Abstract
We adapted and validated a basis function method (BFM) to estimate at the voxel level parameters of the kinetic model of the L-[1-11C]leucine PET method and regional rates of cerebral protein synthesis (rCPS). In simulation at noise levels typical of voxel data, BFM yielded low bias estimates of rCPS; in measured data, BFM and nonlinear least squares parameter estimates were in good agreement. We also examined whether there are advantages to using voxel-level estimates averaged over regions of interest (ROI) in place of estimates obtained by directly fitting ROI time-activity curves (TACs). In both simulated and measured data, fits of ROI TACs were poor, likely due to tissue heterogeneity not taken into account in the kinetic model. In simulation, rCPS determined from fitting ROI TACs was substantially overestimated and BFM-estimated rCPS averaged over all voxels in a ROI was slightly underestimated. In measured data, rCPS determined by regional averaging of voxel estimates was lower than rCPS determined from ROI TACs, consistent with simulation. In both simulated and measured data, intersubject variability of BFM-estimated rCPS averaged over all voxels in a ROI was low. We conclude that voxelwise estimation is preferable to fitting ROI TACs with a homogeneous tissue model.
Keywords: brain, leucine, positron emission tomography, protein synthesis, parametric imaging
INTRODUCTION
Biosynthesis of proteins is a fundamental process necessary for physiological maintenance and functioning of organisms. In the central nervous system, de novo protein synthesis is critical for adaptive responses such as long-term memory formation. Animal studies indicate that regional rates of cerebral protein synthesis (rCPS) are altered in models of various clinical disorders (Qin et al, 2005; Smith and Kang 2000; Widmann et al, 1991, 1992; Collins et al, 1980) and in certain physiologic states such as slow-wave sleep (Nakanishi et al, 1997). rCPS also changes during brain development (Sun et al, 1995) and normal aging (Ingvar et al, 1985).
With the development and validation of the L-[1-11C]leucine PET method (Schmidt et al, 2005; Smith et al, 2005), fully quantitative measurement of rCPS in human subjects is now possible. We have implemented the method in awake, healthy young men and demonstrated that reproducible measurements of rCPS with low variability can be obtained (Bishu et al, 2008). Future applications are expected in studies of normal human brain, neurodegenerative diseases, and neurodevelopmental disorders that might be associated with dysregulation of cerebral protein synthesis (Bear et al, 2008).
Analysis of L-[1-11C]leucine PET data in non-human primates (Smith et al, 2005) and human subjects (Sundaram et al, 2006; Bishu et al, 2008,2009) has to date been carried out only at the region-of-interest (ROI) level. Because ROI data are averaged over large numbers of voxels in the PET image, noise is reduced. Furthermore, computational time is minimal when the number of ROIs is small, and rCPS in each structure can be directly compared across subjects. With ROI analysis, however, any heterogeneity among voxels within the ROI is not taken into account. One consequence is that changes occurring in a subset of voxels within a ROI may be missed. In order to survey the entire volume, methods are needed to analyze each voxel within the image and generate maps of rCPS and other parameters of interest. Application of nonlinear least squares estimation to each of the ~107 voxels in a high resolution PET image is infeasible, however, due to the high computational cost. In the current study we adapted and validated a basis function method (BFM) (Gunn et al, 1997) to estimate parameters of the [11C]leucine kinetic model at the voxel level.
Although parametric maps are useful for surveying the brain volume and developing hypotheses concerning areas that may be affected in a particular condition, they cannot be used as generated for group statistical comparisons; spatial normalization to a common brain structure is required. Particularly with high resolution images, this process introduces uncertainty about the exact location in the brain with which each PET voxel should be associated. An alternative that avoids spatial normalization is to use the parametric maps as the basis for computation of mean ROI parameter estimates. Because the kinetic model parameters are not a linear function of measured activity, the mean of voxelwise estimates for a particular parameter will not, in general, equal the parameter estimated from the mean of the voxel activities, i.e., from the ROI TAC. In order to determine if voxelwise estimation confers any advantage for determining kinetic model parameters and rCPS in a ROI, we compared the mean of the voxelwise-estimated parameters with those determined from the ROI TACs. Comparisons between methods were carried out in simulations and in [11C]leucine PET data acquired in healthy young men. The homogeneous tissue kinetic model for leucine (Schmidt et al, 2005) was used in the analyses.
MATERIALS AND METHODS
PET Studies
Data from previously-reported studies (Bishu et al, 2008, 2009) of six healthy awake male subjects (age 20–24) were used for reanalysis in the current study. The criteria for subject inclusion and the procedure for L-[1-11C]leucine PET studies are described in detail in Bishu et al (2008). All studies were performed on the ECAT High Resolution Research Tomograph (HRRT) (CPS Innovations, Knoxville, TN), which has a spatial resolution of ~2.6 mm full width at half maximum (FWHM) (Weinhard et al, 2002). The 90-min emission scan was initiated coincident with a 2-min intravenous infusion of 25–30 mCi of L-[1-11C]leucine (estimated specific activity 3 mCi/nmol). Images were reconstructed using the motion-compensated 3D ordinary Poisson ordered subset expectation maximization algorithm (Carson et al, 2003) as 42 frames of data (16×15, 4×30, 4×60, 4×150, 14×300 sec); voxel size was 1.21×1.21×1.23 mm. Arterial blood sampling was performed; concentrations of unlabeled and labeled leucine in arterial plasma and total 11C and 11CO2 activities in whole blood were measured according to methods detailed in Bishu et al (2008). All subjects underwent a T1-weighted MRI of the brain for ROI placement.
Kinetic Model
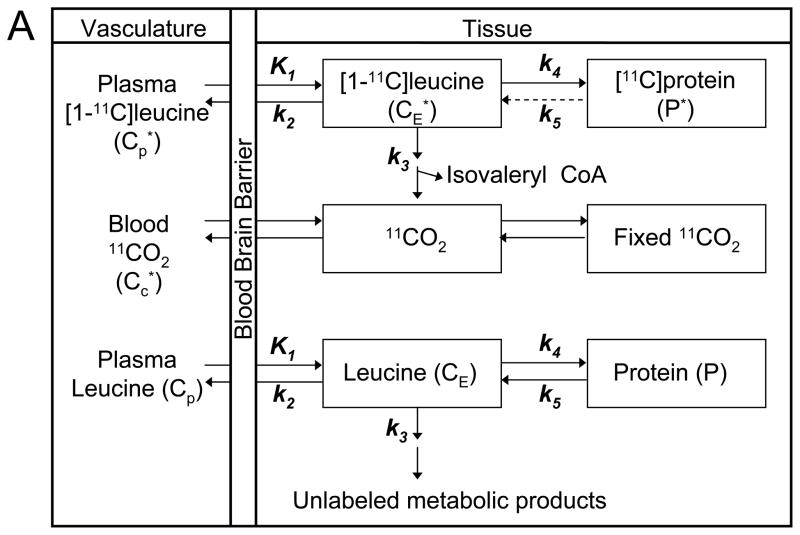
Fig 1A illustrates the kinetic model of the behavior of leucine in brain (Schmidt et al, 2005). Total concentration of 11C in the ROI or voxel (CT*) includes free L-[1-11C]leucine and L-[1-11C]leucine incorporated into tissue protein (CE* and P*, respectively), plus activity in blood (VbCb*, where Vb is the fraction of the ROI or voxel volume occupied by blood and Cb* is the concentration of activity in whole blood). Total 11C also includes labeled products of L-[1-11C]leucine metabolism: 11CO2 and products of 11CO2 fixation. K1 and k2 are the rate constants for transport of leucine from plasma to tissue and back, respectively. k3 is the rate constant for the first two steps in leucine catabolism, transamination and decarboxylation, which yield 11CO2. k4 and k5 are the rate constants for leucine incorporation into protein and release of free leucine from proteolysis, respectively. It is assumed that there is no significant breakdown of labeled product during the experimental interval, i.e., k5P*~0. The model used to describe labeled leucine holds also for unlabeled leucine, except that unlabeled leucine and protein are in steady state, and the steady-state breakdown of unlabeled protein (k5P) is greater than zero. The model assumes the tissue is homogeneous with respect to concentrations of amino acids, blood flow, rates of transport and metabolism of amino acids, and rates of incorporation into protein (Schmidt et al, 2005).
Fig 1. (A) Homogeneous Tissue Model for L-[1-11C]Leucine PET method.
