1993;12:1311–1319. doi: 10.1002/j.1460-2075.1993.tb05776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 11.Bustin M. Chromatin unfolding and activation by HMGN(*) chromosomal proteins. Trends Biochem Sci. 2001;26:431–437. doi: 10.1016/s0968-0004(01)01855-2. [DOI] [PubMed] [Google Scholar]
- 12.Lange SS, Vasquez KM. HMGB1: the jack-of-all-trades protein is a master DNA repair mechanic. Mol Carcinog. 2009;48:571–580. doi: 10.1002/mc.20544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24:4321–4328. doi: 10.1128/MCB.24.10.4321-4328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calogero S, Grassi F, Aguzzi A, Voigtlander T, Ferrier P, Ferrari S, Bianchi ME. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat Genet. 1999;22:276–280. doi: 10.1038/10338. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Bloom O, Zhang M, Vishnubhakat JM, Ombrellino M, Che J, Frazier A, Yang H, Ivanova S, Borovikova L, Manogue KR, Faist E, Abraham E, Andersson J, Andersson U, Molina PE, Abumrad NN, Sama A, Tracey KJ. HMG-1 as a late mediator of endotoxin lethality in mice. Science. 1999;285:248–251. doi: 10.1126/science.285.5425.248. [DOI] [PubMed] [Google Scholar]
- 16.Reddy MC, Christensen J, Vasquez KM. Interplay between human high mobility group protein 1 and replication protein A on psoralen-cross-linked DNA. Biochemistry. 2005;44:4188–4195. doi: 10.1021/bi047902n. [DOI] [PubMed] [Google Scholar]
- 17.Lange SS, Reddy MC, Vasquez KM. Human HMGB1 directly facilitates interactions between nucleotide excision repair proteins on triplex-directed psoralen interstrand crosslinks. DNA Repair (Amst) 2009;8:865–872. doi: 10.1016/j.dnarep.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanuszewska J, Widlak P. High mobility group 1 and 2 proteins bind preferentially to DNA that contains bulky adducts induced by benzo[a]pyrene diol epoxide and N-acetoxy-acetylaminofluorene. Cancer Lett. 2000;158:17–25. doi: 10.1016/s0304-3835(00)00517-6. [DOI] [PubMed] [Google Scholar]
- 19.Pasheva EA, Pashev IG, Favre A. Preferential binding of high mobility group 1 protein to UV-damaged DNA. Role of the COOH-terminal domain. J Biol Chem. 1998;273:24730–24736. doi: 10.1074/jbc.273.38.24730. [DOI] [PubMed] [Google Scholar]
- 20.Giavara S, Kosmidou E, Hande MP, Bianchi ME, Morgan A, d’Adda di Fagagna F, Jackson SP. Yeast Nhp6A/B and mammalian Hmgb1 facilitate the maintenance of genome stability. Curr Biol. 2005;15:68–72. doi: 10.1016/j.cub.2004.12.065. [DOI] [PubMed] [Google Scholar]
- 21.Krynetskaia N, Xie H, Vucetic S, Obradovic Z, Krynetskiy E. High mobility group protein B1 is an activator of apoptotic response to antimetabolite drugs. Mol Pharmacol. 2008;73:260–269. doi: 10.1124/mol.107.041764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lange SS, Mitchell DL, Vasquez KM. High mobility group protein B1 enhances DNA repair and chromatin modification after DNA damage. Proc Natl Acad Sci U S A. 2008;105:10320–10325. doi: 10.1073/pnas.0803181105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prasad R, Liu Y, Deterding LJ, Poltoratsky VP, Kedar PS, Horton JK, Kanno S, Asagoshi K, Hou EW, Khodyreva SN, Lavrik OI, Tomer KB, Yasui A, Wilson SH. HMGB1 is a cofactor in mammalian base excision repair. Mol Cell. 2007;27:829–841. doi: 10.1016/j.molcel.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan F, Gu L, Guo S, Wang C, Li GM. Evidence for involvement of HMGB1 protein in human DNA mismatch repair. J Biol Chem. 2004;279:20935–20940. doi: 10.1074/jbc.M401931200. [DOI] [PubMed] [Google Scholar]
