Abstract
The enzyme organophosphorous hydrolase (OPH) catalyzes the hydrolysis of a wide variety of organophosphorous compounds with high catalytic efficiency and broad substrate specificity. The immobilization of OPH in functionalized mesoporous silica (FMS) surfaces increases significantly its catalytic specific activity compared to the enzyme in solution with important applications for the detection and decontamination of insecticides and chemical warfare agents. Experimental measurements of immobilization efficiency as function of the charge and coverage percentage of different functional groups have been interpreted as electrostatic forces being the predominant interactions underlying the adsorption of OPH onto FMS surfaces. Explicit solvent molecular dynamics simulations have been performed for OPH in bulk solution and adsorbed onto two distinct interaction potential models of the FMS functional groups in order to investigate the relative contributions of non-bonded interactions to the conformational dynamics and adsorption of the protein. Our results support the conclusion that electrostatic interactions are responsible for the binding of OPH to the FMS surface. However, these results also show that van der Waals forces are detrimental for interfacial adhesion. In addition, it is found that OPH adsorption onto the FMS models favors a protein conformation whose active site is fully accessible to the substrate in contrast to the unconfined protein.
Keywords: molecular dynamics simulations, bacterial phosphotriesterase, conformational changes, confined environments, coarse-grain and atomistic models, silanol molecular model
Introduction
The development of new materials has enabled the use of porous solid support for protein immobilization via encapsulation, entrapment, and covalent linking with useful applications in biotechnology, biocatalysis, protein-delivery systems and sensing.1–8 Immobilization on a solid support can enhance enzyme stability while maintaining near-native activity and selectivity.9,10 A range of supports has been described for proteins, each having associated advantages and disadvantages for use in protein biocatalysis.11 Functionalized mesoporous silica (FMS) support has attracted considerable interest as enzyme-immobilization matrix due to the large, rigid and uniform mesopore structure and favorable interaction environment provided by tunable functional groups.11–14 These features allow a much higher protein loading densities and enhanced enzyme activity and stability compared conventional mesoporous silica supports.
It has been previously reported by Lei and co-workers that the immobilization of the metalloenzyme organophosphorous hydrolase (OPH) in FMS enhances its stability and increases its catalytic specific activity by 200% compared to the enzyme in solution.15 Immobilization of other enzymes into FMS supports corroborates these findings and further demonstrates that FMS with appropriated pore sizes and functional groups could result in faster kinetics, greatly enhanced enzymatic activity and stability for target enzymes compared with conventional approaches of enzyme immobilization.16,17 The enzyme organophosphorous hydrolase (OPH; EC 3.1.8.1) from the soil bacteria Brevundimonas diminuta (previously Pseudomonas diminuta) catalyses the hydrolysis of a wide variety of phosphotriester (P-O), phosphonothioate (P-S), phosphofluoridate (P-F) and phosphonocyanate (P-CN) compounds with high catalytic efficiency and broad substrate specificity.18–24 Although the biological role of OPH remains unclear, it is thought to have evolved in the last few decades since phosphotriesters are not naturally occurring compounds and were first synthesized during the Second World War.25,26 Hydrolysis of its most efficient substrate, paraoxon, is remarkably efficient with kcat and kcat/KM values for paraoxon (104 s−1 and 107 s−1 M−1, respectively) that approach the limit of diffusion of the reactants in solution.18 Modifications of the active site were shown to alter the substrate specificity and enhance the catalytic efficiency for slower substrates.19,20,27,28 Variants of OPH have also been shown to be stereoselective for the hydrolysis of chiral organophosphate triesters.29 This is a highly desirable catalyst feature since major chemical warfare agents are racemic mixtures with substantial differences in toxicity of the individual enantiomers. Therefore, the immobilization of OPH in FMS support offers great potential for the development of biosensors for bioremediation and detoxification of toxic pesticides and nerve agents such as soman, sarin and VX.30,31
The maximum amount of enzyme adsorbed on a mesoporous material varies with pH value, ionic strength, pore diameter, pore volume, surface characteristics, isoelectric point (pI), morphology, and particle size.32–34 Protein adsorption can be manipulated via different types of interactions: electrostatic forces, hydrogen bonding, van der Waals forces, conformational entropy, and hydrophobic interactions.10,11 Moreover, surface adsorption can be increased by the gain of conformational entropy upon unfolding at the surface, shift of pH close to the isoelectric point, or/and specific ion binding.35 In addition, the nature of the solid surface, including its hydrophobicity and charge density, has a strong effect on the structure and conformation of the protein layer.1,10,11 Surface modification of mesoporous silica with functional groups that are compatible to those found in the surface of proteins can enhance these interactions and influence the final loading and activity of the immobilized protein.36,37 Hydrophobic and ionic interactions are commonly the most important forces driving protein adsorption to mineral surfaces, but the relative significance of these interactions in a given system depends on the details of the protein structure and the particular surface involved.12,13,34,38–42 However, it is difficult to assess the exact contribution of the different interfacial interactions to the adsorption of proteins onto the FMS surface at the microscopic level. This is in part because of the amorphous nature of the SiO2 walls, which makes it very difficult to extract structural information at atomic resolution from experimental data only.8,12,14 Yet, the molecular characterization of such interfacial interactions is an essential step for the rational design of FMS surface properties that will lead to the efficient immobilization of enzymes for environmental and industrial applications.