Top rows illustrate the model used to describe the behaviour of labeled leucine in brain. K1 and k2 are the rate constants for transport of leucine from plasma to tissue and back, respectively. k3 is the rate constant for the first two steps in leucine catabolism, transamination and decarboxylation, which yields 11CO2. k4 and k5 are the rate constants for leucine incorporation into protein and for the release of free leucine from proteolysis, respectively. Because of the long half-life of protein in brain (Lajtha et al, 1976), it is assumed that there is no significant breakdown of labeled product (P*) during the experimental interval, i.e., k5P*~0. Labeled CO2 arises either through catabolism of labeled leucine in brain or through influx from blood after catabolism in other tissue, and once in brain it may be either transported back to blood or fixed in brain. Assuming no isotope effect, the rate constants for labeled and unlabeled leucine are identical. Thus the model used to describe labeled leucine holds also for unlabeled leucine (bottom part of the figure), except that unlabeled leucine and protein are in steady state, and the steady-state breakdown of unlabeled protein, k5P, is greater than zero. The model assumes that the tissue is homogeneous with respect to concentrations of amino acids, blood flow, rates of transport and metabolism of amino acids, and rates of incorporation into protein. From Schmidt et al, 2005.
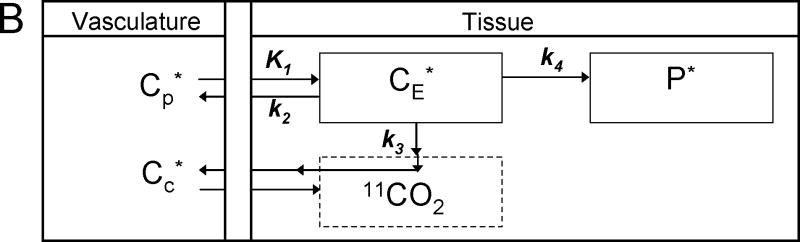
(B) Simplified Homogeneous Tissue Model. Under the assumptions of negligible fixation of 11CO2 during the experimental period (Siesjo and Thompson, 1965) and rapid equilibration of 11CO2 between brain and blood (Buxton et al, 1987), the model reduces to two tissue compartments (CE* and P*) plus the 11 CO2 compartment in which the concentration is known.
Under assumptions of negligible fixation of 11CO2 during the experimental period (Siesjö and Thompson, 1965) and rapid equilibration of diffusible 11CO2 between brain and blood (Buxton et al, 1987), the model simplifies to the homogeneous tissue model used in our analyses (Fig 1B). The concentration of 11CO2 in tissue is approximated by VDCc*, where Cc* is the 11CO2 concentration in whole blood and VD is the brain:blood equilibrium distribution volume of 11CO2. VD was fixed at 0.41, the value measured in rhesus monkeys (Smith et al, 2005), which is in agreement with the mean whole brain:plasma equilibrium distribution volume determined from 11CO2 studies in humans (Brooks et al, 1984). (The blood:plasma equilibrium distribution volume is close to unity (Gunn et al, 2000)).
Therefore
| [1] |
In terms of rate constants
| [2] |
where dependence on the parameter vector ρ=[K1,k2+k3,k4,Vb] is now explicitly shown.
After rate constants have been estimated for a voxel or ROI by one of the methods described below, rCPS can be calculated from the measured plasma concentration of unlabeled leucine, Cp, and estimated values of the rate constants as
| [3] |
Eq [3] takes into account recycling of unlabeled amino acids derived from proteolysis, but effects of recycling can also be examined. The factor λ represents the fraction of unlabeled leucine in the precursor pool for protein synthesis derived from arterial plasma; the fraction derived from tissue proteolysis is (1-λ). Lambda can be computed as
| [4] |
Rate Constant Estimation
To estimate kinetic parameters from each ROI time-activity curve (TAC), weighted nonlinear least squares (NLLS) was used; it minimizes the weighted residual sum of squares (WRSS) objective function
| [5] |
where n is the number of frames of data, CT*(ti;ρ)model is given by Eq [2], si2 is proportional to the variance of decay-corrected measured activity in the ROI, i.e., CT*(ti), and ti is the midpoint of the i-th frame. Variance was modeled, assuming Poisson statistics, as
| [6] |
(Wu and Carson, 2002) where γ is the decay constant for 11C, Δti is the length of Frame i, and α is a proportionality coefficient reflecting the noise level in the data. In PET data, α is not known a priori, but its value does not affect parameter estimates. We also evaluated the effect on parameter estimates of using weights based on whole brain activity instead of activity in the ROI itself for variance calculation by Eq [6]. Results show that the effect is negligible; differences (in absolute value) in estimates of K1,k2+k3,k4, and Vb were 0.4%, 0.8%, 0.6%, and 0.6%, respectively, when averaged over all subjects and ROIs. For comparison with voxel-based estimates weights based on whole brain activity were used. NLLS estimation was performed with the Matlab® function lsqcurvefit (The MathWorks, Inc., Natick, MA, USA) with initial conditions ρ0=[0.0017 ml·g−1sec−1, 0.0033 sec−1, 0.00083 sec−1, 0.05]; iterations stopped when the change in ||ρ|| <10−8 or the change in WRSS<10−6.
To estimate parameters at the voxel level, we implemented a BFM, as described below. For comparison, NLLS was also used for fitting of a subset of voxel data. In voxel-based estimation, activity in whole brain was used to calculate variance by Eq [6]; this variance model was found to perform better than one based on individual voxel values (Tomasi et al, 2008).
The difference between tracer arrival time in brain and arterial sampling site was estimated by shifting blood curves 0–20 sec, fitting the whole brain TAC, and selecting the delay that produced the smallest WRSS. Tracer appearance times in various parts of the brain differ from the mean of the brain as a whole by ±2 sec (Iida et al, 1988); therefore, in each study, for all regions and voxels, whole brain tracer arrival delay was used.
Basis Function Method
The use of NLLS is not feasible when working at the voxel level due its high computational burden. A fast tool to estimate parameters is provided by the BFM. The method is based on a grid search approach and linear least squares estimation that was first introduced for determining regional cerebral blood flow (Koeppe et al, 1985). One observes from Eq [2]: if the value of β=k2+k3+k4 were known, Eq [2] would be linear in the coefficients, which could be quickly estimated using standard weighted linear least squares. The idea of the BFM is to define a grid of values for β in the physiological range. For each β the linear coefficients of Eq [2] are estimated and WRSS computed. The value of β and the coefficients at which WRSS is minimal are used to compute the model parameters. Details are given in the appendix.
For our dataset, 100 values equally spaced between βmin=0.0001 min−1 and βmax=0.5 min−1 were used initially as the grid for β; this widely covers the range of physiological values expected for k2+k3+k4 (Bishu et al, 2008). Dependence of performance on number and distribution of basis functions was tested using grids with 100 or 200 values, uniformly or logarithmically distributed, between βmin and βmax.
With the BFM approach, one or more parameter estimates may be negative for a given voxel. Rather than exclude these voxels from the analysis, which would likely create problems when considering ROI/voxel correlation, we developed the strategy presented in the appendix to reanalyze each voxel that had one or more negative parameters after the initial BFM estimation. A voxel was discarded from further analysis only if reanalysis failed to produce nonnegative parameter estimates.
Performance of BFM for voxel analysis
BFM performance for voxel analysis was assessed in both simulated and measured PET data. For simulation, values of parameters within the range of those previously estimated for cerebellar, cortical, and subcortical regions (Bishu et al, 2008), i.e., K1=0.05 ml·g−1·min−1, k2+k3=0.15 min−1, k4=0.04 min−1, and Vb=0.05, were employed together with measured plasma and blood curves of one subject to generate a noiseless TAC according to Eq [2]. Gaussian noise with zero mean and variance given by Eq [6] was added to obtain simulated noisy TACs at four different noise levels, α=100, 1000, 2000, or 5000. In the same subject from whom we derived the input functions, median estimates of α (WRSS/(n−p), where n is the number of data frames and p the number of parameters (Landaw and DiStefano, 1984)) over all voxels in thalamus, corona radiata, and the central nine slices of frontal cortex, were 1763, 920, and 2591, respectively; this suggests that α=1000 and α=2000 are representative of noise levels found in L-[1-11C]leucine voxel data measured on the HRRT. For each noisy TAC, BFM was employed to estimate kinetic model parameters; weights for estimation were based on the whole brain TAC for this subject. For each noisy TAC, parameters were also estimated by NLLS. We refer to this as “single voxel simulation” to distinguish it from the one described in the next section.
To assess BFM or NLLS performance at the voxel level, percent bias (Bias%) and percent root mean square error (RMSE%) were computed as
| [7] |
and
| [8] |
where pj is the BFM or NLLS estimate of a given rate constant at the jth noise realization and N=1,000 is the number of repetitions. Simulated voxels discarded from BFM analysis, those for which NLLS did not converge, and those for which any estimated parameter>1 were not included in computation of Bias% and RMSE%. In all cases, the fraction of voxels excluded was recorded.
