Abstract
Introduction
Among individuals infected with the human immunodeficiency virus (HIV), biomarkers that predict mortality are also used to determine the time when antiretroviral therapy is initiated. No studies have evaluated the impact of the frequency of marker measurements for either their predictive value of mortality or how they may influence inference of the effect of therapy initiation in analyses from observational data.
Methods
We identified 244 persons who were contemporaneously enrolled in both the AIDS Link to the IntraVenous Experience (an interval cohort) and the Johns Hopkins HIV Clinical Cohort between 1995 and 2004. Data from each study were used separately in 2 ways. We applied time-dependent proportional hazards models to examine the predictive associations between markers and mortality, and marginal structural models to examine the causal inference of therapy on mortality. Biomarkers were used to derive the inverse probability weights.
Results
The timing frequencies of marker measurements in the interval cohort (CD4 interquartile range = 175–194 days) were less heterogeneous than in the clinical cohort (interquartile range = 38–121 days). Despite this, the results were concordant for CD4 (R2 = 0.537 [95% confidence interval = 0.345–0.707] and (R2 = 0.488 [0.297–0.666], respectively). Similar concordance was found for the HIV-1 RNA and hemoglobin analyses. When evaluating the causal effect of highly active antiretroviral therapy (HAART), the relative hazards were 0.34 for the interval cohort study (95% CI = 0.15–0.77) and 0.27 for the clinical cohort study (0.11–0.66).
Conclusion
Utilizing a unique co-enrollment of patients in 2 different types of cohort studies, we find empirical evidence that inferences drawn from these different structures are similar.
Two prospective cohort designs are being used extensively in the HIV research setting–the interval cohort and the clinical cohort.1 The interval cohort is the classic longitudinal cohort in which individuals are followed at specified intervals such that the timing and type of data collected are established by study protocols developed by the researchers. Prime examples of successful HIV interval cohort studies include the AIDS Linked to the Intravenous Experience (ALIVE) study,2 the Amsterdam Cohort Studies,3 the Italian Seroconversion Study,4 the Multicenter AIDS Cohort Study,5 and the Women's Interagency HIV Study.6
In contrast to the interval cohort design, the clinical cohort recruits individuals from those who are actively receiving health care for some condition, and uses data from ongoing patient care as primary study data.1,7 Thus, the type and timing of data collection in clinical cohorts are determined mostly by the nature of the health care services being obtained. Examples of HIV clinical cohort studies include the AIDS Therapy Evaluation in the Netherlands cohort,8 the Chelsea and Westminster HIV cohort,9 the Italian Cohort Naive from Antiretrovirals,10 the Johns Hopkins HIV Clinical Cohort,11 the Swiss Cohort,12 the UK Collaborative HIV Cohort,13 and various US HIV clinical cohorts supported through the Centers for AIDS Research.14–17 The dichotomization of cohorts as either interval or clinical cohorts is not necessarily clear; there may be a blend of the 2 designs in which some information (such as laboratory measurements) are captured from clinical care without any structured visit schedule, yet participants are brought back for interviews at 6 months. This is true for example of the Swiss Cohort Study, which we still classified as a clinical cohort, as the participants are under care at the 7 participating hospitals.12
An interval cohort restricts most of the data collection to a set time interval that must balance logistical and scientific consideration. Thus the data may not capture the fullest extent of clinical and disease information, especially if biologically relevant time points are transient.1 Conversely, clinical cohort data come from ongoing health care delivery and reflect the actual treatment decisions made in the health care setting. With data collected more frequently but irregularly among patients with greater health care needs, selection bias is a potential concern. Whether similar frequency of measurements on healthier participants is needed to ensure equitability with the possibly increased frequency for sicker patients is not clear. Such comparisons may depend upon the data being collected. If the longitudinal data being collected are unlikely to change while a participant remains relatively healthy, the frequency of data collection may be irrelevant for healthier patients, as misclassification of exposures are unlikely to occur. However, increased frequency of data collection on sicker participants may be necessary if the data being collected (such as biologic markers) are likely to fluctuate as the individual progresses in disease status. In addition to severity of disease, other factors such as access to health care may also influence an individual's frequency of visits, further contributing to the potential for selection biases.18
Questions have been raised about the comparability of inferences from interval- and clinical-based cohort studies.19 If differences exist, then one might question which design is more “valid.” Few studies have compared interval and clinical cohort study designs, especially with regard to frequency of data collection. Previously, Griffin et al20 showed that analyses of time to a specific biomarker level (eg, time to undetectable HIV-1 RNA) may be biased when the visit frequency is not standardized.
An issue not addressed is whether a heterogeneous frequency of marker measurement within a cohort is likely to affect either prediction or causal inference. In the present study, we examined the potential for frequency of biomarker data collection to affect these 2 types of analyses. Utilizing data collected on persons co-enrolled in the AIDS Linked to the Intravenous Experience study (ALIVE, an interval cohort) and the Johns Hopkins HIV Clinical Cohort, the association of HIV markers with all-cause mortality was examined in each study. In a second analysis, the causal effect of highly active antiretroviral therapy (HAART) on mortality was explored using HIV markers as predictors of HAART initiation.