It has been shown that the adsorption of OPH (pI = 8.3) to FMS functionalized with negative -CH2-CH2-COO− groups at low coverage (2%) and pH 7.5 results in optimal affinity and immobilization efficiency whereas its adsorption to FMS functionalized with positive -CH2-CH2-NH3+ groups at high coverage (20%) leads to significantly lower immobilization efficiency.15 OPH was also shown to adsorb to unfunctionalized mesoporous silica (UMS) whose silanol surface is negatively charged, although the entrapped enzyme exhibited low specific enzymatic activity.15 From these results, electrostatic interactions have been proposed to be the predominant forces governing the adsorption of OPH onto FMS surfaces. However, such hypothesis is not fully compatible with the observation that OPH adsorbs to FMS functionalized with a low coverage (2%) of positively charged -CH2-CH2-NH3+ groups whereas an increase in the coverage of negatively charged CH2-CH2-COO− groups from 2% to 20% leads the a sharp decrease of immobilization efficiency.15 These results appear to indicate that other interactions play also an important role modulating OPH adsorption to FMS surfaces. Indeed, previous immobilization studies for different enzymes have suggested that hydrophobic interactions play a more important role in protein-FMS surface adsorption than it has been assumed.13,36,43 For instance, it was demonstrated that the immobilization of penicillin acylase (pI = 7.0) onto mesoporous silica surfaces (SBA-15) functionalized with the hydrophobic groups phenyltrimethoxysilane and vinyltriethoxysilane resulted in enhanced adsorption rates and adsorption capacity compared to the pure SBA-15 which is negatively charged.36
In this context, molecular dynamics (MD) simulations can provide a microscopic level characterization of the role of different interactions in the adsorption of proteins to FMS surfaces.44–46 As a basis for the understanding of the nature of these interactions in the adsorption and confinement of OPH into FMS support, simulations have been performed for the enzyme OPH free in solution as well as adsorbed to two coarse-grain models of the FMS functional carboxyl groups. In one model, van der Waals interactions are made negligible by representing the functional group as an oxyanion atom. In the second model, the van der Waals parameters for the ethyl acetate functional groups were used to fully account for these contributions. In order to reproduce the spatial arrangement of functional groups on the silicate wall, the FMS surface is represented as a cylinder whose curvature and functional group coverage was estimated from experimental measurements.47 Each sphere representing a functional group is spaced within 1 nm from each other and a ratio protein/functional groups of 1:57. The effect of the different representations of the FMS surface on the structural dynamics and interfacial diffusion properties of OPH is investigated. These properties are characterized through time-dependent analysis of atom-positional root-mean-square deviation (RMSD) and root-mean-square atom-positional fluctuations (RMSF) from the X-ray structure of OPH, mean-square displacement (MSD) and inter-atomic distances of OPH with respect to the FMS surfaces, solvent-accessible surface and secondary structure content. Principal component analysis of the MD simulations have been undertaken to investigate the mechanisms by which FMS confinement steers local and global motions of OPH towards conformations of enhanced stability and catalytic competence.
Computational Methodology
OPH Model
High-resolution crystallographic structures of OPH from Brevundimonas diminuta were used as initial coordinates in the MD simulations (PDB ID 1EZ2 and 1HZY).48,49 The substrate analogue diisopropylmethyl phosphonate was removed from the X-ray structure 1EZ2 removed prior to simulations. Atom positional root-mean-square deviation (RMSD) between the two crystallographic structures was 0.02 nm and demonstrates that the two conformations are nearly indistinguishable. Missing atoms in the crystallographic structures were verified and added if necessary with the WHAT IF web server.50 Atoms additions were necessary only to the terminal residues. Protonation states were assigned accordingly to pKa calculations with the program propKa 2.0.51 All simulations were carried out using the GROMOS force field force parameter set 53A6.52,53 The active site of OPH contains two zinc ions bridged by a hydroxide anion and a carbamylated lysine. The two zinc and hydroxide ions were treated as a non-bonded model. The entire subsystem presents a formal charge of +3 e. Charges for the hydroxide ion and carbamylated lysine were assigned via a restrained hyperbolic fit of the electrostatic potential (RESP)54 on the nuclei positions of each atom after geometry optimizations at the HF/6-31G* level using the NWCHEM software55 as described in ref.56 This procedure has been shown to be fully compatible with the parameter set 53A6 of the GROMOS force field.57–59
FMS Model
The interactions between OPH and the FMS surface have two components: steric due to confinement nature of the mesoporous surface, and non-bonded due to the functionalization of the mesopore. To study the confinement we assume that these interactions can be approximated by a coarse representation modeled as a cylindrical, uniform array of particles, each corresponding to a given functional group and spaced apart to represent the percentage of functional group coverage.15,47 In this coarse-grain model each particle incorporates the atomic attributes (van der Waals and Coulomb parameters) characteristic of the given functional group derived from a molecular mechanics force field. The coarse-grain representation of the functional groups allows for a decrease of the degrees of freedom of the system, and consequently longer sampling of the protein configurational space. In order to evaluate the relative contributions of electrostatics and vdW interactions to the adsorption of the OPH onto the FMS, two models were developed to represent the negative functional groups commonly used in experimental studies of OPH immobilization onto FMS. In the model FMScoul the pore surface was represented by point-charged particles with charge −1 e and vdW parameters corresponding to an oxyanion. In the model FMSvdW, the pore surface was represented by particles with charge −1 e and vdW parameters corresponding to acetate groups from the Martini force field atom type P3.60 Structure-based characterization of the electrostatic potential surface of OPH demonstrates that the surface opposed to the active site exhibit a very positive electrostatic potential whereas the surface containing the entrance to the active site exhibit a mostly negative electrostatic potential.17 From the charge distribution along the OPH surface, it has been proposed that the positively charged surface of OPH is attracted via electrostatic interactions to the negatively charged surface of FMS, leaving the entrance to the active sites exposed to the lumen of the mesoporous and freely accessible to substrates and products.17 In the present work, the OPH structure was docked to the FMS pore wall based on the complementarity of their electrostatic potential surfaces calculated with the program APBS as previously described in ref.17 The system is illustrated in Figure 1 and described in Table 1.
Figure 1.
Cartoon representation of the organophosphorous hydrolase structure adsorbed onto the functionalized mesoporous silica surface. (A) Top view of the OPH dimer. Regions of large atomic fluctuations in the MD simulations are indicated in violet (L1, residues 155-165), blue (L2, residues 172-182), yellow (L3, residues 201-215), orange (L4, residues 230-240), purple (L5, residues 255-275) and green (L6, residues 306-325) for only one monomer. (B) Close-up view of the active site pocket with residues represented in licorice and Zn2+ cations in the Corey–Pauling–Koltun (CPK) model (yellow). The residue Y309 in the gateway to the active site pocket is highlighted. (C) Side view of the OPH dimer. Positively charged residues in the protein that interact with the functionalized mesoporous silica are represented in licorice. The functionalized mesoporous silica is represented in CPK (red). The explicit model water molecules are omitted for clarity of visualization.
Table 1.
Description of the simulated systems.