In a subset of measured data (one image plane each from two PET studies, ~20,000 total voxels), parametric maps generated by BFM and NLLS were compared. In this case, in absence of “true” values, NLLS estimates were regarded as the gold standard. Average Bias% and RMSE% were computed for each parameter estimate as
| [9] |
and
| [10] |
where pjTRUE and pj indicate the estimate of a given rate constant at the jth voxel determined by NLLS and BFM, respectively; M is the total number of voxels. Notice that Eq [10] can be derived as the mean value of RMSE% given in Eq [8] when N=1. To avoid having outliers unduly influence results, (pj-pjTRUE)/pjTRUE in Eq[9] was bounded between −1 and 1, and the threshold for the upper bound of |pj-pjTRUE|/pjTRUE in Eq[10] was set at 1. Furthermore, to avoid division by zero or near zero, voxels in which the NLLS estimate of either Vb or k4<10−4 were excluded. This avoids, for example, including in the average a 100% bias for a voxel in which k4(NLLS)=10−6 and k4(BFM)=10−5, both estimates effectively zero.
ROI analysis
For use with measured PET data, ROIs were placed on each subject’s MRI by visually identifying anatomic landmarks, manually outlining the region, and constructing a binary mask to identify voxels in the region. The set of voxels in the PET image belonging, wholly or fractionally, to a given ROI was defined in the following way. The MRI was aligned to the average 30–60 min PET image by use of Vinci software (Volume Imaging in Neurological Research, Max-Planck-Institute for Neurological Research, Cologne, Germany) with a 3D rigid body transformation. This transformation was then applied to each ROI volume mask to effect its alignment with the PET data. Note that fractional values in the resliced mask indicate fractional inclusion of the PET voxel in the ROI. Eighteen ROIs and whole brain were analyzed for each subject. The average TAC for each ROI was computed by summing, over all voxels, the product of activity in each frame of PET data multiplied by the resliced mask for the ROI, and dividing by the total number of voxels. NLLS was used to estimate rate constants from each ROI TAC; weights were based on the whole brain TAC. Performance of the model for fitting ROI TACs was assessed by using the Akaike Information Criterion (AIC) in its small-sample formulation (Hurvich and Tsai, 1989; Sugiura, 1978), i.e.,
| [11] |
where p=4; n and WRSS have been defined above. ROI TAC analyses were performed on both simulated and measured PET data.
Simulated region of interest PET data were generated to test simultaneously effects of noise and heterogeneity. To create a realistic data set for all voxels within a region, three ROIs (thalamus, corona radiata, and central slices of frontal cortex, comprised of 11209, 22777, and 19942 voxels, respectively) were identified in the PET data of one subject. For each voxel in each of the three ROIs, BFM was used to estimate kinetic model parameters from the measured PET data. For every voxel j, the noise level αj was also estimated as WRSS/(n−p), where n=42 and p=4. This initial set of BFM-estimated parameters became “true” parameters for the voxels in the simulation. While it is recognized that these parameters are not the “true” parameters of the underlying tissue (due to noise in the data, potential biases in the estimates, etc.), for simulation purposes we need only assume that the collection of parameters taken together resemble those found in a typical ROI. Under this assumption, simulated [11C]leucine images were derived by generating TACs for each voxel employing Eq [2], measured plasma and blood curves, and “true” parameters. Gaussian noise with zero mean and variance defined by Eq [6] (with α =αj) was then added to each voxel TAC. A noisy ROI TAC was computed as the average of noisy voxel TACs. Parameters were then estimated in two ways: 1) by use of BFM at the voxel level and averaging estimates over all voxels in the ROI, and 2) by use of NLLS to fit the ROI TAC. Bias% and RMSE%, as defined in Eqs [7]–[8], were computed for each rate constant and each estimation method. This process was repeated 100 times, and average Bias% and RMSE% were computed. Due to the large number of voxels included in each region, average Bias% and RMSE% exhibited little variation between repetitions; hence 100 repetitions were deemed sufficient. We refer to this as “multiple-voxel ROI simulation.”
In both simulated and measured data, parameter estimates derived from average ROI TACs by NLLS were compared with the average of the corresponding parameter estimates derived for each voxel in the ROI by BFM, with outliers excluded as defined above, i.e., K1,k2+k3,k4, or Vb>1.
Both in “single voxel simulation” and “multiple-voxel ROI simulation”, adding Gaussian noise with variance described by Eq [6] in some instances gave rise to negative values in the simulated noisy TAC. This was almost exclusively in the first few frames of data when activity is low. Because negative values of concentration have no physical meaning, they were set to zero so that simulated noisy TACs were strictly nonnegative.
RESULTS
Performance of BFM for voxel analysis
Results obtained at the voxel level with BFM with the four different grids (100 and 200 values for β, uniform and logarithmic distributions) were indistinguishable in terms of Bias% and RMSE% in comparisons between BFM and NLLS voxel estimates on two image planes of measured PET data. Therefore the simplest grid with 100 uniformly distributed values of β was used in all further analyses.
In measured PET data, the percentage of voxels in a given ROI that had at least one negative initial BFM estimate, averaged over all subjects, ranged from 9–18%, depending on the ROI; the mean was ~13%. The algorithm developed to address the problem of negative estimates (Appendix) restored most voxels. Increases in voxel WRSS due to the restoration process were undetectable for those voxels with initial estimates of Vb<0 and ~13% for those voxels with initial estimates of k4<0. After application of the restoration algorithm, the percentage of analyzed voxels requiring exclusion due to non-physiological (negative) parameter estimates was ≪1% in most regions and did not exceed 2% in any region or subject. The fraction of excluded voxels, averaged over the six subjects, was <1.1%. Additional voxels excluded as outliers (K1,k2+k3,k4, or Vb>1) did not exceed 0.01% in any region or subject. The percentage of voxels included in the analysis, therefore, remained >98% for all subjects and regions.
Results for single voxel simulation are presented in Table 1. BFM performance was excellent at low noise levels (α=100): bias in all parameter estimates was <1% in absolute value. At noise levels representative of those found in measured PET data (α=1000–2000), biases in the estimates became nonnegligible. In particular, K1 and k2+k3 were characterized by positive biases ranging from moderate (K1) to severe (k2+k3). k4 was slightly underestimated. λ was overestimated by <1% and rCPS underestimated by <4%. NLLS performance was also excellent at low noise levels, but at higher noise levels NLLS biases in K1 and k2+k3 were higher than with BFM, and k4 was slightly overestimated. NLLS biases in λ and rCPS were comparable to those found with BFM. As expected, with both estimation methods RMSE% increased as noise levels increased.
Table 1.
Performance of algorithms in single voxel simulation*
| Basis Function Method | |||||||
|---|---|---|---|---|---|---|---|
| Mean Estimated Parameter Value | |||||||
| Noise level α | K1 (ml g−1min−1) | k2+k3 (min−1) | k4 (min−1) | Vb | λ | rCPS (nmol g−1min−1) | % voxels included |
| 100 | 0.050 | 0.151 | 0.040 | 0.050 | 0.789 | 1.45 | 100 |
| 1000 | 0.052 | 0.164 | 0.039 | 0.050 | 0.79 | 1.43 | 100 |
| 2000 | 0.053 | 0.170 | 0.039 | 0.052 | 0.80 | 1.40 | 99.9 |
| 5000 | 0.054 | 0.174 | 0.038 | 0.065 | 0.81 | 1.34 | 99.6 |
| Bias % | |||||||
| α | K1 | k2+k3 | k4 | Vb | λ | rCPS | |
| 100 | 0.3 | 0.5 | −0.3 | 0.1 | 0.0 | −0.1 | |
| 1000 | 4.9 | 9.2 | −1.5 | 0.8 | 0.3 | −1.8 | |
| 2000 | 6.8 | 13.6 | −2.4 | 4.6 | 0.8 | −3.8 | |
| 5000 | 7.6 | 15.7 | −4.7 | 29.4 | 2.1 | −7.6 | |
| RMSE % | |||||||
| α | K1 | k2+k3 | k4 | Vb | λ | rCPS | |
| 100 | 7.7 | 13.2 | 8.3 | 14.2 | 2.0 | 4.1 | |
| 1000 | 29.6 | 53.7 | 29.2 | 44.1 | 6.8 | 15.1 | |
| 2000 | 37.9 | 69.3 | 40.4 | 57.8 | 9.0 | 23.2 | |
| 5000 | 51.6 | 88.6 | 62.4 | 86.0 | 13.9 | 45.2 | |
| Nonlinear Least Squares | |||||||
|---|---|---|---|---|---|---|---|
| Mean Estimated Parameter Value | |||||||
| Noise level α | K1 (ml g−1min−1) | k2+k3 (min−1) | k4 (min−1) | Vb | λ | rCPS (nmol g−1min−1) | % voxels included |
| 100 | 0.050 | 0.151 | 0.040 | 0.050 | 0.790 | 1.45 | 100 |
| 1000 | 0.053 | 0.167 | 0.040 | 0.050 | 0.79 | 1.43 | 99.7 |
| 2000 | 0.055 | 0.182 | 0.040 | 0.051 | 0.80 | 1.40 | 99.1 |
| 5000 | 0.059 | 0.206 | 0.042 | 0.059 | 0.81 | 1.36 | 95.5 |
| Bias % | |||||||
| α | K1 | k2+k3 | k4 | Vb | λ | rCPS | |
| 100 | 0.4 | 0.8 | −0.1 | −0.1 | 0.0 | −0.1 | |
| 1000 | 6.0 | 11.4 | −1.0 | −0.1 | 0.4 | −1.8 | |
| 2000 | 10.5 | 21.3 | 0.1 | 1.4 | 1.1 | −3.6 | |
| 5000 | 18.4 | 37.1 | 4.5 | 18.8 | 2.5 | −6.7 | |
| RMSE % | |||||||
| α | K1 | k2+k3 | k4 | Vb | λ | rCPS | |
| 100 | 7.7 | 13.2 | 8.3 | 14.2 | 2.0 | 4.1 | |
| 1000 | 31.4 | 59.1 | 29.3 | 44.0 | 6.8 | 15.0 | |
| 2000 | 42.6 | 83.0 | 42.5 | 57.6 | 8.7 | 21.5 | |
| 5000 | 66.8 | 129.0 | 101.9 | 80.5 | 13.0 | 38.1 | |
True values were K1=0.05 ml·g −1·min−1, k2+k3=0.15 min−1, k4=0.04 min−1, Vb=0.05, λ=0.79, rCPS=1.45 nmol g−1 min−1; number of noise realizations=1,000.