Methods
Study Design and Population
We identified persons who were co-enrolled in 2 prospective cohort studies. The AIDS Link to IntraVenous Experience study is a prospective cohort study of HIV-infected and -uninfected individuals in Baltimore, Maryland with a history of injection drug use. At its inception in 1988, this study recruited 2946 persons with further enrollment of 735 subjects through recruitment in 1994–1995, 1998, and 2000. All participants were required to be at least 18 years of age and free of AIDS at entry. All participants provided written informed consent before study participation; the study protocol has been continually reviewed and approved by the Institutional Review Board at Johns Hopkins. Participants were scheduled for semiannual visits including structured interviews, a computer-based risk questionnaire, clinical examination, and collection of blood specimens. The interview and questionnaire collect information on demographic variables, medical history, HIV risk behavior (including illicit drug use), and use of HIV therapies. Details on the study design have been published elsewhere.2
The Johns Hopkins AIDS Service provides longitudinal primary and subspecialty care for a large proportion of HIV-infected individuals in the Baltimore metropolitan area. In 1990, the Johns Hopkins HIV Clinical Cohort was established to understand and quantify the processes and the outcomes of care for HIV-infected patients seen in clinical practice. Enrollment into this dynamic cohort coincides with first enrollment at the HIV clinic. Information is collected from outpatient and inpatient medical records, the Johns Hopkins Health System automated databases, supplemental medical records from outside facilities, and vital records. Additionally, patients provide written informed consent for study access to additional information from other research studies being conducted within the Johns Hopkins Medical Institution. Information is reviewed and abstracted by trained medical record technicians, and includes comprehensive demographic, clinical, laboratory, pharmaceutical, and psychosocial data. Mortality data are abstracted from record review of Johns Hopkins and outside hospital records, death certificates, family reports, and linkage to both the National Death Index and the national Social Security Death Index. The details of the study design and follow-up have been described previously.11
For both cohort studies, CD4+ T-lymphocyte levels were measured in peripheral blood and analyzed by 2-color flow cytometry stained with monoclonal antibodies specific for CD4+ lymphocytes.21,22 HIV-1 RNA level was quantified by in vitro nucleic acid amplification using the Amplicor RT-PCR assay (version 1.0 before 2002 and version 1.5 after 2002 [Roche Molecular Diagnostics, Pleasanton, CA]). Hemoglobin was obtained from complete blood counts via automated Coulter blood count. We used Social Security numbers, names, birthdates, sex, and race to identify persons enrolled in both cohorts.
Statistical Methods
Data were restricted to measurements taken from the time an individual had biologic markers assessed in both cohorts until either death or censoring. Thus, only those persons who had data collected with contemporaneous visits in both cohorts were included, although someone might initially contribute data to both cohorts and then, due to loss to follow-up, not contribute data in one or the other of the cohorts. To ensure comparability of the datasets, a person was considered co-enrolled in both cohorts by taking the later of the 2 cohort-entry CD4 measurements (one from each cohort). From this time point, marker measurements were carried forward until we identified visits in each cohort that did not have missing marker measurements (either from being measured on this date or carried forward from a previous date). The later visit (from either study) with all 3 markers was defined as a common baseline visit date.
The distribution of visits was examined to compare the frequency of visits between the 2 studies. Furthermore, the medians of either the participants' most extreme measurements (nadir value for CD4 and hemoglobin; maximum value for HIV-1 RNA) occurring during follow-up, or overall median values during follow-up, were compared between cohorts via Wilcoxon signed rank test (as the distribution of the individual differences between cohort measurements were not normally distributed). We used a negative binomial model to examine whether more advanced HIV disease progression (defined by baseline CD4, baseline log10 HIV-1 RNA, baseline hemoglobin, and progression to clinical AIDS during follow-up) was associated with the number of marker measurements in the clinical cohort study.
We conducted 2 analyses to examine the effect of visit structure of study designs between the outcome and exposures. With these analyses, we wanted to remove any study differences, other than the structure of the visit data, that could influence analytical inferences. In the first set of analyses, we investigated the association and predictive value of established biologic markers, CD4 counts,23–25 HIV-1 RNA levels,23,25,26 and hemoglobin concentration27–31 with mortality. For these analyses, no adjustments were made for potential confounders because the focus was not to evaluate the “true” association but rather to compare results from each cohort. The only potential covariate that was included (other than the biomarker of interest) was the time elapsed from measurement of the marker until the risk set was defined by an event occurrence. The 3 different markers have varying patterns over the course of HIV disease.32–34
Cox proportional hazards were used to examine the relationship between time-varying CD4+ counts, HIV-1 RNA levels, and hemoglobin concentration with mortality. Person-time began accumulating from the baseline date up to one of the following events: death, censored as loss-to-follow-up at last marker visit, or administratively censored on 31 December 2004. Furthermore, to address the time gap between when a person's biomarker was last measured and an event, the time elapsed since the last visit was included in the model, as described by de Bruijne et al.35 Both the biomarkers and the time-elapsed variables were allowed to be nonlinear by applying fractional polynomials.36–38 The CD4 count plus one was modeled to allow for the logarithm transformation of CD4. To assess differences in inferences, we compared the ratio of the relative hazards between the 2 studies. To evaluate the predictive value of each biomarker with mortality, an estimate of the “explained randomness” (which is analogous to explained variability, R2) was determined utilizing methods outlined by O'Quigley et al.39,40 Furthermore, we determined percentile-based nonparametric bootstrap 95% confidence intervals (CIs) for the ratio of relative hazards and for the R2 estimates based on 1500 full samples with replacement.41
In a second set of analyses, the association between HAART initiation and mortality was assessed using marginal structural models.42,43 This analysis was restricted to individuals who were not on HAART at baseline, such that the probability of treatment initiation could be evaluated for the inverse probability of treatment weights.42,43 HAART was defined as the receipt of a minimum of 3 antiretroviral drugs to capture early definitions of HAART.
The data were restructured into a person-month dataset in which values were carried forward until a new marker measurement was made. Specifically, we used a pooled logistic regression model that was weighted by inverse-probability-of-treatment and inverse-probability-of-censor to approximate a Cox proportional hazards model.42,43 Persons who did not die during follow-up were censored at either 31 December 2004 or 1 year after their last visit, whichever occurred first. For a monthly interval in which a visit did not occur, the last marker value was carried forward. We used stabilized weights to increase the efficiency of the estimator.42,43 Furthermore, because the use of weights induces within-subject correlation, generalized estimating equations44 were used to fit the weighted logistic model by utilizing the “repeated” option within the genmod procedure in SAS (SAS Institute, Cary, NC), to produce robust variance estimators.42,43
For the inverse probability of treatment weights, the model from which the numerator is derived included the following variables: time (as a restricted cubic spline with knots at the 5, 27.5, 50, 72.5, and 95th percentiles), baseline CD4 categories (≤200, 201–350, and >350 cells/mL), baseline HIV RNA categories (≤500, 501–10000, and > 10000 copies/mL), and baseline hemoglobin categories (≤9.5, 9.6–11.4, 11.5–12.7, and > 12.7 g/dL). For the denominator, the model also included the time-varying marker levels (CD4, HIV RNA, and hemoglobin, each as restricted cubic spline with knots at the 5, 35, 65, and 95th percentiles). Similar models were used for the inverse probability of censoring weights. For the marginal structural model, we examined a univariate model in which only time and an indicator of HAART use were included as covariates. A multivariate model was also examined that included the baseline marker categories. Similar standard pooled logistic models (without inverse weights) were also examined, except that time-varying marker levels were included within the multivariate model.