- 25.Stros M, Cherny D, Jovin TM. HMG1 protein stimulates DNA end joining by promoting association of DNA molecules via their ends. Eur J Biochem. 2000;267:4088–4097. doi: 10.1046/j.1432-1327.2000.01450.x. [DOI] [PubMed] [Google Scholar]
- 26.van Gent DC, Hiom K, Paull TT, Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang JC, Zamble DB, Reardon JT, Lippard SJ, Sancar A. HMG-domain proteins specifically inhibit the repair of the major DNA adduct of the anticancer drug cisplatin by human excision nuclease. Proc Natl Acad Sci U S A. 1994;91:10394–10398. doi: 10.1073/pnas.91.22.10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malina J, Kasparkova J, Natile G, Brabec V. Recognition of major DNA adducts of enantiomeric cisplatin analogs by HMG box proteins and nucleotide excision repair of these adducts. Chem Biol. 2002;9:629–638. doi: 10.1016/s1074-5521(02)00134-5. [DOI] [PubMed] [Google Scholar]
- 29.Moggs JG, Szymkowski DE, Yamada M, Karran P, Wood RD. Differential human nucleotide excision repair of paired and mispaired cisplatin-DNA adducts. Nucleic Acids Res. 1997;25:480–491. doi: 10.1093/nar/25.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, Gu L, Li GM. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 31.Yumoto Y, Shirakawa H, Yoshida M, Suwa A, Watanabe F, Teraoka H. High mobility group proteins 1 and 2 can function as DNA-binding regulatory components for DNA-dependent protein kinase in vitro. J Biochem. 1998;124:519–527. doi: 10.1093/oxfordjournals.jbchem.a022143. [DOI] [PubMed] [Google Scholar]
- 32.Sawchuk DJ, Weis-Garcia F, Malik S, Besmer E, Bustin M, Nussenzweig MC, Cortes P. V(D)J recombination: modulation of RAG1 and RAG2 cleavage activity on 12/23 substrates by whole cell extract and DNA-bending proteins. J Exp Med. 1997;185:2025–2032. doi: 10.1084/jem.185.11.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swanson PC. Fine structure and activity of discrete RAG-HMG complexes on V(D)J recombination signals. Mol Cell Biol. 2002;22:1340–1351. doi: 10.1128/mcb.22.5.1340-1351.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 35.Bonaldi T, Talamo F, Scaffidi P, Ferrera D, Porto A, Bachi A, Rubartelli A, Agresti A, Bianchi ME. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J. 2003;22:5551–5560. doi: 10.1093/emboj/cdg516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ugrinova I, Pashev IG, Pasheva EA. Post-synthetic acetylation of HMGB1 protein modulates its interactions with supercoiled DNA. Mol Biol Rep. 2009;36:1399–1404. doi: 10.1007/s11033-008-9327-z. [DOI] [PubMed] [Google Scholar]
- 37.Lavrik OI, Prasad R, Sobol RW, Horton JK, Ackerman EJ, Wilson SH. Photoaffinity labeling of mouse fibroblast enzymes by a base excision repair intermediate. Evidence for the role of poly(ADP-ribose) polymerase-1 in DNA repair. J Biol Chem. 2001;276:25541–25548. doi: 10.1074/jbc.M102125200. [DOI] [PubMed] [Google Scholar]
- 38.Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orimo H, Tokura Y, Hino R, Kasai H. Formation of 8-hydroxy-2′-deoxyguanosine in the DNA of cultured human keratinocytes by clinically used doses of narrowband and broadband ultraviolet B and psoralen plus ultraviolet A. Cancer Sci. 2006;97:99–105. doi: 10.1111/j.1349-7006.2006.00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horton JK, Joyce-Gray DF, Pachkowski BF, Swenberg JA, Wilson SH. Hypersensitivity of DNA polymerase beta null mouse fibroblasts reflects accumulation of cytotoxic repair intermediates from site-specific alkyl DNA lesions. DNA Repair (Amst) 2003;2:27–48. doi: 10.1016/s1568-7864(02)00184-2. [DOI] [PubMed] [Google Scholar]
- 41.Horton JK, Prasad R, Hou E, Wilson SH. Protection against methylation-induced cytotoxicity by DNA polymerase β-dependent long patch base excision repair. J Biol Chem. 2000;275:2211–2218. doi: 10.1074/jbc.275.3.2211. [DOI] [PubMed] [Google Scholar]