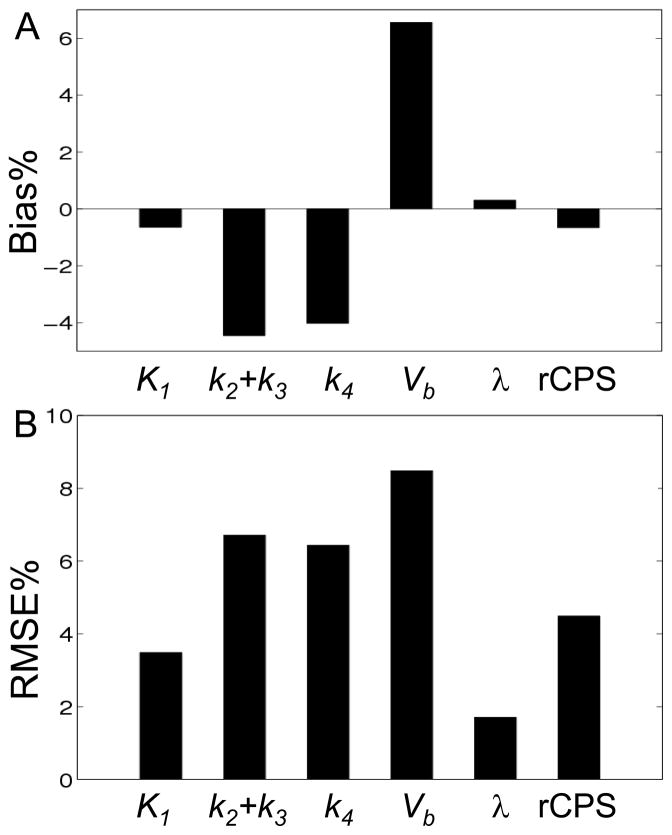
Comparisons between NLLS and BFM parameter estimates on a subset of voxels in the measured PET data are displayed in Fig 2, where NLLS estimates were taken as the “gold standard”. Bias% and RMSE% of BFM estimates, relative to NLLS estimates, are shown. As noted previously, comparisons exclude all voxels for which NLLS estimates of Vb or k4<10−4. Excluding these voxels, the total number of voxels analyzed in the two slices decreased 9%, from 19468 to 17727. Biases of −4% were obtained for k2+k3 and k4, whereas K1 showed almost no bias and lower RMSE%. Estimates of the most important parameters from a physiological point of view, λ and rCPS, had negligible biases (2% and 4.5%, respectively). Because these comparisons assume that NLLS estimates represent true voxel parameter values, low biases in λ and rCPS reflect agreement between estimation methods; they do not address whether both methods might have inherent biases.
Fig 2.
Average Bias% (A) and RMSE% (B) of K1, k2+k3, k4, Vb, λ, and rCPS estimated with the basis function method. Parameters of the homogeneous tissue model were estimated for all voxels in one image plane from each of two subjects by both nonlinear least squares (NLLS) and BFM. Results were computed using Eqs [9]–[10] with the NLLS estimate regarded as the true value. Only voxels in which the NLLS estimates of k4 and Vb ≥104 were included in the statistics.
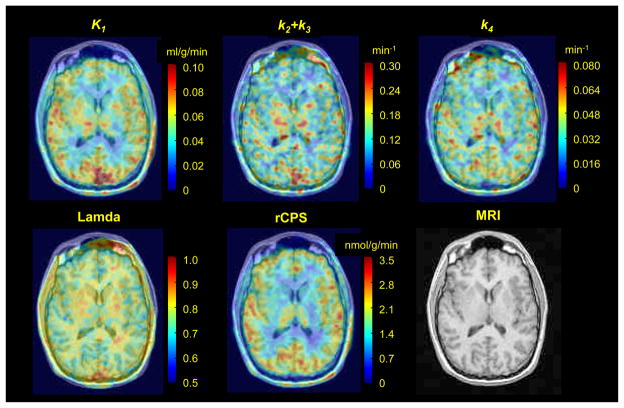
Parametric maps from the voxel BFM estimates of K1,k2+k3,k4, λ and rCPS for one transverse slice from a [11C]leucine PET study of a 23 year old male are displayed in Fig 3.
Fig 3.
Parametric maps of K1, k2+k3, k4, λ, and rCPS obtained with the basis function method for one transverse image plane from a L-[1-11C]leucine PET study of an awake 23 year old male subject. A Gaussian filter (FWHM 3.9 mm) was employed to smooth each of the parametric images in three dimensions before visualization. The same image plane of the co-registered MRI is shown in the bottom right.
Comparison of Voxel-based and ROI TAC-based analyses
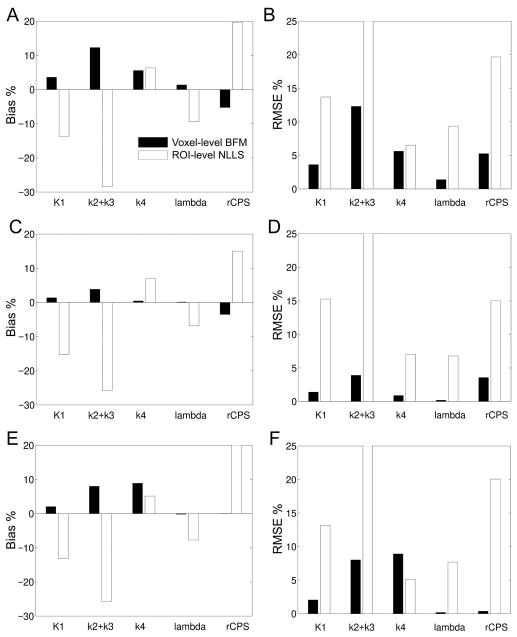
Bias% and RMSE% for multiple-voxel ROI simulations are displayed in Fig 4. For each of the regions simulated (thalamus, corona radiata, frontal cortex), the mean of the parameter values estimated in each voxel with BFM was compared with parameter estimates derived from the ROI TAC with NLLS. Results were quite consistent across ROIs in terms of direction and relative magnitude of biases. The voxel BFM method, consistent with single voxel simulation, overestimated both K1 and k2+k3, but unlike single voxel simulation, estimates of k4 were (on average) positively biased in the three regions. Regional means of voxel BFM estimates of λ were excellent, but rCPS was underestimated by an average of 6%, 4%, and <1% in simulated frontal cortex, thalamus, and corona radiata, respectively. Performance of NLLS applied to the ROI TACs was worse. In particular, a substantial underestimation of k2+k3 was found; this, together with a moderate overestimation of k4, led to underestimation of λ by 6–10% and overestimation of rCPS by 15–20%.
Fig 4.
Bias% and RMSE% for multiple-voxel ROI simulation for central slices of frontal cortex (A and B), thalamus (C and D), and corona radiata (E and F). Parameter estimates were computed in two ways: by use of basis function method (BFM) at the simulated voxel level and averaging the parameter estimates over all voxels within the ROI (voxel-level BFM), and by use of nonlinear least squares to fit the simulated ROI TAC (ROI-level NLLS). Bias% and RMSE% were computed by Eqs [7]–[8] for each rate constant and each estimation method. See text for details.