| system | solute | number of molecules |
total atoms | dimension (nm) | ||
|---|---|---|---|---|---|---|
| water | ions | FMS | ||||
| OPHfree | 6,472 | 27,765 | 0 | 0 | 89,769 | 0.8 × 0.90 × 13.0 |
| FMScoul | 6,444 | 51,611 | 166 | 168 | 161,611 | 15.0 × 14.0 × 0.8 |
| FMSvdW | 6,444 | 51,611 | 166 | 168 | 161,611 | 15.0 × 14.0 × 0.8 |
Simulation Setup
The systems were placed in a rectangular box, treated for periodic boundary conditions and solvated with explicit SPC model water molecules.61 The systems were neutralized with Na+ counter ions where necessary. Simulations were carried out in the NPT ensemble and a time step of 2 fs was used to integrate the equations of motion based on the Leap-Frog algorithm.62 The temperature of the solute and solvent were separately coupled to the velocity-rescale thermostat at 298.15 K with a relaxation time of 0.1 ps. The pressure was maintained as 1 atm by isotropic coordinate scaling with a relaxation of 1 ps. The bond lengths and angles were constrained by using the P-LINCS algorithm63 and the geometry of the water molecules was constrained using the SETTLE algorithm.64 A twin-range cutoff of 1.0 and 1.2 nm was used for vdW interactions, and long-range electrostatic interactions were treated by the Particle Mesh Ewald method65. The systems were initially minimized through 20.000 iterations of the steepest descent algorithm. Solvent molecules were relaxed during 500 ps at 298.15 K with positional restraints applied to the heavy atoms of the protein. The full system was equilibrated for 10 ns followed by the production phase of 40 ns. Configurations of the system were recorded as trajectory files at every 1.0 ps. The software package GROMACS v.4.04 was used for the simulations and property analyses compiled in double precision.66 Protein structures were visualized with the software VMD 1.86.67 The interface between monomers was computed using our in-house software SurfinMD. Calculated properties such as atom positional root-mean-square deviation (RMSD) and fluctuation (RMSF), principal component analysis (PCA), time-dependent secondary structure pattern, solvent number density, radial distribution function, and solvent accessible surface area (SASA) were computed using the implemented algorithms in the GROMACS package.66
Results
Structural characterization of OPH in bulk water and adsorbed onto FMS surfaces
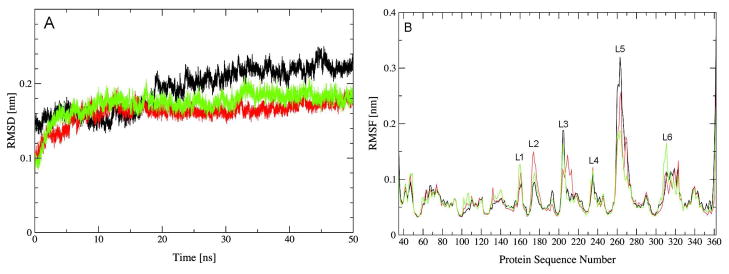
Atom positional root-mean-square deviation (RMSD) was calculated from the MD simulations OPHfree, FMSCoul and FMSvdW with respect to the substrate bound conformation of the X-ray structure (PDB ID 1EZ2 and 1HZY)48,49 (Figure 2). The RMSD from the three simulations converged to a plateau of 0.15 nm during the first 15 ns of simulations, indicating that OPH initially adopted a similar conformation when in bulk water or adsorbed to the FMS surface. These RMSD values remain fairly stable for the FMSCoul and FMSvdW ensembles during the remaining of simulation time. Conversely, the RMSD values from the OPHfree ensemble increases further before converging to 0.2 nm after 20 ns of simulation. Therefore, the OPH average conformation deviates more from the substrate-bound conformation in the X-ray structure on bulk water than adsorbed to FMS surfaces. The atom positional root-mean-square fluctuations (RMSF) from the three simulations exhibit similar profiles (Figure 2). The regions of highest atomic fluctuations are the same for OPHfree, FMSCoul and FMSvdW, and they correspond to loop motifs. The loops L3 (residues 201-215), L5 (residues 255-275) and L6 (residue 305-325) are situated at the entrance of the active site whereas loop L1 (residues 155-165) is located at both monomer-monomer and protein-FMS interfaces (Figure 2). Loop L2 (residues 172-182) is also at the monomer-monomer interface, but on a region that is not in contact with the FMS surface. Several trends can be observed when comparing the atom positional RMSF from the three simulations. First, FMS-adsorbed OPH exhibits smaller atomic fluctuations in loops L3 and L5, which together delimit the entrance of the active site. Second, it also displays larger atomic fluctuations for residues in the monomer-monomer interface (residues 100-110, 130-140, loops L1 and L2) compared to OPH in bulk water. Since loop L2 is not close to the FMS surface, the increased atomic displacement of this region suggests that interfacial interactions have a long-range influence on the structural dynamics OPH. Lastly, RMSF values are particularly large for residues S308, Y309 and V310 in the FMSvdW simulation. Previous computational simulations of wild type and mutant variants of OPH bound to paraoxon, sarin and soman have also reported an increased atomic displacement of Y309.56,68 These simulations shown that the displacement of Y309 leads to the opening/closing of a gateway to the active site pocket and it has been proposed to assist the exit of the leaving group from the hydrophobic pocket.68 However, experimental studies of the mutation Y309F failed to find any significant difference in the magnitude of either kcat or kcat/KM when compared to the wild-type enzyme for the hydrolysis of paraoxon or diethyl p-chlorophenyl phosphate.69 No significant changes in the secondary structure content were observed in the MD simulations (Table 2). This is in agreement with Fourier transform infrared spectroscopy experiments for OPH entrapped in FMS.12 Changes in structural dynamics were more significant in the monomer-monomer interface whose contact area decreases significantly in the FMSvdW ensemble while exhibiting an identical profile in the OPHfree and FMSCoul simulations (Table 2).
Figure 2.
Root-mean-square deviation (RMSD) of backbone atoms of OPH from the X-ray structure (1HYZ) (A) and root-mean-square fluctuation (RMSF) of Cα atoms of OPH as function of residue sequence number (B). OPHfree (black), FMScoul (red) and FMSvdW (green). Rotational and translational fitting of pairs of structures was applied using all backbone atoms. RMSF is averaged for the two monomers over the final 40 ns and a time window of 10 ps.
Table 2.
Averaged structural properties for OPH in the MD simulations.