Results
The study population comprised 244 individuals enrolled in both cohort studies. The population was primarily African American (95%) and male (67%), with a median age of 41 years (interquartile range [IQR] = 37–45) years. The baseline marker characteristics are shown in Table 1. The marker measurements at the baseline visit were similar for the 2 studies. The 244 participants contributed 1250 person-years of data from July 1995 through December 2004; 81 (33%) individuals died. The standard of care during this time period called for HIV patients to be seen approximately every 3 months; thus, it is not surprising that the clinical cohort had many more measurements of CD4, HIV-1 RNA, and hemoglobin than the interval cohort. The number of measurements of hemoglobin in the clinical cohort greatly exceeded that of any other marker measurements.
TABLE 1.
Median Biomarker Levels at Baseline and Total Number of Biomarker Measurements for 244 Persons Co-enrolled in an Interval Cohort Study and a Clinical Cohort Study
| Interval Cohort | Clinical Cohort | |||||
|---|---|---|---|---|---|---|
| Median Biomarker Level (IQR) | Median No. Measurements per Individual Over Entire Follow-Up (IQR) | No. Measurements Over Entire Follow-Up | Median Biomarker Level (IQR) | Median No. Measurements per Individual Over Entire Follow-Up (IQR) | No. Measurements Over Entire Follow-Up | |
| CD4 count (cells/uL) | 238 (123, 377) | 7 (4, 12) | 1951 | 285 (141, 432) | 9 (4, 20) | 3387 |
| Log10 HIV-1 RNA | 4.38 (3.45, 4.99) | 8 (4, 12) | 1938 | 4.40 (3.10, 4.97) | 9 (4, 19) | 3232 |
| Hemoglobin (g/dL) | 12.8 (11.6, 13.9) | 8 (4, 12) | 1949 | 12.6 (11.1, 13.7) | 24 (9, 47) | 8096 |
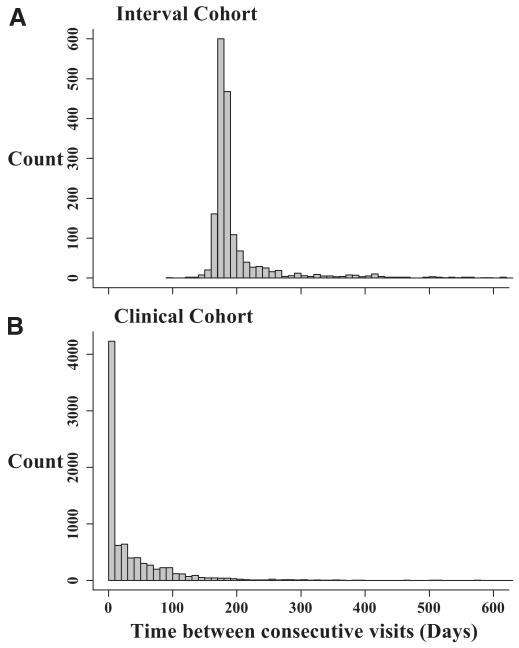
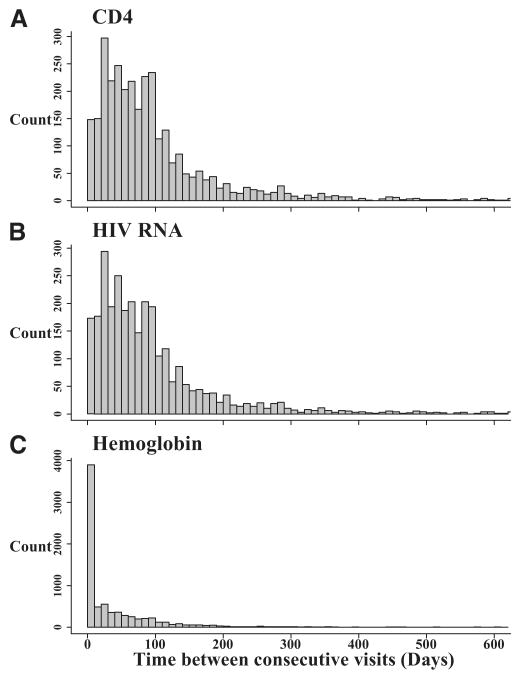
Figure 1 shows the distribution of the time between consecutive visits for each cohort study. By design, the interval cohort study had visits that occurred every 6 months (Fig. 1A; median = 182 [IQR = 175–194 days]). However, the clinical cohort study data collection was heavily skewed to the right (Fig. 1B) with a wide range of times between visits (median = 11 days [IQR = 1–54 days]). The time between consecutive visits differed in the clinical cohort by the biomarker being collected (Fig. 2). The visits in which CD4 (Fig. 2A) and HIV-1 RNA (Fig. 2B) were collected had similar time between consecutive visits, as expected (median of 77 days [38–121] and 72 days, [35–120], respectively). For both CD4 and HIV-1 RNA, participants had a median of 9 measurements during follow-up (for CD4, IQR = 4–20; for HIV-1 RNA, IQR = 4–19). The time between consecutive hemoglobin measurements, however, was more skewed (Fig. 2C), as the median time between consecutive visits was 11 days (IQR = 1–57), with a median of 24 measurements per person (IQR = 9–47).
FIGURE 1.
Time between consecutive visits for 244 persons co-enrolled in the, A, interval cohort and, B, clinical cohort studies, between 1995 and 2004.
FIGURE 2.
Time between consecutive, A, CD4 count, B, HIV-1 RNA, and, C, hemoglobin measurements, for the clinical cohort data collected on 244 persons between 1995 and 2004.