Table 2 compares mean voxel BFM estimates with estimates found by applying NLLS to the ROI TACs from measured PET data. Means and standard deviations for the six subjects are reported for 18 ROIs and whole brain. Consistent with simulation results, estimates of K1 were higher with the voxel BFM method than with NLLS applied to the ROI TACs. Estimates of k2+k3 were substantially lower, and those of k4 slightly higher, with ROI-level NLLS than with voxel BFM. Again consistent with simulation, λ estimates were ~5–10% lower and rCPS estimates ~20% higher with ROI-level NLLS than voxel BFM.
Table 2.
Regional estimates of kinetic model parameters, lambda, and rCPS1
| Voxelwise Estimation with BFM3 | ||||||
|---|---|---|---|---|---|---|
| K1 (ml g−1min−1) | k2+k3 (min−1) | k4 (min−1) | Vb | Lambda | rCPS (nmol g−1min−1) | |
| Whole Brain (1403 ± 161 cc) | 0.050 ± 0.008 | 0.130 ± 0.008 | 0.037 ± 0.003 | 0.066 ± 0.006 | 0.77 ± 0.02 | 1.61 ± 0.12 |
| Cerebellum (119 ± 12 cc) | 0.069 ± 0.012 | 0.175 ± 0.015 | 0.045 ± 0.004 | 0.078 ± 0.008 | 0.78 ± 0.02 | 1.95 ± 0.15 |
| Vermis2 (5 ± 1 cc) | 0.065 ± 0.012 | 0.174 ± 0.017 | 0.047 ± 0.007 | 0.060 ± 0.013 | 0.78 ± 0.03 | 1.92 ± 0.19 |
| Cortical | ||||||
| Frontal cortex (114 ± 27 cc) | 0.053 ± 0.009 | 0.122 ± 0.007 | 0.038 ± 0.003 | 0.066 ± 0.008 | 0.75 ± 0.02 | 1.85 ± 0.13 |
| Temporal cortex (53 ± 9 cc) | 0.048 ± 0.007 | 0.116 ± 0.007 | 0.037 ± 0.003 | 0.069 ± 0.006 | 0.75 ± 0.02 | 1.73 ± 0.10 |
| Occipical cortex (33 ± 15 cc) | 0.066 ± 0.009 | 0.148 ± 0.010 | 0.043 ± 0.004 | 0.078 ± 0.010 | 0.76 ± 0.03 | 2.13 ± 0.23 |
| Parietal cortex (14 ± 4 cc) | 0.056 ± 0.008 | 0.137 ± 0.009 | 0.042 ± 0.003 | 0.070 ± 0.009 | 0.76 ± 0.02 | 1.91 ± 0.20 |
| Supraorbital gyrus (4 ± 1 cc) | 0.050 ± 0.009 | 0.126 ± 0.014 | 0.040 ± 0.003 | 0.057 ± 0.006 | 0.75 ± 0.03 | 1.77 ± 0.09 |
| Precentral gyrus (3 ± 2 cc) | 0.057 ± 0.009 | 0.142 ± 0.012 | 0.043 ± 0.003 | 0.063 ± 0.009 | 0.76 ± 0.02 | 1.91 ± 0.16 |
| Postcentral gyrus (2 ± 1 cc) | 0.061 ± 0.009 | 0.153 ± 0.019 | 0.042 ± 0.001 | 0.065 ± 0.010 | 0.77 ± 0.02 | 1.93 ± 0.16 |
| Subcortical | ||||||
| Thalamus (19 ± 3 cc) | 0.051 ± 0.009 | 0.151 ± 0.010 | 0.039 ± 0.002 | 0.063 ± 0.007 | 0.78 ± 0.02 | 1.48 ± 0.11 |
| Putamen (9 ± 1 cc ) | 0.052 ± 0.011 | 0.126 ± 0.013 | 0.031 ± 0.002 | 0.054 ± 0.005 | 0.80 ± 0.02 | 1.35 ± 0.09 |
| Caudate (8 ± 1 cc) | 0.038 ± 0.007 | 0.109 ± 0.009 | 0.029 ± 0.004 | 0.047 ± 0.006 | 0.79 ± 0.02 | 1.04 ± 0.05 |
| Hippocampus2 (5 ± 2 cc) | 0.037 ± 0.007 | 0.098 ± 0.019 | 0.033 ± 0.004 | 0.084 ± 0.013 | 0.76 ± 0.01 | 1.38 ± 0.17 |
| Amygdala2 (5 ± 1 cc) | 0.035 ± 0.004 | 0.104 ± 0.013 | 0.036 ± 0.003 | 0.066 ± 0.012 | 0.75 ± 0.02 | 1.33 ± 0.09 |
| Hypothalamus (0.6 ± 0.3 cc) | 0.046 ± 0.006 | 0.112 ± 0.019 | 0.030 ± 0.004 | 0.048 ± 0.007 | 0.78 ± 0.02 | 1.33 ± 0.20 |
| White Matter | ||||||
| Corona radiata (47 ± 6 cc) | 0.029 ± 0.006 | 0.098 ± 0.009 | 0.027 ± 0.004 | 0.033 ± 0.004 | 0.79 ± 0.02 | 0.83 ± 0.06 |
| Centrum semiovale (41 ± 17 cc) | 0.031 ± 0.006 | 0.105 ± 0.008 | 0.029 ± 0.003 | 0.034 ± 0.004 | 0.79 ± 0.02 | 0.93 ± 0.09 |
| Cerebellar peduncles (13 ± 2 cc) | 0.044 ± 0.008 | 0.159 ± 0.024 | 0.037 ± 0.004 | 0.045 ± 0.007 | 0.80 ± 0.02 | 1.14 ± 0.10 |
| ROI TAC-based Estimation with NLLS4 | ||||||
|---|---|---|---|---|---|---|
| K1 (ml g−1min−1) | k2+k3 (min−1) | k4 (min−1) | Vb | Lambda | rCPS (nmol g−1min−1) | |
| Whole Brain | 0.043 ± 0.007 | 0.092 ± 0.008 | 0.040 ± 0.003 | 0.073 ± 0.007 | 0.70 ± 0.03 | 1.93 ± 0.11 |
| Cerebellum | 0.059 ± 0.011 | 0.137 ± 0.017 | 0.051 ± 0.004 | 0.086 ± 0.008 | 0.73 ± 0.03 | 2.30 ± 0.15 |
| Vermis2 | 0.058 ± 0.011 | 0.142 ± 0.020 | 0.052 ± 0.006 | 0.064 ± 0.014 | 0.73 ± 0.03 | 2.24 ± 0.19 |
| Cortical | ||||||
| Frontal cortex | 0.046 ± 0.008 | 0.086 ± 0.007 | 0.040 ± 0.003 | 0.072 ± 0.009 | 0.69 ± 0.03 | 2.22 ± 0.11 |
| Temporal cortex | 0.042 ± 0.007 | 0.080 ± 0.007 | 0.038 ± 0.002 | 0.075 ± 0.007 | 0.68 ± 0.02 | 2.08 ± 0.09 |
| Occipical cortex | 0.057 ± 0.009 | 0.110 ± 0.010 | 0.046 ± 0.003 | 0.085 ± 0.011 | 0.71 ± 0.03 | 2.49 ± 0.25 |
| Parietal cortex | 0.048 ± 0.007 | 0.097 ± 0.008 | 0.044 ± 0.003 | 0.076 ± 0.010 | 0.69 ± 0.03 | 2.28 ± 0.18 |
| Supraorbital gyrus | 0.044 ± 0.008 | 0.089 ± 0.011 | 0.041 ± 0.004 | 0.062 ± 0.007 | 0.68 ± 0.03 | 2.14 ± 0.10 |
| Precentral gyrus | 0.049 ± 0.008 | 0.104 ± 0.017 | 0.045 ± 0.005 | 0.068 ± 0.012 | 0.69 ± 0.03 | 2.29 ± 0.16 |
| Postcentral gyrus | 0.053 ± 0.007 | 0.113 ± 0.021 | 0.046 ± 0.002 | 0.071 ± 0.013 | 0.71 ± 0.03 | 2.28 ± 0.14 |
| Subcortical | ||||||
| Thalamus | 0.044 ± 0.008 | 0.113 ± 0.011 | 0.042 ± 0.002 | 0.068 ± 0.007 | 0.73 ± 0.02 | 1.70 ± 0.13 |
| Putamen | 0.046 ± 0.010 | 0.101 ± 0.012 | 0.031 ± 0.002 | 0.059 ± 0.006 | 0.76 ± 0.02 | 1.52 ± 0.09 |
| Caudate | 0.033 ± 0.007 | 0.081 ± 0.006 | 0.029 ± 0.004 | 0.051 ± 0.006 | 0.74 ± 0.02 | 1.24 ± 0.08 |
| Hippocampus2 | 0.031 ± 0.006 | 0.057 ± 0.017 | 0.031 ± 0.005 | 0.091 ± 0.014 | 0.64 ± 0.04 | 1.81 ± 0.18 |
| Amygdala2 | 0.029 ± 0.003 | 0.064 ± 0.014 | 0.035 ± 0.005 | 0.072 ± 0.013 | 0.64 ± 0.04 | 1.70 ± 0.08 |
| Hypothalamus | 0.042 ± 0.005 | 0.089 ± 0.023 | 0.030 ± 0.005 | 0.052 ± 0.008 | 0.75 ± 0.03 | 1.50 ± 0.20 |
| White Matter | ||||||
| Corona radiata | 0.025 ± 0.005 | 0.070 ± 0.008 | 0.027 ± 0.004 | 0.037 ± 0.004 | 0.72 ± 0.03 | 1.00 ± 0.05 |
| Centrum semiovale | 0.027 ± 0.005 | 0.075 ± 0.006 | 0.029 ± 0.003 | 0.037 ± 0.005 | 0.72 ± 0.03 | 1.12 ± 0.06 |
| Cerebellar peduncles | 0.038 ± 0.007 | 0.121 ± 0.024 | 0.041 ± 0.005 | 0.050 ± 0.008 | 0.74 ± 0.03 | 1.36 ± 0.07 |
Values are mean ± SD for 6 subjects, except where indicated.