| system | secondary structure [%] | SASA [nm2] | diffusion coefficients [cm2s−1] | |||
|---|---|---|---|---|---|---|
| α-helix | β-strand | turn+coil | total | monomer interface | estimated | |
| OPHfree | 45.0 | 12.6 | 33.1 | 455.2 | 140.7 | 3.69 × 10−7 |
| FMScoul | 45.0 | 13.1 | 32.3 | 448.8 | 138.5 | 2.93 ×10−7 |
| FMSvdW | 44.8 | 13.4 | 32.0 | 439.8 | 118.6 | 1.3 × 10−8 |
| experimentala | ||||||
| lysozyme | - | - | - | - | - | ~10−7/~10−11 |
Conformational dynamics of OPH in bulk water and adsorbed onto FMS surfaces
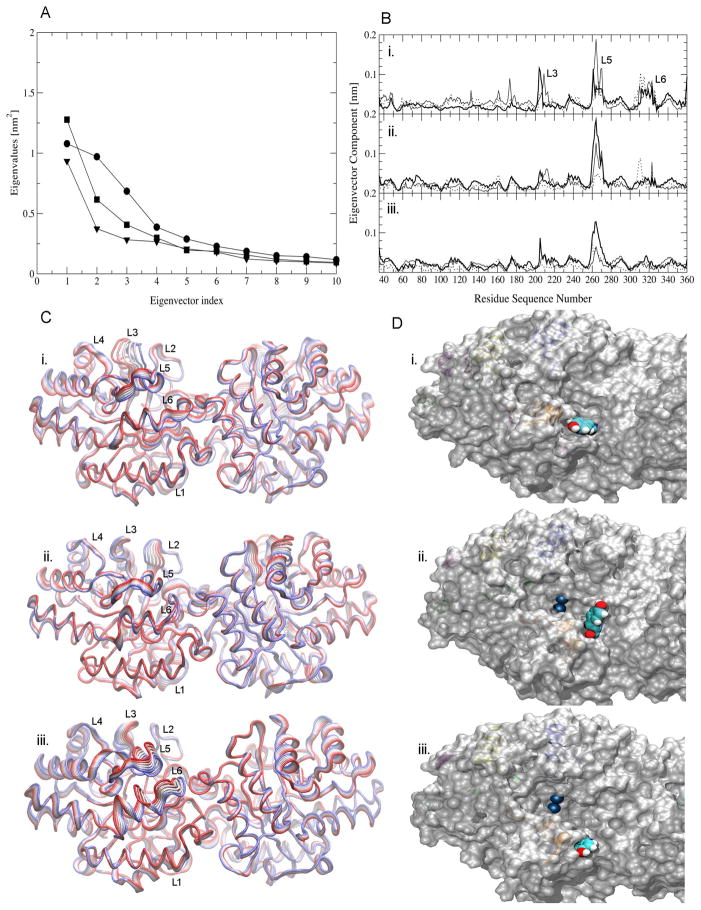
PCA of the atom positional displacement from the corresponding average structure was performed for OPH in the three MD simulations.70,71 This mathematical treatment enables the separation of low-frequency motions that typically determine the kinetics of enzymatic activity from the much larger number of remaining high-frequency motions. The separation in low- and high-frequency components is obtained via a change of coordinate system from Cartesian to eigenvectors of the covariance matrix, which allows the extrapolation of motions in the direction of selected eigenvectors.70,71 PCA of a molecular dynamics simulation trajectory does not rest on the assumption of a harmonic potential and therefore modes are usually sorted according to variance rather than frequency. Nevertheless, the largest-amplitude modes of a PCA usually also represent the slowest dynamical transitions. In this study, PCA is used to describe low frequency, persistent motions of OPH during the MD simulations. The eigenvalue amplitudes calculated from the MD trajectories indicate that only a few modes are required to account for most of the large-amplitude motions of OPH in these ensembles (Figure 3). The first three eigenvalues account for 40%, 34% and 24% of the total low frequency motions in OPHfree, FMSCoul and FMSvdW, respectively. Projection of the corresponding eigenvectors onto the protein sequence reveals that residues in the loops L3, L5 and/or L6 are the primary contributors to the motions (Figure 3). In the OPHfree and FMSCoul ensembles, the regions of highest atomic displacement are confined to loops L3 and L5 in the entrance of the active site. These loops, however, do not contribute significantly to the low frequency motions in the FMSvdW ensemble, where loop L6 exhibits the largest atomic displacement. The persistence of large displacements of residues in loop L3 along the three eigenvectors in the OPHfree simulation indicates that the loop moves in a more disordered way than in the FMSCoul or most notably in the FMSvdW simulations (Figure 3). In the latter simulation, loops L3 and L5 appear to adopt one predominant conformation in which the active site is more exposed to the solvent and presumably to OPH substrates (Figure 3).
Figure 3.
Eigenvalues and atomic displacement along the most representative eigenvectors calculated from the MD simulations OPHfree (circle or bold line), FMScoul (square or solid line) and FMSvdW (triangle or dashed line). (A) Largest eigenvalues. (B) Atomic displacement along the first (i.), second (ii.) and third (iii.) eigenvectors. (C) Projection of atomic displacements along the first eigenvector onto the three-dimensional structure of (i.) OPHfree, (ii.) FMScoul and (iii.) FMSvdW. The width of the ribbons illustrates the amplitude of the atomic displacement. Only eigenvector components larger than 0.05 ps are shown for clarity. (D) Solvent accessibility surface of the trajectory frame at 50 ns for OPH in (i.) OPHfree, (ii.) FMScoul and (iii.) FMSvdW. The residue Y309 and Zn2+ cations are represented in CPK.
Surface-protein interfacial interactions
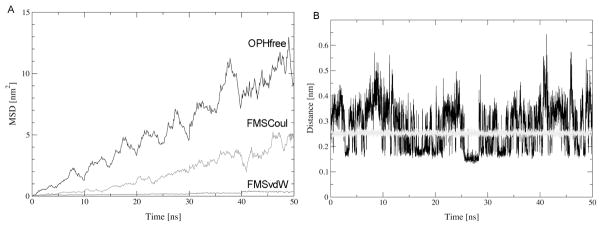
The mean square displacement (MSD) of all protein atoms from their initial position for OPH in bulk water and in the different FMS surface models was calculated from the MD trajectories (Figure 4). These MSD values do not exhibit a linear dependence of time, indicating that this property is not fully converged after 50 ns of simulation. Nevertheless, the qualitative inference of these values is still useful for the proposal of comparing the relative differences in diffusion behavior of OPH in bulk solution and adsorbed to different representations of the FMS surface at the microscopic level. The MSD curves differ significantly among the simulations (Figure 4). In the presence of the FMS surface, OPH exhibits predominantly a lateral diffusion along the surface plane (axes x-y) whereas in bulk water, the protein diffuses randomly along all the Cartesian axes. The MSD of OPH along the FMSCoul surface was found to be larger than along the FMSvdW surface. Diffusion coefficients calculated from the MSD of the protein in OPHfree, FMSCoul and FMSvdW are 3.69 × 10−7 cm2s−1, 2.93 × 10−7 cm2s−1 and 1.3 × 10−8 cm2s−1, respectively (Table 2). Experimentally determined diffusion coefficients for OPH in bulk water or adsorbed to inorganic surfaces are not available. However, diffusion coefficients estimated for the enzyme lysozyme adsorbed onto oxidized silica and glass surfaces are more than three orders of magnitude smaller than those in bulk solution, i.e. ~10−7 cm2s−1.72,73 Lysozyme is quite distinct from OPH with respect to size (163 a.a. versus 662 a.a.), pI (pH 11 versus pH 8.3) and conformational flexibility (lysozyme is stabilized by intramolecular disulfide bridges). Therefore, the diffusion coefficients for the two proteins adsorbed to silicate surfaces are expected to differ in absolute value. However, it can be reasoned from the diffusion behavior of lysozyme that proteins generally exhibit much slower diffusion when adsorbed than in bulk water. The neglect of van der Waals interactions in the FMSCoul surface model results in a diffusion coefficient for OPH of the same order of magnitude of those in bulk water.
Figure 4.
Mean-square displacement of OPH atoms from their initial positions over 50 ns of simulation (A) and minimum atom-atom distances between OPH atoms and the FMS surface (B) in simulations FMScoul (black) and FMSvdW (gray).