Fitting a negative binomial model to the clinical cohort data, persons with a lower baseline CD4 lymphocyte count (incidence rate ratio [IRR] = 0.91 [95% CI = 0.827–1.007] per 100 cells) and higher baseline log10 HIV-1 RNA (IRR = 1.17 [1.003–0.368]) had a greater number of CD4 measurements. When the number of hemoglobin measurements were considered, these trends were slightly stronger (for baseline CD4 levels, IRR = 0.89 [0.793–0.993] per 100 cells and 1.36 [1.100–1.671] per log10 HIV-1 RNA). Taken together, these data support our hypothesis that the rate of marker measurements was associated with HIV disease severity.
The different distributions of time between consecutive visits for the 2 studies resulted in substantial differences in the values of the marker data. The median of an individual's most extreme and median measurement over the entire follow-up period are shown in Table 2. Although the overall distribution for CD4 was similar for the 2 studies, the difference between extreme or median values obtained from each cohort paired by individual showed that the interval cohort values tended to be lower (and the only comparison that was not statistically significant was for the paired difference of the extreme measurement for those who had died; Table 2). Furthermore, higher hemoglobin measurements were seen in the interval cohort as compared with the clinical cohort, both overall and for the individually paired measurements.
TABLE 2.
Median and Interquartile Range of Individual's Most Extreme and Median Marker Measurements During Follow-Up for 244 Persons Co-enrolled in 2 Cohort Studies, Stratified by Vital Status
| Extreme Valuea | Median Value | |||||
|---|---|---|---|---|---|---|
| Biomarker Level | Median Paired Differenceb (P) | Biomarker Level | Median Paired Differenceb (P) | |||
| Interval Cohort | Clinical Cohort | Interval Cohort | Clinical Cohort | |||
| CD4 count (cells/uL) | ||||||
| Alive (n = 163) | 135 (46, 257) | 138 (36, 300) | − 8 (0.002) | 274 (144, 408) | 296 (180, 436) | − 34 (<0.0001) |
| Died (n = 81) | 56 (15, 157) | 52 (12, 234) | 1.0 (0.60) | 136 (44, 244) | 154 (52, 320) | − 12 (0.005) |
| log10 HIV-1 RNA | ||||||
| Alive (n = 163) | 4.87 (4.27, 5.35) | 4.86 (4.26, 5.44) | 0.00 (0.37) | 4.02 (2.90, 4.62) | 3.54 (2.60, 4.42) | 0.11 (<0.0001) |
| Died (n = 81) | 5.32 (4.59, 5.70) | 5.20 (4.59, 5.83) | −0.02 (0.81) | 4.72 (3.98, 5.22) | 4.63 (3.98, 5.09) | 0.06 (0.075) |
| Hemoglobin (g/dL) | ||||||
| Alive (n = 163) | 11.4 (10.2, 12.7) | 10.7 (8.3, 12.2) | 0.8 (<0.0001) | 13.0 (12.1, 13.9) | 12.7 (11.5, 13.7) | 0.4 (<0.0001) |
| Died (n = 81) | 10.6 (9.4, 11.6) | 8.0 (7.0, 9.7) | 2.0 (<0.0001) | 12.0 (11.0, 13.2) | 10.6 (9.7, 12.1) | 0.9 (<0.0001) |
Extreme value is defined as the minimum marker measurement occurring during follow-up for CD4 and hemoglobin; for HIV-1 RNA, the extreme value was the highest measurement occurring during follow-up.
Median paired difference is the median of the differences between an individual's pair of extreme or pair of median marker values obtained from each cohort (interval cohort marker value-clinical cohort marker value); P value is by Wilcoxon signed rank test.
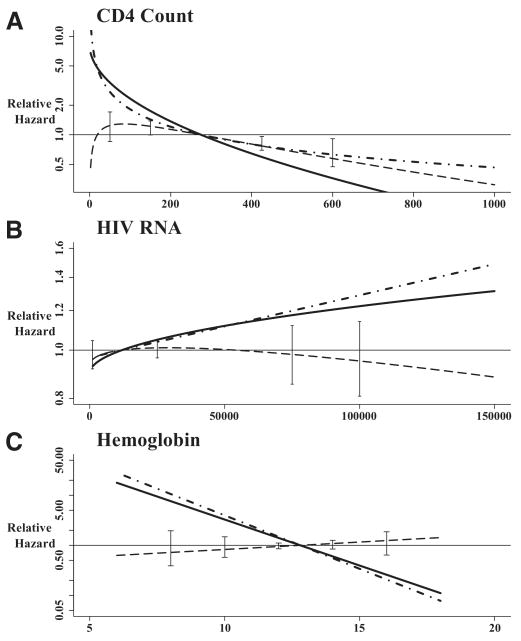
Given the differences in frequency of data collection and in the value of markers collected over the follow-up period, the associations and predictive values of CD4 counts, HIV-1 RNA levels, and hemoglobin concentration with mortality were assessed. Table 3 shows the model formula and regression coefficients for the associations between biomarkers and mortality. The time elapsed between last biomarker measurement and time of event was included only in the models assessing HIV-1 RNA (clinical cohort only) and hemoglobin (both cohorts). As shown in the table, the relative hazards for mortality were not linear with higher biomarker levels. Therefore, the relative hazard for mortality associated with each biomarker is shown relative to the median marker level across all risk sets (Fig. 3). As can be seen by the curve representing the ratio of the relative hazard for the 2 cohorts (Fig. 3, dashed line), the estimates between the 2 cohorts were remarkably similar for HIV RNA and for hemoglobin, but not for CD4 counts. For CD4 counts, the relative hazard curve for the interval cohort suggested a greater protective association for higher CD4counts than does the curve for the clinical cohort. Below the reference of 275 cells, there were no major differences in the ratio, as suggested by the bootstrap 95% CIs at 50 and 150 cells/uL. However, for the clinical cohort study, a model with a square root transformation of CD4 counts did not fit significantly worse than the slightly better logarithm transformation (−2 × log-likelihood: 772.4 vs. 770.7, respectively; P = 0.2). The beta coefficient for this model was −0.129 (SE = 0.018), which essentially removes all differences between the 2 cohorts. Similarly, for the interval cohort data, the square root transformation (−2 × log-likelihood) was 779.7 and the logarithm transformation (−2 × log-likelihood) was 783.0 (P = 0.07). The beta coefficient for the logarithm transformation of CD4 counts was −0.506 (SE= 0.064), which is similar to the value obtained in the clinical cohort study (Table 3).