Values are mean ± SD for 5 subjects.
BFM Estimates based on the homogeneous tissue model and voxel time activity curves; weights based on whole brain TAC; parameters averaged over volume of interest; number in parenthesis is volume of ROI.
NLLS Estimates based on the homogeneous tissue model and ROI time activity curves; weights based on whole brain TAC.
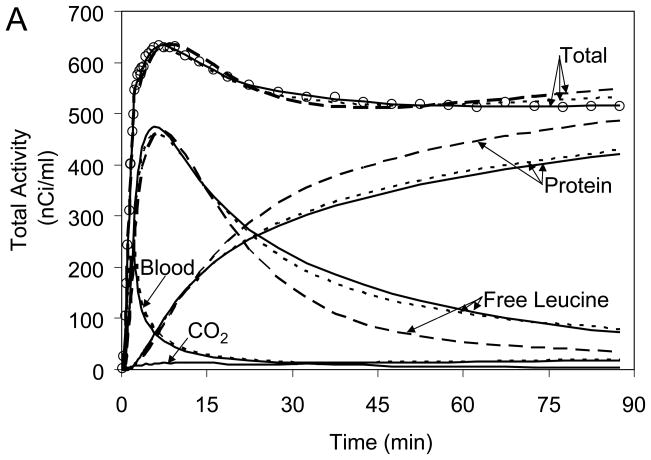
Fig 5A illustrates the NLLS fit of a cortical ROI TAC measured in one subject. Predicted time courses of labeled protein and free leucine in tissue, derived from estimated parameters, are also represented. ROI-level NLLS clearly overestimates measured activity in early and late frames of data and underestimates it in the intermediate interval. Fig 5A also shows the average of the TACs determined in each voxel from BFM parameter estimates for that voxel. The slower clearance of labeled leucine from the tissue and slower accumulation of labeled protein estimated with voxel BFM are consistent with its lower rCPS determinations. Note that the average of the BFM voxel TACs fits more closely the measured data, but it still shows some systematic underestimation at intermediate times and overestimation in the late frames of data. This is evidence that the current kinetic model for leucine is not fully adequate to describe ROI TACs and at least some voxel TACs. The assumption of tissue homogeneity, in particular, is not always a good approximation for a given region. Rates of blood flow, transport, metabolism, and incorporation of amino acids into protein vary regionally throughout the brain; rates in gray and white matter are particularly different. Due to limited spatial resolution of even the HRRT (~2.6mm FWHM), thinness of cortical areas (~3–5 mm), and white matter tracts that run through subcortical gray matter, it is likely that activity measured in most ROIs, and possibly also in many voxels, derives from a heterogeneous mixture of tissues. For comparison, therefore, we illustrate activities estimated by fitting the ROI TAC with a kinetic model that specifically takes tissue heterogeneity into account; this model assumes each tissue is composed of two homogeneous subregions, e.g., gray and white matter (for Model details, see Supplemental File). The fit provided by the tissue heterogeneity model is excellent, and activities in the free leucine and protein pools in brain more closely resemble those found with voxel BFM than those derived by fitting the ROI TAC with the homogeneous tissue kinetic model for leucine.
Fig 5.
Kinetic model fits of whole brain and regions of interest. (A) Open circles indicate activity measured in the frontal cortex of one subject, dashed lines represent activities estimated by fitting the ROI TAC with the homogeneous tissue kinetic model for leucine, and solid lines represent activities estimated by fitting the ROI TAC with a model that explicitly takes tissue heterogeneity into account. For comparison, the average of the TACs determined in each voxel from the BFM parameter estimates for that voxel are shown (dotted line). The predicted time courses of labeled protein and of free leucine in tissue, derived from the estimated parameters, are also represented. The predicted time courses of activity in blood within the ROI volume were approximately equal for all three methods. Tissue 11CO2 (bottom solid line) was very low throughout the study (<15 nCi/ml). Note the substantially better fit of the heterogeneity model and the remarkable differences between the homogeneity and heterogeneity models applied to the ROI TACs in the predicted time courses of free [11C]leucine and labeled protein in tissue. The average of the voxel BFM-estimated time courses of free [11C]leucine and labeled protein resemble most closely those determined from the ROI TAC with the tissue heterogeneity model. Total activity predicted voxelwise with BFM was intermediate between the ROI-level homogeneity and heterogeneity model values; both voxel BFM and ROI-level homogeneity model NLLS overestimated the measured data at late times. Kinetic model fits exhibited similar patterns in all but the smallest and noisiest regions.
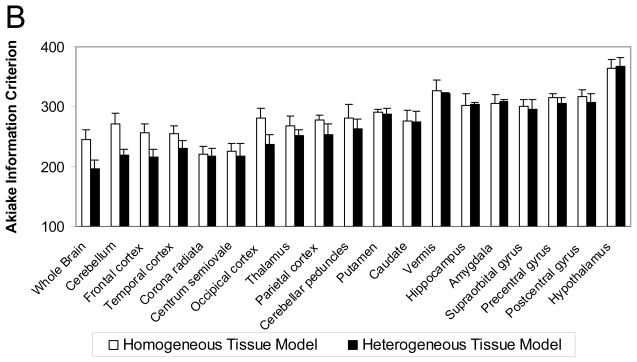
(B) Goodness-of-fit, as measured by the Akaike Information Criterion (AIC), in whole brain and 18 ROIs. Values, computed by Eq [11] and averaged over six subjects, are displayed for the homogeneous tissue model (p=4; white bars) and for the heterogeneous tissue model (p=6; black bars) fits of the ROI TACs; ROIs are sorted in order of decreasing size. Error bars indicate standard deviation among subjects. Lower AIC values indicate better fits. A clear trend is visible with the tissue heterogeneity model yielding much lower mean AIC values for the largest ROIs; this indicates substantial improvements in fit over the homogeneous tissue model.
To further investigate the poor fits of the ROI TACs with the homogeneous tissue kinetic model, all ROI TACs were fit with the kinetic model that explicitly takes tissue heterogeneity into account. Fig 5B shows the goodness-of-fit of ROI TACs by the two kinetic models for 18 ROIs and whole brain. AIC values, averaged over six subjects, are shown with ROIs sorted in order of decreasing size. In whole brain, cerebellum, and the largest cortical ROIs, one sees the greatest improvement in fit by use of a model that explicitly takes tissue heterogeneity into account, as indicated by much lower AIC values. In white matter and smaller ROIs, the two models produced more similar AIC values. Estimates of rCPS based on applying the tissue heterogeneity model to ROI TACs, however, were considerably more variable than those produced by either voxelwise BFM estimation or fitting the homogeneous tissue model to the ROI TACs (data not shown). Furthermore, the heterogeneity model estimation did not always converge, probably due to the presence of six parameters in the model. For these reasons, the tissue heterogeneity model was not pursued further as a practical alternative for estimation of rCPS.
DISCUSSION
Prior to this study, estimation of kinetic model parameters of the L-[1-11C]leucine PET method has been carried out only at the ROI level. In the present work, we addressed our attention to generation of parametric maps. Such maps provide important information concerning heterogeneity within a region and take full advantage of the high spatial resolution of the scanner for visualization of parameters. Our purpose was not to be able to compare subjects at the voxel level; this type of analysis would require spatial normalization with its inherent uncertainties about the actual anatomical location of a voxel. Rather, we are using a voxel-based estimation method to compute mean estimates of parameters for an anatomically defined ROI. A voxel-based estimation method applied to high spatial resolution images may mitigate some of the problems of tissue heterogeneity inherent in PET data.