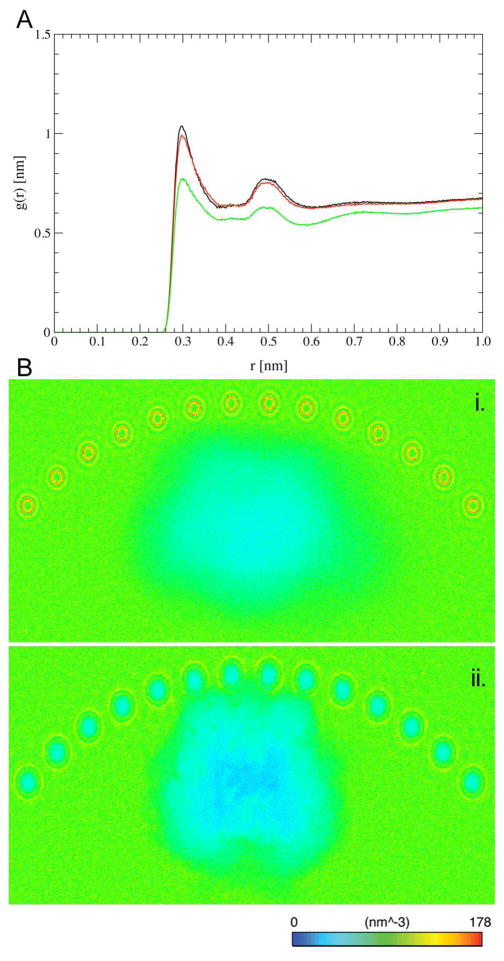
The difference in the diffusional behavior of OPH in the FMSCoul and FMSvdW simulations is further reflected in the average FMS-OPH interfacial inter-atomic distances. Comparison of the minimum distance between any pair of atoms in OPH and in the FMS surface displays a remarkably distinct behavior in the two systems (Figure 4). The minimum interfacial atom-atom distance is nearly invariable at 0.25 nm for the FMSvdW ensemble whereas it fluctuates between 0.09 nm and 0.64 nm in the FMSCoul ensemble within a 50-ns timescale. These findings indicate that although only electrostatic interactions suffice for the binding of OPH to the FMS surface, van der Waals interactions play a major role in its adhesion. The different OPH-FMS binding regimes are correlated with distinguishable hydration patterns in the interfacial region between OPH and the two surface models (Figure 5). The solvent structure in the protein-FMS interface was inferred through the calculation of the number density of water molecules as well as the intermolecular radial distribution function (RDF) for atom pairs from the water molecules and the protein surface (Figure 5). The RDF of N-O atom pairs from residues on the FMS binding surface of OPH and water molecules shows that the OPH-FMS interfacial surface is less hydrated in the FMSvdW model. Projection of the solvent number density onto the three-dimensional structure of the solute further demonstrates that the differences in hydration are restricted to the protein-FMS interface region. Within a cutoff of 0.45 nm, there are a total of 139, 144 and 122 water molecules coordinated to side-chain nitrogen atoms on the FMS-protein interface in the OPHfree, FMSCoul and FMSvdW simulations, respectively. Hence, local hydration patterns of OPH adsorbed to the FMSCoul surface model and in bulk solvent are comparable, though distinct from that observed for the FMSvdW surface model.
Figure 5.
(A) Radial distribution function g(r) of intermolecular N-O pairs in the side-chains of residues at the FMS interface of OPH and in the water molecules and (B) the average solvent number density around OPH in the FMScoul (a) and FMSvdW (b) ensembles. Solvent density representation along the longitudinal y-axis. OPHfree (black), FMScoul (red) and FMSvdW (green).
Discussion
The representation of FMS surfaces by the means of coarse-grain models with distinct interaction potentials can provide valuable insight into the effect of non-bonded interactions with the FMS surface on protein dynamics. In this approach, interactions between OPH and the FMS surface can be generically grouped in two components: steric due to the confining nature of the mesoporous surface, and electrostatic/van der Waals due to the functionalization of the mesopore. It is found that the steric contact to the FMS surface as well as the nature of the interfacial interactions modulate the conformational dynamics of OPH towards different ensembles of structures. The steric effect imposed by the curvature of the FMS surface induces mainly local structural rearrangements whereas the interaction potentials influence local and non-local structural rearrangements. The local rearrangements are predominantly confined to loops in the entrance of the active site (Figure 4) that become more accessible to the solvent and presumably to substrates. The rate-determining step for second-order catalytic rate of enzymes with extremely efficient reaction rates (1010 M−1s−1 to 108M−1s−1) corresponds to the association of the free enzyme and the free substrate. OPH hydrolyses paraoxon at nearly diffusion-controlled rates (~107 M−1s−1),74 although it has been demonstrated that the rate-limiting step of the reaction changes from chemical to physical (diffusional) events as the pKa of the leaving group of a series of paraoxon analogues is decreased.18 It has been argued that the slower reaction rate of OPH compared to super-efficient enzymes may be due to a conformational rearrangement upon substrate binding.18 Indeed, our findings suggest that FMS-confined OPH adopts a conformational state that may facilitate substrate binding compared to OPH free in solution.
Local structural rearrangements are also influenced by the nature of the OPH-FMS interfacial interactions as evidenced by the distinct atomic displacement patterns of eigenvectors projection along the protein sequence in the MD ensembles (Figure 4). The regions of largest atomic displacement correspond to loops L3 and L5 in the OPHfree and FMSCoul ensembles and loop L6 in the FMSvdW ensemble. In the latter, loops L3 and L5 are stabilized in one predominant conformation where the active site is more exposed to the solvent. Residues S308, Y309 and V310 are the major contributors to the atomic displacement of the loop L6, leading to the opening/closing of the gateway to the active site pocket as previously proposed in refs.56,68 The different potential interactions describing the FMS surface affect also the structural dynamics of the quaternary structure of OPH. The strong protein-surface adhesion in the FMSvdW ensemble results in the rearrangement of the two monomers with respect to each other which produces a decrease of 20% and 18% in the SASA of the dimer interface with respect to the OPHfree and FMSCoul ensembles, respectively (Table 2). These rearrangements of quaternary structure were not found to produce any significant change in the secondary structure content of OPH, in agreement with Fourier transform infrared spectroscopy measurements for OPH entrapped in FMS.12
The role of non-bonded interactions in the adsorption of OPH to FMS surfaces has also been investigated. Although electrostatic interactions suffice for the binding of OPH to the FMS surface, the neglect of van der Waals contributions in the representation of the FMS pore model leads to a significant decrease in protein-surface adhesion and increase in desorption events (Figure 4). Diffusion coefficients calculated from the FMSCoul and OPHfree simulations are one order of magnitude faster than that in the FMSvdW simulation (Table 2). The latter is more consistent with experimental measurements of diffusion coefficients for lysozyme adsorbed onto oxidized silica and glass surfaces which are more than three orders of magnitude smaller than those for the protein in bulk solution, i.e. ~10−7 cm2 s−1.72,73 The different adhesion regimes of the two surface models are correlated with distinguishable hydration patterns in the interfacial region between OPH and FMS surface (Figure 5). The protein-FMS interface is significantly less hydrated in the FMSvdW ensemble than in the OPHfree, FMSCoul ones whose hydration patterns are comparable. The binding of OPH to the FMS surface in aqueous solution involves a tradeoff between the unfavorable desolvation penalty due to the removal of favorable interactions with water molecules, and the generally favorable intermolecular interactions made in the bound state. The comparatively lower hydration of the FMSvdW surface model implies a smaller desolvation penalty to overcome during the protein adsorption process. This makes protein adhesion to the FMSvdW surface stronger than to the FMSCoul surface.