- 42.Liuzzi M, Talpaert-Borle M. A new approach to the study of the base-excision repair pathway using methoxyamine. J Biol Chem. 1985;260:5252–5258. [PubMed] [Google Scholar]
- 43.Rosa S, Fortini P, Karran P, Bignami M, Dogliotti E. Processing in vitro of an abasic site reacted with methoxyamine: a new assay for the detection of abasic sites formed in vivo. Nucleic Acids Res. 1991;19:5569–5574. doi: 10.1093/nar/19.20.5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maher RL, Bloom LB. Pre-steady-state kinetic characterization of the AP endonuclease activity of human AP endonuclease 1. J Biol Chem. 2007;282:30577–30585. doi: 10.1074/jbc.M704341200. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y, Beard WA, Shock DD, Prasad R, Hou EW, Wilson SH. DNA polymerase beta and flap endonuclease 1 enzymatic specificities sustain DNA synthesis for long patch base excision repair. J Biol Chem. 2005;280:3665–3674. doi: 10.1074/jbc.M412922200. [DOI] [PubMed] [Google Scholar]
- 46.Beard WA, Prasad R, Wilson SH. Activities and mechanism of DNA polymerase beta. Methods Enzymol. 2006;408:91–107. doi: 10.1016/S0076-6879(06)08007-4. [DOI] [PubMed] [Google Scholar]
- 47.Kovtun IV, Liu Y, Bjoras M, Klungland A, Wilson SH, McMurray CT. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Prasad R, Beard WA, Hou EW, Horton JK, McMurray CT, Wilson SH. Coordination between polymerase beta and FEN1 can modulate CAG repeat expansion. J Biol Chem. 2009;284:28352–28366. doi: 10.1074/jbc.M109.050286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gibb CL, Cheng W, Morozov VN, Kallenbach NR. Effect of nuclear protein HMG1 on in vitro slippage synthesis of the tandem repeat dTG × dCA. Biochemistry. 1997;36:5418–5424. doi: 10.1021/bi962037v. [DOI] [PubMed] [Google Scholar]
- 50.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 51.Hamada H, Bustin M. Hierarchy of binding sites for chromosomal proteins HMG 1 and 2 in supercoiled deoxyribonucleic acid. Biochemistry. 1985;24:1428–1433. doi: 10.1021/bi00327a022. [DOI] [PubMed] [Google Scholar]
- 52.Liu Y, Bambara R. Analysis of human flap endonuclease 1 mutants reveals a mechanism to prevent triplet repeat expansion. J Biol Chem. 2003;278:13728–13739. doi: 10.1074/jbc.M212061200. [DOI] [PubMed] [Google Scholar]
- 53.Liu Y, Zhang H, Veeraraghavan J, Bambara RA, Freudenreich CH. Saccharomyces cerevisiae flap endonuclease 1 uses flap equilibration to maintain triplet repeat stability. Mol Cell Biol. 2004;24:4049–4064. doi: 10.1128/MCB.24.9.4049-4064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Owen BA, Yang Z, Lai M, Gajec M, Badger JD, 2nd, Hayes JJ, Edelmann W, Kucherlapati R, Wilson TM, McMurray CT. (CAG)(n)-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat Struct Mol Biol. 2005;12:663–670. doi: 10.1038/nsmb965. [DOI] [PubMed] [Google Scholar]
- 55.Agresti A, Bianchi ME. HMGB proteins and gene expression. Curr Opin Genet Dev. 2003;13:170–178. doi: 10.1016/s0959-437x(03)00023-6. [DOI] [PubMed] [Google Scholar]
- 56.Bonaldi T, Langst G, Strohner R, Becker PB, Bianchi ME. The DNA chaperone HMGB1 facilitates ACF/CHRAC-dependent nucleosome sliding. Embo J. 2002;21:6865–6873. doi: 10.1093/emboj/cdf692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ditsworth D, Zong WX, Thompson CB. Activation of poly(ADP)-ribose polymerase (PARP-1) induces release of the pro-inflammatory mediator HMGB1 from the nucleus. J Biol Chem. 2007;282:17845–17854. doi: 10.1074/jbc.M701465200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]