As the cost of standard nonlinear least squares estimation at the voxel level is prohibitive, a basis function method was employed as a computationally feasible alternative. The time required for the analysis of one image plane decreased from ~2 hours with NLLS to ~1 minute with BFM. Further savings in computational time could be achieved if estimation were performed first on a coarse grid of βs and then a finer one, as proposed by Koeppe et al (1985). In a subset of measured voxels from two PET studies, BFM gave estimates in very good agreement with NLLS estimates. In single voxel simulation BFM provided excellent performance at low noise levels, but at higher noise levels, including those typical of measured voxel data, the method was characterized by high RMSE% and nonnegligible positive biases that were moderate for K1 and much higher for k2+k3. The most important parameters from a physiological point of view, λ and rCPS, were only slightly biased, except at the very highest voxel noise levels. Simulation results and the good agreement between BFM and NLLS suggest that, in this case, it is the high noise level inherent in voxel data, rather than our choice of algorithm, that is responsible for observed biases. We are currently investigating whether improvements in performance can be achieved by pre-processing data to reduce the noise of voxel TACs.
One remarkable outcome of the analysis was the poor performance of the homogeneous tissue kinetic model for leucine when applied to ROI TACs. Simulations showed a substantial underestimation of k2+k3 (−20% to −30%) which, along with a moderate overestimation of k4, led to underestimation of λ by ~8% and to overestimation of rCPS by as much as 15–20%. Differences of a similar magnitude were observed in measured data between ROI-level homogeneous tissue model estimates and mean voxel-level BFM estimates. Additionally, fits of the homogeneous tissue model to ROI TACs were generally poor: in 73% of ROIs examined in 6 subjects, AIC showed that a kinetic model that explicitly takes tissue heterogeneity into account provided the better fit. This strongly suggests the need for a model that takes tissue heterogeneity into account for analysis of L-[1-11C]leucine at the ROI level.
Effects of tissue heterogeneity have been previously investigated for [14C]deoxyglucose and [18F]fluorodeoxyglucose (Schmidt et al 1991; 1992; 1995). From the similarities between the model for deoxyglucose/fluorodeoxyglucose and the model for leucine, overestimation of k2+k3 and k4 was predicted if a model that assumes tissue homogeneity is used. In the present study k4 was somewhat overestimated under these conditions, but k2+k3 was substantially underestimated, contrary to expectation. To overcome limitations of the homogeneous tissue model we evaluated a heterogeneous tissue model comprised of two subregions for each ROI. The heterogeneity model fit ROI TACs extremely well. For estimating parameters, however, it performed poorly on measured PET data. It is our view that more work is needed to develop a useful tissue heterogeneity model for analysis of ROI TACs, or alternative approaches that do not require homogeneity assumptions are required.
We did not apply the heterogeneity model at the voxel level due to the high number of parameters to be estimated. With voxel BFM we did observe some systematic misfit of data: measured total activity was underestimated at intermediate times and overestimated at later times. The direction of the systematic overestimation/underestimation was similar to that found when the ROI TAC was fit with the tissue homogeneity model, but the magnitude of the disparity was less with voxel BFM. This suggests that the homogeneous tissue model may not be fully adequate for at least some voxels. These errors in model specification will be reflected in errors in parameter estimates. Furthemore, measurement noise interacts with model specification error in affecting final parameter estimates. Other scanners have different noise properties, and those with lower spatial resolution may be more affected by tissue heterogeneity; hence voxel parameter estimates may exhibit different bias and RMSE than found in the current study.
Differences in parameter values estimated with different procedures are not unique to L-[1-11C]leucine studies. Each of the factors examined in the current study (choice of model to define relationships among parameters, noise level in the data, algorithm used for parameter estimation) affects the estimates. It has long been recognized that employing different kinetic models gives rise to differences in both micro- and macroparameter estimates that can be substantial, even when each model is applied to the same TAC (see, e.g., Lammertsma et al, 1987). Ultimately, all models oversimplify underlying tissue kinetics, and some degree of model specification error occurs. Even in the absence of model specification error, however, estimation biases can occur due to measurement noise in the data or error in measurement noise characterization. Thus we should expect differences between parameter estimates at the voxel level (potentially greater homogeneity and less model misspecification, but higher noise levels) and at the ROI level (lower noise levels, but potentially more heterogeneity and model specification error). Yet few studies have quantitatively compared parameter estimates at the voxel and ROI levels. In one such study, several methods for estimating binding potentials for dopamine and 5-HT1A receptors were compared; even when the same analysis procedure was used, estimates differed substantially between ROI-based and voxel-based methods (Cselényi et al, 2006). In the present study we also found substantial differences between ROI-based and voxel-based estimates of kinetic model parameters when the same homogeneous tissue model was used in both analyses. Since each method has its own set of limitations, we used simulation to look at bias and RMSE of each procedure under consideration. We also examined how the different methods compare when applied to measured data, taking into account each method’s limitations.
Of the methods examined for determining kinetic model parameters for L-[1-11C]leucine in identified ROIs, the voxel BFM method provided the best overall performance. This method requires estimation of parameters, λ and rCPS for each voxel, and averaging values over all voxels within the region. Simulations suggest that resultant biases in λ and rCPS are small. Furthermore, intersubject variability of parameters estimated with voxel BFM in measured PET data was low, comparable to that achieved by ROI-based application of the homogeneous tissue model. Minimally biased, low variance estimates are important for tracking changes in rCPS and/or λ across regions and over time, particularly as changes may be relatively small in magnitude. Optimal estimates will also be required for rCPS to be used as an objective means by which to identify abnormalities or as an outcome measure to evaluate potential treatments. Ultimately, these quantitative measurements in human subjects may give new insights into mechanisms underlying pathologies in neurodevelopmental, cognitive and neurodegenerative disorders. BFM analysis of L-[1-11C]leucine data at the voxel level is a useful tool for quantitative determination of rCPS.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program, National Institute of Mental Health.
APPENDIX
Implementation of Basis Function Method
Eq [2], repeated below, relates measured concentration of 11C (CT*) in a generic voxel at time t to measured [11C]leucine concentration in plasma (Cp*), total 11C in whole blood (Cb*), and 11CO2 concentration in tissue (VDCc*) as:
| [A1] |
where β=k2+k3+k4. Eq [A1] can be rewritten as:
| [A2] |
with
| [A3] |
| [A4] |
| [A5] |
If the value of β were known, Eq [A2] would be linear in the parameter vector θ=[θ1,θ2,θ3] which could be quickly estimated using standard weighted linear least squares. The idea of BFM is to define a grid of values for β in the physiological range, β1,β2,…βm, solve for each the corresponding linear problem expressed by Eq [A2], and select the value β* which gives rise to the smallest WRSS. Let θ*=[θ1*,θ2*,θ3*] be the estimated parameter vector at β= β*. Eqs [A3–A5] can then be solved to yield final values of the rate constants (K1,k2+k3,k4) and blood volume (Vb) as
| [A6] |
| [A7] |
| [A8] |
| [A9] |
Eqs [A6–A9] may provide one or more negative parameter estimates for a given voxel, while all rate constants and Vb should be nonnegative. Rather than constrain the estimation and substantially increase computation time, we developed the following algorithm to re-estimate parameters when one or more initial BFM estimates was negative:
| Algorithm |
| Let θ*=[θ1*,θ2*,θ3*] be the estimated parameter vector at β=β*. |
| Calculate K1, k2+k3, k4 and Vb from Eqs [A6]–[A9]. |
| If K1≤0 or k2+k3≤0: |
| Discard voxel. |
| Else if Vb<0: |
| Set θ3=Vb=0 in Eq [A2] and re-estimate θ*=[θ1*,θ2*]. |
| Calculate K1, k2+k3, k4 from Eqs [A6]–[A8]. |
| If k4<0: |
| Set θ2=θ3=0 in Eq [A2] and re-estimate θ*=[θ1*]. |
| Set k4=0 and calculate K1, k2+k3 from Eqs [A6]–[A7]. |
| If K1≤0 or k2+k3≤0 discard voxel; otherwise retain voxel. |
| Else if k4<0: |
| Set θ2=0 in Eq [A2] and re-estimate θ*=[θ1*,θ3*]. |
| Set k4=0 and calculate K1, k2+k3, Vb from Eqs [A6], [A7], [A9]. |
| If Vb<0: |
| Set θ2=θ3=0 in Eq [A2] and re-estimate θ*=[θ1*]. |
| Set Vb=0 and calculate K1, k2+k3 from Eqs [A6]–[A7]. |
| If K1≤0 or k2+k3≤0 discard voxel; otherwise retain voxel. |
| Else |
| Retain voxel. |
| End |
With this algorithm a set of nonnegative parameters is attributed to almost every voxel. In practice it happened rarely that K1 and/or k2+k3 was nonpositive and the voxel had to be discarded from the parametric map.