It has been shown that OPH is optimally immobilized onto FMS surfaces functionalized with negatively charged groups and low coverage (2%).15 In consonance with that, OPH immobilization efficiency is very low for FMS surfaces functionalized with positively charged groups at high coverage (20%).15 Based on these findings, a predominantly electrostatic-driven mechanism has been proposed to underlie the immobilization of OPH onto functionalized FMS surfaces. However, such mechanism is not entirely reconcilable with the observation that under low surface coverage by positively charged groups, OPH is also immobilized onto FMS surfaces with significant efficiency, and that high surface coverage by negatively charged groups causes a sharp decrease in the immobilization efficiency of the system. 15 Our simulations of OPH adsorbed onto FMS surface models described by distinct interaction potentials show that interfacial adhesion is significantly enhanced by an increase in the van der Waals contributions and support an alternative interpretation of the aforementioned macroscopic measurements. Therefore, we propose that the adsorption of OPH onto FMS surfaces functionalized with either negatively or positively charged groups is modulated not only by attraction between opposite charges but also from an increase in surface hydrophobicity due to the presence of methylene groups linking the functional groups to the mesoporous wall. In summary, while charge-charge interactions are detrimental to the binding of OPH to the FMS surface, a balance between hydrophobic and electrostatic interactions appears necessary to facilitate the desolvation of the FMS functional groups and OPH adhesion to the surface.
Conclusion
The successful design of mesoporous support for protein immobilization requires a detailed understanding of the conformational and adsorption properties of the target protein.10,11 It can provide important insights into the enhancement of protein loading, and specific activity as well as into the modulation of the hydrophobicity and/or amphiphilicity of the support for effective substrate diffusion through the pores for instance. Explicit solvent MD simulations have been performed for the enzyme OPH free in solution as well as adsorbed onto two models of the FMS functional carboxyl groups to investigate the role of non-bonded interactions in the conformational dynamics and adsorption of the protein. In the FMSCoul model, van der Waals interactions were made negligible by representing the functional group by interaction potentials corresponding to an oxyanion whereas in the FMSvdW model, van der Waals interactions were accounted for by interaction potentials representative of an acetate group. It has been found that the steric contact to the FMS surface as well as the nature of the interfacial interactions modulate the structural dynamics of OPH towards different ensembles of structures. Structural rearrangements of loops in the entrance of the active site increase its accessibility to the solvent and presumably to substrates, and could facilitate productive binding between substrate and enzyme. In fact, the hydrolysis rate of paraoxon by OPH appears to be limited by chemical events among which substrate-induced conformational changes as proposed by Caldwell et al.18 The different FMS surface models affect also the structural dynamics of the quaternary structure of OPH, with a significant decrease in the SASA of the dimer interface for the FMSvdW ensembles (Table 2). These rearrangements of quaternary structure were not found to produce any significant change in the secondary structure content of OPH, in agreement with Fourier transform infrared spectroscopy measurements for OPH entrapped in FMS.12 It has also been found that although electrostatic interactions sufficed for the binding of OPH to the FMS surface, the neglect of van der Waals contributions in the representation of the FMS pore model led to a significant decrease in protein-surface adhesion and increase in desorption events. The comparatively lower hydration of the FMSvdW surface model implies a smaller desolvation penalty to overcome during the protein adsorption process and stronger protein adhesion to the FMSvdW compared to the FMSCoul surface models. Furthermore, the diffusion coefficient calculated from the FMSvdW simulation is consistent with experimental measurements of diffusion for lysozyme adsorbed onto oxidized silica and glass surfaces.73 In addition, when fully adsorbed onto the FMSvdW surface model, OPH displays a conformational dynamics in which the active site of the enzyme is unobstructed by loop L6. This finding suggests that the observed enhanced catalytic activity of FMS-confined OPH may sprawl from a combined effect of confinement and the adoption of a conformation induced upon adsorption. It has also been argued, with basis on the presented simulations, that the adsorption of OPH onto FMS surfaces + functionalized with either CH2-CH2-COO− or CH2-CH2-NH 3 groups is modulated not only by attraction between opposite charges but also from an increase in surface hydrophobicity due to the presence of methylene (-CH2) groups linking the functional groups to the mesoporous.
Acknowledgments
This work was supported by the NIH National Institute of General Medical Sciences (grant number R01GM080987), the Laboratory Directed Research & Development program “Modeling Protein-Nanomaterial Interactions” funded by the Pacific Northwest national Laboratory and the Brazilian National Council for Research and Development (CNPq). Computational resources were provided by the Environmental Molecular Sciences Laboratory at Pacific Northwest National Laboratory. Pacific Northwest National Laboratory is operated for DOE by Battelle.