TABLE 3.
Cox Proportional Hazards Models Examining the Association of CD4, HIV-1 RNA, and Hemoglobin With Mortality Among 244 Persons Co-enrolled in 2 Cohort Studies
| Interval Cohorta | Clinical Cohorta | |||||||
|---|---|---|---|---|---|---|---|---|
| Formb | β1 SE | β2 SE | R2c 95% CI | Formb | β1 SE | β2 SE | R2c (95% CI) | |
| CD4 countsd | β1(CD4 + l)0.5 | −0.128(0.019) | — | 0.488 (0.297–0.666) | β1 log(CD4) | −0.592 (0.068) | — | 0.537 (0.345–0.707) |
| HIV-1 RNAe | β1RNA0.5 | 9.8 × 10−4 | — | 0.168 (0.030–0.404) | β1RNA + β2RNA × e−0.5×tel | 4.4 × 10−6 | −1.38 × 10−6 | 0.254 (0.084–0.466) |
| (2.0 × 10−4) | (1.3 × 10−6) | (9.81 × 10−7) | ||||||
| Hemoglobinf | β1Hgb × e(0.5×tel) + β2tel3 | −0.492 (0.063) | −5.07 (0.744) | 0.510g (0.325–0.674) | β1Hgb + β2tel0.5 | −0.492 (0.060) | 0.144(0.017) | 0.585g (0.386–0.749) |
In the interval cohort, 81 deaths; in the clinical cohort, 81 deaths.
Form is how the marker was transformed to be used within the model, thus allowing for non-linearity.
The time elapsed (tel) between when the marker was measured and when the event occurred did not contribute to models with CD4.
For the HIV-1 RNA model using the clinical cohort data, only time elapsed (tel) was included of this form RNA × e−0.5×tel and therefore the effect of HIV-1 RNA was dependent on time elapsed.
For the interval cohort data, the main effect of hemoglobin was dropped in favor of a more parsimonious model that included time elapsed and is of the form shown in the table (see de Bruijne et al.35).
For Hemoglobin, the R2 in the table is a partial R2 for β1 after the inclusion of β2 in the model. The R2 for the full hemoglobin model is 0.512 (95% CI = 0.332–0.678) for the interval cohort and 0.867 (0.742–0.937) for the clinical cohort; however this may not be a fair comparison because the time-elapsed in the clinical setting provides much more information beyond that of the marker. The difference between R2 (R2clinical − R2interval) was not statistically significant at the 0.05 significance level using difference between each bootstrap replicate as the 2.5 and 97.5 percentiles crossed 0.
SE indicate standard error.
FIGURE 3.
The nonlinear relative hazard for mortality according to level of, A, CD4 count (cells/uL), B, HIV RNA (cps/uL), and, C, hemoglobin (g/dL). Interval cohort study represented by solid line; clinical cohort study represented by dot-dash. The dashed line with bootstrap 95% confidence interval bars is the interval cohort curve divided by the clinical cohort curve. For HIV RNA (clinical cohort model only) and hemoglobin, the time elapsed was set at 0.3 years (Table 3), as the median time between measurement and event was 0.29 years ([IQR] = 0.11–1.02) for the clinical cohort study HIV RNA, 0.21 (0.08–0.72) years for the clinical cohort study hemoglobin, and 0.31 (0.15–0.52) years for the interval cohort study hemoglobin.
The proportion of explained variability for each of the models is also shown in Table 3. The confidence intervals for each of the time-updated models are wide (reflecting the limited number of deaths). Note that the R2 for the hemoglobin models is the partial R2 for the β1 coefficient, to provide a fair comparison of hemoglobin marker after accounting for most of the association of time elapsed. For the full model R2, there is a difference in predictive value of the model (for the interval cohort, R2 = 0.512 [95% CI = 0.332–0.678]; for the clinical cohort, R2 = 0.867 [0.742–0.937]) suggesting that the timing of hemoglobin measurements in the clinical setting does contribute to the predictive value.
The relative hazard odds ratios for the analyses of the causal effect of HAART on mortality are shown in Table 4. Because these analyses were restricted to those who were not on HAART at baseline, the population consists of 160 people. At baseline, the majority were not on therapy (81%), with 14% on antiretroviral monotherapy and 6% on non-HAART antiretroviral combination therapy. During follow-up, 77 participants initiated a HAART regimen, with a median initiation date of May 1999 (interquartile range: December 1997–April 2001). The results for both the standard pooled logistic model and the marginal-structural-model pooled logistic model are shown in Table 4, with and without adjustment for the marker levels (baseline and time-varying values for the multivariate standard pooled logistic model; baseline values only for the multivariate marginal structural model). The mean of the stabilized weights were 0.98 for the interval cohort and 1.03 for the clinical cohort. The relative hazard odds ratios were similar between the 2 cohorts at approximately 0.30, providing evidence of a 70% decreased risk of death after HAART initiation in both cohorts. The marginal structural model resulted in a larger relative change from the multivariate standard pooled logistic model for the clinical cohort than for the interval cohort.
TABLE 4.
The Relative Hazard OR for the Pooled Logistic Models Examining the Causal Effect of HAART on Mortality
| Interval Cohorta Relative Hazard OR (95% CI) | Clinical Cohorta Relative Hazard OR (95% CI) | |
|---|---|---|
| Standard pooled logistic model | ||
| Univariate | 0.72 (0.36–1.40) | 0.72 (0.37–1.40) |
| Multivariateb | 0.47 (0.22–1.04) | 0.58 (0.26–1.26) |
| Weighted pooled logistic modelc | 0.27 (0.11–0.66) | 0.34 (0.15–0.77) |
In the interval cohort, 38 deaths; in the clinical cohort, 39 deaths. Difference is due to censoring individuals 1 year after follow-up.