References
- 1.Kane RS, Stroock AD. Biotechnol Progr. 2007;23:316. doi: 10.1021/bp060388n. [DOI] [PubMed] [Google Scholar]
- 2.Rusmini F, Zhong ZY, Feijen J. Biomacromolecules. 2007;8:1775. doi: 10.1021/bm061197b. [DOI] [PubMed] [Google Scholar]
- 3.Penn SG, He L, Natan MJ. Curr Opin Chem Biol. 2003;7:609. doi: 10.1016/j.cbpa.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 4.Zhao YN, Trewyn BG, Slowing II, Lin VSY. J Am Chem Soc. 2009;131:8398. doi: 10.1021/ja901831u. [DOI] [PubMed] [Google Scholar]
- 5.Slowing II, Trewyn BG, Lin VSY. J Am Chem Soc. 2007;129:8845. doi: 10.1021/ja0719780. [DOI] [PubMed] [Google Scholar]
- 6.Trewyn BG, Giri S, Slowing II, Lin VSY. Chem Commun. 2007:3236. doi: 10.1039/b701744h. [DOI] [PubMed] [Google Scholar]
- 7.Slowing II, Trewyn BG, Giri S, Lin VSY. Adv Funct Mater. 2007;17:1225. [Google Scholar]
- 8.Weitkamp J, Hunger M, Rymsa U. Micropor Mesopor Mat. 2001;48:255. [Google Scholar]
- 9.Eggers DK, Valentine JS. Protein Science. 2001;10:250. doi: 10.1110/ps.36201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanefeld U, Gardossi L, Magner E. Chem Soc Rev. 2009;38:453. doi: 10.1039/b711564b. [DOI] [PubMed] [Google Scholar]
- 11.Hudson S, Cooney J, Magner E. Angew Chem Int Edit. 2008;47:8582. doi: 10.1002/anie.200705238. [DOI] [PubMed] [Google Scholar]
- 12.Lei CH, Shin Y, Magnuson JK, Fryxell G, Lasure LL, Elliott DC, Liu J, Ackerman EJ. Nanotechnology. 2006;17:5531. doi: 10.1088/0957-4484/17/22/001. [DOI] [PubMed] [Google Scholar]
- 13.Deere J, Magner E, Wall JG, Hodnett BK. J Phys Chem B. 2002;106:7340. [Google Scholar]
- 14.Liu J, Shin Y, Nie ZM, Chang JH, Wang LQ, Fryxell GE, Samuels WD, Exarhos GJ. J Phys Chem A. 2000;104:8328. [PubMed] [Google Scholar]
- 15.Lei C, Shin Y, Liu J, Ackerman EJ. J Am Chem Soc. 2002;124:11242. doi: 10.1021/ja026855o. [DOI] [PubMed] [Google Scholar]
- 16.Lei C, Shin Y, Liu J, Ackerman EJ. Nano Lett. 2007;7:1050. doi: 10.1021/nl070255b. [DOI] [PubMed] [Google Scholar]
- 17.Lei C, Soares TA, Shin Y, Liu J, EJEJA Nanotechnology. 2008;19:125102. doi: 10.1088/0957-4484/19/12/125102. [DOI] [PubMed] [Google Scholar]
- 18.Caldwell SR, Newcomb JR, Schlechtand KA, Raushel FM. Biochemistry-Us. 1991;30:7438. doi: 10.1021/bi00244a010. [DOI] [PubMed] [Google Scholar]
- 19.Chen-Goodspeed M, Sogorb MA, Wu FY, Hong SB, Raushel FM. Biochemistry-Us. 2001;40:1325. doi: 10.1021/bi001548l. [DOI] [PubMed] [Google Scholar]
- 20.Chen-Goodspeed M, Sogorb MA, Wu FY, Raushel FM. Biochemistry-Us. 2001;40:1332. doi: 10.1021/bi001549d. [DOI] [PubMed] [Google Scholar]
- 21.Gopal S, Rastogi V, Ashman W, Mulbry W. Biochem Biophys Res Comm. 2000;279:516. doi: 10.1006/bbrc.2000.4004. [DOI] [PubMed] [Google Scholar]
- 22.Li WA, Aubert SD, Raushel FM. J Am Chem Soc. 2003;125:7526. doi: 10.1021/ja035625m. [DOI] [PubMed] [Google Scholar]
- 23.Li WA, Lum KT, Chen-Goodspeed M, Sogorb MA, Raushel FM. Bioorganic Medicinal Chemistry. 2001;9:2083. doi: 10.1016/s0968-0896(01)00113-4. [DOI] [PubMed] [Google Scholar]
- 24.Omburo GA, Kuo JM, Mullins LS, Raushel FM. J Biol Chem. 1992;267:13278. [PubMed] [Google Scholar]
- 25.Raushel FM. Current Opinion in Microbiology. 2002;5:288. doi: 10.1016/s1369-5274(02)00314-4. [DOI] [PubMed] [Google Scholar]
- 26.Roodveldt C, Tawfik DS. Protein Engineering Design and Selection. 2005;18:51. doi: 10.1093/protein/gzi005. [DOI] [PubMed] [Google Scholar]
- 27.Cho CMH, Mulchandani A, Chen W. Applied and Environmental Microbiology. 2004;68:2026. doi: 10.1128/AEM.68.4.2026-2030.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho CMH, Mulchandani A, Chen W. Protein Engineering Design and Selection. 2006;19:99. doi: 10.1093/protein/gzj007. [DOI] [PubMed] [Google Scholar]
- 29.Hill CM, Li WS, Thoden JB, Holden HM, Raushel FM. J Am Chem Soc. 2004;125:8990. doi: 10.1021/ja0358798. [DOI] [PubMed] [Google Scholar]
- 30.Lei CH, Valenta MM, Saripalli KP, Ackerman EJ. J Environ Qual. 2007;36:233. doi: 10.2134/jeq2006.0216. [DOI] [PubMed] [Google Scholar]
- 31.Ghanem E, Raushel FM. Toxicol Appl Pharmacol. 2005;207:459. doi: 10.1016/j.taap.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 32.Deere J, Serantoni M, Edler KJ, Hodnett BK, Wall JG, Magner E. Langmuir. 2004;20:532. doi: 10.1021/la035358y. [DOI] [PubMed] [Google Scholar]
- 33.Essa H, Magner E, Cooney J, Hodnett BK. J Mol Catal B-Enzym. 2007;49:61. [Google Scholar]
- 34.Goradia D, Cooney J, Hodnett BK, Magner E. J Mol Catal B-Enzym. 2005;32:231. [Google Scholar]
- 35.Haynes CA, Norde W. J Colloid Interf Sci. 1995;169:313. [Google Scholar]
- 36.Chong ASM, Zhao XS. Appl Surf Sci. 2004;237:398. [Google Scholar]
- 37.He J, Liu ZJ, Hai CX. Aiche J. 2008;54:2495. [Google Scholar]
- 38.Vinu A, Miyahara M, Ariga K. J Phys Chem B. 2005;109:6436. doi: 10.1021/jp050454o. [DOI] [PubMed] [Google Scholar]
- 39.Vinu A, Streb C, Murugesan V, Hartmann M. J Phys Chem B. 2003;107:8297. [Google Scholar]
- 40.Katiyar A, Ji L, Smirniotis P, Pinto NG. J Chromatogr A. 2005;1069:119. doi: 10.1016/j.chroma.2004.10.077. [DOI] [PubMed] [Google Scholar]
- 41.Katiyar A, Ji L, Smirniotis PG, Pinto NG. Micropor Mesopor Mat. 2005;80:311. [Google Scholar]
- 42.Vinu A, Murugesan V, Tangermann O, Hartmann M. Chem Mater. 2004;16:3056. [Google Scholar]
- 43.Diaz JF, Balkus KJ. J Mol Catal B-Enzym. 1996;2:115. [Google Scholar]
- 44.Adcock SA, McCammon JA. Chem Rev. 2006;106:1589. doi: 10.1021/cr040426m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Gunsteren WF, Bakowies D, Baron R, Chandrasekhar I, Christen M, Daura X, Gee P, Geerke DP, Glattli A, Hunenberger PH, Kastenholz MA, Ostenbrink C, Schenk M, Trzesniak D, van der Vegt NFA. Angewandte Chemie - International Edition. 2006;45:4064. doi: 10.1002/anie.200502655. [DOI] [PubMed] [Google Scholar]
- 46.van Gunsteren WF, Bakowies D, Burgi R, Chandrasekhar I, Christen M, Daura X, Gee P, Glattli A, Hansson T, Oostenbrink C, Peter C, Pitera J, Schuler L, Soares TA, Yu HB. Chimia. 2001;55:856. [Google Scholar]
- 47.Feng X, Fryxell GE, Wang LQ, Kim AY, Liu J, Kemner KM. Science. 1997;276:923. [Google Scholar]
- 48.Benning MM, Hong SB, Raushel FM, Holden HM. J Biol Chem. 2000;275:30556. doi: 10.1074/jbc.M003852200. [DOI] [PubMed] [Google Scholar]
- 49.Benning MM, Shim H, Raushel FM, Holden HM. Biochemistry-Us. 2001;40:2712. doi: 10.1021/bi002661e. [DOI] [PubMed] [Google Scholar]
- 50.Rodriguez R, Chinea G, Lopez N, Pons T, Vriend G. Bioinformatics. 1998;14:523. doi: 10.1093/bioinformatics/14.6.523. [DOI] [PubMed] [Google Scholar]
- 51.Bas D, Rogers DM, Jensen JH. Proteins: Structure, Function and Bioinformatics. 2008;73:765. doi: 10.1002/prot.22102. [DOI] [PubMed] [Google Scholar]
- 52.Oostenbrink C, Soares TA, van der Vegt NFA, van Gunsteren WF. Eur Biophys J Biophy. 2005;34:273. doi: 10.1007/s00249-004-0448-6. [DOI] [PubMed] [Google Scholar]
- 53.Soares TA, Daura X, Oostenbrink C, Smith LJ, van Gunsteren WF. J Biomol Nmr. 2004;30:407. doi: 10.1007/s10858-004-5430-1. [DOI] [PubMed] [Google Scholar]
- 54.Cornell WD, Cieplak P, Bayly CI, Kollman PA. J Am Chem Soc. 1993;115:9620. [Google Scholar]
- 55.Bylaska EJ, Jong WAd, Kowalski K, Straatsma TP, Valiev M, Wang D, Aprà E, Windus TL, Hirata S, Hackler MT, Zhao Y, Fan P-D, Harrison RJ, Dupuis M, Smith DMA, Nieplocha J, Tipparaju V, Krishnan M, Auer AA, Nooijen M, Brown E, Cisneros G, Fann GI, Frücht H, Garza J, Hirao K, Kendall R, Nichols JA, Tsemekhman K, KWolinsk Anchell J, Bernholdt D, Borowski P, Clark T, Clerc D, Dachsel H, Deegan M, Dyall K, Elwood D, Glendening E, Gutowski M, Hess A, Jaffe J, Johnson B, Ju J, Kobayashi R, Kutteh R, Lin Z, Littlefield R, Long X, Meng B, Nakajima T, Niu S, Pollack L, Rosing M, Sandrone G, Stave M, Taylor H, Thomas G, Lenthe Jv, Wong A, Zhang Z. NWChem, A Computational Chemistry Package for Parallel Computers, Version 5.0. Pacific Northwest National Laboratory; Richland, Washington 99352–0999, USA: 2006. A modified version. [Google Scholar]
- 56.Soares TA, Osman M, Straatsma TP. J Chem Theory Comput. 2007;3:1569. doi: 10.1021/ct700024h. [DOI] [PubMed] [Google Scholar]
- 57.Chandrasekhar I, Kastenholz M, Lins RD, Oostenbrink C, Schuler LD, Tieleman DP, van Gunsteren WF. Eur Biophys J Biophy. 2003;32:67. doi: 10.1007/s00249-002-0269-4. [DOI] [PubMed] [Google Scholar]
- 58.Lins RD, Hunenberger PH. J Comput Chem. 2005;26:1400. doi: 10.1002/jcc.20275. [DOI] [PubMed] [Google Scholar]
- 59.Soares TA, Hunenberger PH, Kastenholz MA, Krautler V, Lenz T, Lins RD, Oostenbrink C, van Gunsteren WF. J Comput Chem. 2005;26:725. doi: 10.1002/jcc.20193. [DOI] [PubMed] [Google Scholar]
- 60.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. J Phys Chem B. 2007;111:7812. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 61.Berendsen HJC, Postma JPM, van Gunsteren WF. J., H. Interaction models for water in relation to protein hydration. In: Pullman B, editor. Intermolecular Forces. Reidel; Dordrecht: 1981. p. 331. [Google Scholar]
- 62.van Gunsteren WF, Berendsen HJC. Molecular Simulation. 1988;1:173. [Google Scholar]
- 63.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. J Comput Chem. 1997;18:1463. [Google Scholar]
- 64.Miyamoto S, Kollman PA. J Comput Chem. 1992;13:952. [Google Scholar]
- 65.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. Journal of Chemical Physics. 1995;103:8577. [Google Scholar]
- 66.Hess B, Kutzner C, van der Spoel D, Lindahl E. J Chem Theory Comput. 2008;4:435. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 67.Humphrey W, Dalke A, Schulten K. J Mol Graphics. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 68.Koça J, Zhan CG, Rittenhouse RC, Ornstein RL. J Am Chem Soc. 2001;123:817. doi: 10.1021/ja000439r. [DOI] [PubMed] [Google Scholar]
- 69.Aubert SD, Li YC, Raushel FM. Biochemistry-Us. 1994;43:5707. doi: 10.1021/bi0497805. [DOI] [PubMed] [Google Scholar]
- 70.Amadei A, Linssen AB, Berendsen HJ. Proteins. 1993;17:412. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 71.García AE. Physical Review Letters. 1992;68:2696. doi: 10.1103/PhysRevLett.68.2696. [DOI] [PubMed] [Google Scholar]
- 72.Dubin SB, Clark NA, Benedek GB. Journal of Chemical Physics. 1971;54:5158. [Google Scholar]
- 73.Hanasaki I, Takahashi H, Sazaki G, Nakajima K, Kawano S. J Phys D Appl Phys. 2008:41. [Google Scholar]
- 74.Dumas DP, Caldwell SR, Wild JR, Raushel FM. J Biol Chem. 1989;264:19659. [PubMed] [Google Scholar]
- 75.Nag A, Dinner AR. Biophysical Journal. 2006;90:896. doi: 10.1529/biophysj.105.072785. [DOI] [PMC free article] [PubMed] [Google Scholar]