Baseline and time-varying marker levels (CD4, HIV RNA, and hemoglobin) were included the multivariate standard pooled logistic model.
Only baseline marker levels are included in the weighted pooled logistic model.
DISCUSSION
Despite a difference in the frequency of data collection between the interval and clinical cohorts, and a tendency for the more immunocompromised patients in the clinical cohort study to have measurements more frequently, similar inferences could be made about the associations and predictive values of CD4 count, HIV-1 RNA level, and hemoglobin concentration with mortality. It is reassuring to see that the inferences regarding CD4 counts and HIV-1 RNA levels were similar despite study design differences, especially considering that the analyses are being conducted on the same people. This study provides evidence that the pattern of measurement has little practical effect on analyses of the biomarkers routinely used for monitoring HIV patients, and suggests that prediction inferences from both types of cohort studies are comparable. Rather than using only the most recent marker value, the predictive power of the models would probably be increased by using the 2 most recent values, or by including the baseline value as a time-fixed value. Furthermore, it is possible that the inclusion of previous marker values would have greater predictive capabilities within a clinical cohort as compared with an interval cohort. For example, we have previously shown that a rapid decline in hemoglobin levels is suggestive of HIV disease progression to AIDS or death.28,33,34 However, these analyses were conducted within an interval cohort; if infected persons have a rapid decline in a short time interval, it is possible that this pattern would not be observed in data from an interval cohort. A clinical cohort may be more likely to capture enough data points to detect this decline and thus have improved predictive ability.28
Furthermore, the relationship of HAART with mortality was similar between the 2 cohort studies. The 70% reduction in the relative hazard odds ratio for mortality is between the estimate for HAART versus no therapy (relative hazard = 0.14 [95% CI = 0.07–0.29]) and for HAART compared with dual therapy (relative hazard = 0.49 [0.31–0.79]) seen in a marginal structural analysis among the Swiss HIV Cohort Study.19 However, their outcome variable was a composite of either progression to AIDS or death, whereas ours was only mortality. Nevertheless, our results are consistent, as our population is a mix of individuals initiating HAART, some monotherapy and non-HAART combination therapy, and individuals not on any antiretrovirals. Another study found the causal effect for HAART with progression to AIDS or death was estimated to be a relative hazard of 0.54 (95% CI = 0.38–0.78).45 However, this study population had a greater proportion on either single or combination therapy (40% vs. 20% in our cohort). One might therefore expect a greater attenuation in the estimate. Our results are also congruent with the period-analysis of Detels et al46 in which calendar period was used as a surrogate for HAART exposure in the Multicenter AIDS Cohort Study population. A relative hazard of 0.35 (95% CI = 0.20–0.61) for AIDS or death was seen for the period after HAART was introduced.
The exposures in these analyses were focused on comparing biologic markers between cohort studies. This was by design because the laboratory methods for measuring these biomarkers allow for comparability between cohort studies, whereas data collected through interviews and questionnaires may not be comparable. Whether these types of data would provide similar inferences depending on the structure of cohort visit design would depend on the specific exposure being measured by the interviewer or questionnaire, and on the outcome. A limitation of our study, however, was that these types of data were not included in this analysis because different protocols were in place for these data.
Marginal structural models have several assumptions. One of the strongest assumptions is that the covariates used to determine the inverse probability of weights are sufficient to adjust for the confounding.42 It is possible that there are unmeasured confounders such that the correction by the inverse probability weights is not adequate. However, CD4 counts and HIV-1 RNA levels are the standard biomarkers that clinicians use to initiate treatment. Furthermore, despite being examined on a small sample of persons who were co-enrolled in both studies, the results are similar to what has previously been seen in this clinical cohort.47 In 1995, the incidence of all-cause mortality was 112 deaths/1000 person-years. Since 1996, mortality has been relatively stable at approximately 40/1000 person-years, which would be a relative reduction of 0.36.47 This analysis relies on a different set of assumptions compared with a marginal structural model, and yet has similar inferences.
In summary, our results give empirical evidence that in analyses of markers measured on the same individuals, inferences were similar whether examining the markers as an exposure or as a time-dependent confounder in assessing the causal effect of HAART on mortality. It is encouraging that the inferences between the 2 cohort designs for CD4 counts and HIV-1 RNA were so similar, as these markers are the standard for assessing HIV disease progression in patients. Given that time-dependent confounding may be greater in clinical cohorts, appropriate methodologic tools are vital to properly assess the causal effect of clinical interventions.
Acknowledgments
Supported by National Institutes of Health (RO1-DA11602; R01-DA04334; R01-DA12546; U01-AI069918; K01-AI071754).
References
- 1.Lau B, Gange SJ, Moore RD. Interval and clinical cohort studies: epidemiological issues. AIDS Res Hum Retroviruses. 2007;23:769–776. doi: 10.1089/aid.2006.0171. [DOI] [PubMed] [Google Scholar]
- 2.Vlahov D, Anthony JC, Munoz A, et al. The ALIVE study, a longitudinal study of HIV-1 infection in intravenous drug users: description of methods and characteristics of participants. NIDA Res Monogr. 1991;109:75–100. [PubMed] [Google Scholar]
- 3.De Wolf F, Lange JM, Houweling JT, et al. Numbers of CD4+ cells and the levels of core antigens of and antibodies to the human immunodeficiency virus as predictors of AIDS among seropositive homosexual men. J Infect Dis. 1988;158:615–622. doi: 10.1093/infdis/158.3.615. [DOI] [PubMed] [Google Scholar]
- 4.Rezza G, Lazzarin A, Angarano G, et al. The natural history of HIV infection in intravenous drug users: risk of disease progression in a cohort of seroconverters. AIDS. 1989;3:87–90. doi: 10.1097/00002030-198902000-00006. [DOI] [PubMed] [Google Scholar]
- 5.Kaslow RA, Ostrow DG, Detels R, Phair JP, Polk BF, Rinaldo CR., Jr The Multicenter AIDS Cohort Study: rationale, organization, and selected characteristics of the participants. Am J Epidemiol. 1987;126:310–318. doi: 10.1093/aje/126.2.310. [DOI] [PubMed] [Google Scholar]
- 6.Barkan SE, Melnick SL, Preston-Martin S, et al. The Women's Interagency HIV Study. WIHS Collaborative Study Group. Epidemiology. 1998;9:117–125. [PubMed] [Google Scholar]
- 7.Liddell FD. The development of cohort studies in epidemiology: a review. J Clin Epidemiol. 1988;41:1217–1237. doi: 10.1016/0895-4356(88)90027-3. [DOI] [PubMed] [Google Scholar]
- 8.van Sighem AI, van de Wiel MA, Ghani AC, et al. Mortality and progression to AIDS after starting highly active antiretroviral therapy. AIDS. 2003;17:2227–2236. doi: 10.1097/00002030-200310170-00011. [DOI] [PubMed] [Google Scholar]
- 9.Portsmouth S, Stebbing J, Gill J, et al. A comparison of regimens based on non-nucleoside reverse transcriptase inhibitors or protease inhibitors in preventing Kaposi's sarcoma. AIDS. 2003;17:F17–F22. doi: 10.1097/00002030-200307250-00001. [DOI] [PubMed] [Google Scholar]
- 10.d'Arminio MA, Lepri AC, Rezza G, et al. Insights into the reasons for discontinuation of the first highly active antiretroviral therapy (HAART) regimen in a cohort of antiretroviral naive patients. I.CO.N.A. Study Group. Italian Cohort of Antiretroviral-Naive Patients. AIDS. 2000;14:499–507. doi: 10.1097/00002030-200003310-00005. [DOI] [PubMed] [Google Scholar]
- 11.Moore RD. Understanding the clinical and economic outcomes of HIV therapy: the Johns Hopkins HIV clinical practice cohort. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;17(suppl 1):S38–S41. doi: 10.1097/00042560-199801001-00011. [DOI] [PubMed] [Google Scholar]
- 12.Sudre P, Rickenbach M, Taffe P, Janin P, Volkart AC, Francioli P. Clinical epidemiology and research on HIV infection in Switzerland: the Swiss HIV Cohort Study 1988-2000. Schweiz Med Wochenschr. 2000;130:1493–1500. [PubMed] [Google Scholar]
- 13.The UK Collaborative HIV Cohort Steering Committee. The creation of a large UK-based multicentre cohort of HIV-infected individuals: The UK Collaborative HIV Cohort (UK CHIC) Study. HIV Med. 2004;5:115–124. doi: 10.1111/j.1468-1293.2004.00197.x. [DOI] [PubMed] [Google Scholar]
- 14.Hooshyar D, Napravnik S, Miller WC, Eron JJ., Jr Effect of hepatitis C coinfection on discontinuation and modification of initial HAART in primary HIV care. AIDS. 2006;20:575–583. doi: 10.1097/01.aids.0000210612.37589.12. [DOI] [PubMed] [Google Scholar]
- 15.Kitahata MM, Rodriguez B, Haubrich R, et al. Cohort profile: the Centers for AIDS Research Network of Integrated Clinical Systems. Int J Epidemiol. 2008;37:948–955. doi: 10.1093/ije/dym231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.National Institutes of Health. Centers for AIDS Research. [National Institutes of Health] 2008 Available at: http://www3.niaid.nih.gov/research/cfar/
- 17.Rodriguez B, Sethi AK, Cheruvu VK, et al. Predictive value of plasma HIV RNA level on rate of CD4 T-cell decline in untreated HIV infection. JAMA. 2006;296:1498–1506. doi: 10.1001/jama.296.12.1498. [DOI] [PubMed] [Google Scholar]
- 18.Celentano DD, Galai N, Sethi AK, et al. Time to initiating highly active antiretroviral therapy among HIV-infected injection drug users. AIDS. 2001;15:1707–1715. doi: 10.1097/00002030-200109070-00015. [DOI] [PubMed] [Google Scholar]
- 19.Sterne JA, Hernan MA, Ledergerber B, et al. Long-term effectiveness of potent antiretroviral therapy in preventing AIDS and death: a prospective cohort study. Lancet. 2005;366:378–384. doi: 10.1016/S0140-6736(05)67022-5. [DOI] [PubMed] [Google Scholar]
- 20.Griffin JT, Fraser C, Gras L, De Wolf F, Ghani AC. The effect on treatment comparisons of different measurement frequencies in human immunodeficiency virus observational databases. Am J Epidemiol. 2006;163:676–683. doi: 10.1093/aje/kwj083. [DOI] [PubMed] [Google Scholar]
- 21.Giorgi JV, Cheng HL, Margolick JB, et al. Quality control in the flow cytometric measurement of T-lymphocyte subsets: the multicenter AIDS cohort study experience. The Multicenter AIDS Cohort Study Group. Clin Immunol Immunopathol. 1990;55:173–186. doi: 10.1016/0090-1229(90)90096-9. [DOI] [PubMed] [Google Scholar]
- 22.Schenker EL, Hultin LE, Bauer KD, Ferbas J, Margolick JB, Giorgi JV. Evaluation of a dual-color flow cytometry immunophenotyping panel in a multicenter quality assurance program. Cytometry. 1993;14:307–317. doi: 10.1002/cyto.990140311. [DOI] [PubMed] [Google Scholar]
- 23.Anastos K, Kalish LA, Hessol N, et al. The relative value of CD4 cell count and quantitative HIV-1 RNA in predicting survival in HIV-1-infected women: results of the women's interagency HIV study. AIDS. 1999;13:1717–1726. doi: 10.1097/00002030-199909100-00016. [DOI] [PubMed] [Google Scholar]
- 24.Fahey JL, Taylor JM, Detels R, et al. The prognostic value of cellular and serologic markers in infection with human immunodeficiency virus type 1. N Engl J Med. 1990;322:166–172. doi: 10.1056/NEJM199001183220305. [DOI] [PubMed] [Google Scholar]
- 25.Mellors JW, Munoz A, Giorgi JV, et al. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann Intern Med. 1997;126:946–954. doi: 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- 26.Mellors JW, Rinaldo CR, Jr, Gupta P, White RM, Todd JA, Kingsley LA. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 27.Creagh-Kirk T, Doi P, Andrews E, et al. Survival experience among patients with AIDS receiving zidovudine. Follow-up of patients in a compassionate plea program. JAMA. 1988;260:3009–3015. [PubMed] [Google Scholar]
- 28.Lau B, Gange SJ, Phair JP, Riddler SA, Detels R, Margolick JB. Use of total lymphocyte count and hemoglobin concentration for monitoring progression of HIV infection. J Acquir Immune Defic Syndr. 2005;39:620–625. [PubMed] [Google Scholar]
- 29.Lundgren JD, Mocroft A, Gatell JM, et al. A clinically prognostic scoring system for patients receiving highly active antiretroviral therapy: results from the EuroSIDA study. J Infect Dis. 2002;185:178–187. doi: 10.1086/338267. [DOI] [PubMed] [Google Scholar]
- 30.Moore RD, Keruly JC, Chaisson RE. Anemia and survival in HIV infection. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;19:29–33. doi: 10.1097/00042560-199809010-00004. [DOI] [PubMed] [Google Scholar]
- 31.Sullivan PS, Hanson DL, Chu SY, Jones JL, Ward JW. Epidemiology of anemia in human immunodeficiency virus (HIV)-infected persons: results from the multistate adult and adolescent spectrum of HIV disease surveillance project. Blood. 1998;91:301–308. [PubMed] [Google Scholar]
- 32.Gange SJ, Mellors JW, Lau B, et al. Longitudinal patterns of HIV type 1 RNA among individuals with late disease progression. AIDS Res Hum Retroviruses. 2001;17:1223–1229. doi: 10.1089/088922201750461276. [DOI] [PubMed] [Google Scholar]
- 33.Gange SJ, Lau B, Phair J, Riddler SA, Detels R, Margolick JB. Rapid declines in total lymphocyte count and hemoglobin in HIV infection begin at CD4 lymphocyte counts that justify antiretroviral therapy. AIDS. 2003;17:119–121. doi: 10.1097/00002030-200301030-00016. [DOI] [PubMed] [Google Scholar]
- 34.Lau B, Gange SJ, Phair JP, Riddler SA, Detels R, Margolick JB. Rapid declines in total lymphocyte counts and hemoglobin concentration prior to AIDS among HIV-1-infected men. AIDS. 2003;17:2035–2044. doi: 10.1097/00002030-200309260-00004. [DOI] [PubMed] [Google Scholar]
- 35.de Bruijne MH, le Cessie S, Kluin-Nelemans HC, van Houwelingen HC. On the use of Cox regression in the presence of an irregularly observed time-dependent covariate. Stat Med. 2001;20:3817–3829. doi: 10.1002/sim.1083. [DOI] [PubMed] [Google Scholar]
- 36.Royston P, Altman DG. Regression using fractional polynomials of continuous covariates-parsimonious parametric modeling. J Royal Stat Soc Ser C Appl Stat. 1994;43:429–467. [Google Scholar]
- 37.Royston P, Ambler G, Sauerbrei W. The use of fractional polynomials to model continuous risk variables in epidemiology. Int J Epidemiol. 1999;28:964–974. doi: 10.1093/ije/28.5.964. [DOI] [PubMed] [Google Scholar]
- 38.Sauerbrei W, Meier-Hirmer C, Benner A, Royston P. Multivariable regression model building by using fractional polynomials: Description of SAS, STATA, and R programs. Comput Stat Data Anal. 2006;50:3464–3485. [Google Scholar]
- 39.O'Quigley J, Xu RH, Stare J. Explained randomness in proportional hazards models. Stat Med. 2005;24:479–489. doi: 10.1002/sim.1946. [DOI] [PubMed] [Google Scholar]
- 40.O'Quigley J, Flandre P. Quantification of the prentice criteria for surrogate endpoints. Biometrics. 2006;62:297–300. doi: 10.1111/j.1541-0420.2006.00538.x. [DOI] [PubMed] [Google Scholar]
- 41.Efron B. The Jackknife, the Bootstrap, and Other Resampling Plans. Philadelphia: Society for Industrial and Applied Mathematics; 1982. [Google Scholar]
- 42.Hernan MA, Brumback B, Robins JM. Marginal structural models to estimate the causal effect of zidovudine on the survival of HIV-positive men. Epidemiology. 2000;11:561–570. doi: 10.1097/00001648-200009000-00012. [DOI] [PubMed] [Google Scholar]
- 43.Hernan MA, Brumback B, Robins JM. Marginal structural models to estimate the joint causal effect of nonrandomized treatments. J Am Stat Assoc. 2001;96:440–448. [Google Scholar]
- 44.Zeger SL, Liang KY. Longitudinal data-analysis for discrete and continuous outcomes. Biometrics. 1986;42:121–130. [PubMed] [Google Scholar]
- 45.Cole SR, Hernan MA, Robins JM, et al. Effect of highly active antiretroviral therapy on time to acquired immunodeficiency syndrome or death using marginal structural models. Am J Epidemiol. 2003;158:687–694. doi: 10.1093/aje/kwg206. [DOI] [PubMed] [Google Scholar]
- 46.Detels R, Munoz A, McFarlane G, et al. Effectiveness of potent antiretroviral therapy on time to AIDS and death in men with known HIV infection duration. Multicenter AIDS Cohort Study Investigators. JAMA. 1998;280:1497–1503. doi: 10.1001/jama.280.17.1497. [DOI] [PubMed] [Google Scholar]
- 47.Lau B, Gange SJ, Moore RD. Risk of non-AIDS-related mortality may exceed risk of AIDS-related mortality among individuals enrolling into care with CD4+ counts greater than 200 cells/mm3. J Acquir Immune Defic Syndr. 2007;44:179–187. doi: 10.1097/01.qai.0000247229.68246.c5. [DOI] [PubMed] [Google Scholar]