Footnotes
DISCLOSURE
The authors have no duality of interest to declare.
References
- Bear MF, Dölen G, Osterweil E, Nagarajan N. Fragile X: Translation in Action. Neuropsychopharmacology. 2008;33:84–87. doi: 10.1038/sj.npp.1301610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishu S, Schmidt KC, Burlin T, Channing M, Conant S, Huang T, Liu Z-H, Qin M, Vuong B, Unterman A, Xia Z, Zametkin A, Herscovitch P, Smith CB. Regional rates of cerebral protein synthesis measured with L-[1–11C]leucine and PET in conscious, young adult men: normal values, variability, and reproducibility. J Cereb Blood Flow Metab. 2008;28:1502–1513. doi: 10.1038/jcbfm.2008.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishu S, Schmidt KC, Burlin T, Channing M, Horowitz L, Huang T, Liu Z-H, Qin M, Vuong B, Unterman A, Xia Z, Zametkin A, Herscovitch P, Quezado Z, Smith CB. Propofol anaesthesia does not alter regional rates of cerebral protein synthesis measured with L-[1–11C]leucine and PET in healthy male subjects. J Cereb Blood Flow Metab. 2009 February 18;29:1035–1047. doi: 10.1038/jcbfm.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DJ, Lammertsma AA, Beaney RP, Leenders KL, Buckingham PD, Marshall J, Jones T. Measurement of regional cerebral pH in human subjects using continuous inhalation of 11CO2 and positron emission tomography. J Cereb Blood Flow Metab. 1984;4:458–65. doi: 10.1038/jcbfm.1984.65. [DOI] [PubMed] [Google Scholar]
- Buxton RB, Alpert NM, Babikian V, Weise S, Correia JA, Ackerman RH. Evaluation of the 11CO2 positron emission tomographic method for measuring brain pH. I. pH changes measured in states of altered PCO2. J Cereb Blood Flow Metab. 1987;7:709–719. doi: 10.1038/jcbfm.1987.125. [DOI] [PubMed] [Google Scholar]
- Carson R, Barker W, Liow J-S, Johnson C. Design of a motion-compensation OSEM list-mode algorithm for resolution-recovery reconstruction for the HRRT. IEEE Trans Nucl Sci. 2003;5:3281–5. [Google Scholar]
- Collins RC, Nandi N, Smith CB, Sokoloff L. Focal seizures inhibit brain protein synthesis. Trans Am Neurol Assoc. 1980;105:43–46. [PubMed] [Google Scholar]
- Cselényi Z, Olsson H, Halldin C, Gulyás B, Farde L. A comparison of recent parametric neuroreceptor mapping approached based on measurements with the high affinity PET radioligands [11C]FLB 457 and [11C]WAY 100635. Neuroimage. 2006;32:1690–1708. doi: 10.1016/j.neuroimage.2006.02.053. [DOI] [PubMed] [Google Scholar]
- Gunn R, Lammertsma A, Hume S, Cunningham V. Parametric Imaging of Ligand-Receptor Binding in PET Using a Simplified Reference Region Model. Neuroimage. 1997;6:279–287. doi: 10.1006/nimg.1997.0303. [DOI] [PubMed] [Google Scholar]
- Gunn RN, Yap JT, Wells P, Osman S, Price P, Jones T, Cunningham VJ. A general method to correct PET data for tissue metabolites using a dual-scan approach. J Nucl Med. 2000;41:706–11. [PubMed] [Google Scholar]
- Hurvich CM, Tsai C-L. Regression and time series model selection in small samples. Biometrika. 1989;76:297–307. [Google Scholar]
- Iida H, Higano S, Tomura N, Shishido F, Kanno I, Miura S, Murakami M, Takahashi K, Sasaki H, Uemura K. Evaluation of regional differences of tracer appearance time in cerebral tissues using [15O]water and dynamic positron emission tomography. J Cereb Blood Flow Metab. 1988;8:285–288. doi: 10.1038/jcbfm.1988.60. [DOI] [PubMed] [Google Scholar]
- Ingvar MC, Maeder P, Sokoloff L, Smith CB. Effects of ageing on local rates of cerebral protein synthesis in Sprague-Dawley rats. Brain. 1985;108 (Pt 1):155–170. doi: 10.1093/brain/108.1.155. [DOI] [PubMed] [Google Scholar]
- Koeppe RA, Holden JE, Ip WR. Performance comparison of parameter estimation techniques for quantitation of local cerebral blood flow by dynamic positron computed tomography. J Cereb Blood Flow Metab. 1985;5:224–234. doi: 10.1038/jcbfm.1985.29. [DOI] [PubMed] [Google Scholar]
- Lajtha A, Latzkovits L, Toth J. Comparison of turnover rates of proteins of the brain, liver and kidney in mouse in vivo following long term labeling. Biochim Biophys Acta. 1976;425:511–520. doi: 10.1016/0005-2787(76)90015-0. [DOI] [PubMed] [Google Scholar]
- Lammertsma AA, Brooks DJ, Frackowiak RSJ, Beaney RP, Herold S, Heather JD, Palmer AJ, Jones T. Measurement of glucose utilisation with [18F]2-fluoro-2-deoxy-D-glucose: A comparison of different analytical methods. J Cereb Blood Flow Metab. 1987;7:161–172. doi: 10.1038/jcbfm.1987.39. [DOI] [PubMed] [Google Scholar]
- Landaw EM, DiStefano JJ. Multiexponential, multicompartmental, and noncompartmental modeling. II. Data analysis and statistical considerations. Am J Physiol. 1984;246:R665–R677. doi: 10.1152/ajpregu.1984.246.5.R665. [DOI] [PubMed] [Google Scholar]
- Nakanishi H, Sun Y, Nakamura RK, Mori K, Ito M, Suda S, Namba H, Storch FI, Dang TP, Mendelson W, Mishkin M, Kennedy C, Gillin JC, Smith CB, Sokoloff L. Positive correlations between cerebral protein synthesis rates and deep sleep in Macaca mulatta. Eur J Neurosci. 1997;9:271–279. doi: 10.1111/j.1460-9568.1997.tb01397.x. [DOI] [PubMed] [Google Scholar]
- Qin M, Kang J, Burlin TV, Jiang C, Smith CB. Postadolescent changes in regional cerebral protein synthesis: an in vivo study in the FMR1 null mouse. J Neurosci. 2005;25:5087–5095. doi: 10.1523/JNEUROSCI.0093-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K, Mies G, Sokoloff L. Model of kinetic behavior of deoxyglucose in heterogeneous tissues in brain: a reinterpretation of the significance of parameters fitted to homogeneous tissue models. J Cereb Blood Flow Metab. 1991;11 :10–24. doi: 10.1038/jcbfm.1991.2. [DOI] [PubMed] [Google Scholar]
- Schmidt K, Lucignani G, Moresco RM, Rizzo G, Gilardi MC, Messa C, Colombo F, Fazio F, Sokoloff L. Errors introduced by tissue heterogeneity in estimation of local cerebral glucose utilization with current kinetic models of the [18F]fluorodeoxyglucose method. J Cereb Blood Flow Metab. 1992;12:823–834. doi: 10.1038/jcbfm.1992.114. [DOI] [PubMed] [Google Scholar]
- Schmidt KC, Mies G, Dienel GA, Cruz NF, Crane AM, Sokoloff L. Analysis of time courses of metabolic precursors and products in heterogeneous rat brain tissue: limitations of kinetic modeling for predictions of intracompartmental concentrations from total tissue activity. J Cereb Blood Flow Metab. 1995;15:474–484. doi: 10.1038/jcbfm.1995.59. [DOI] [PubMed] [Google Scholar]
- Schmidt KC, Cook MP, Qin M, Kang J, Burlin TV, Smith CB. Measurement of regional rates of cerebral protein synthesis with L-[1–11C]leucine and PET with correction for recycling of tissue amino acids: I. Kinetic modeling approach. J Cereb Blood Flow Metab. 2005;25:617–628. doi: 10.1038/sj.jcbfm.9600067. [DOI] [PubMed] [Google Scholar]
- Siesjo BK, Thompson WO. The rate of incorporation of gaseous 14CO2 into brain tissue constituents. Experientia. 1965;20:98–99. doi: 10.1007/BF02151265. [DOI] [PubMed] [Google Scholar]
- Smith CB, Kang J. Cerebral protein synthesis in a genetic mouse model of phenylketonuria. Proc Natl Acad Sci USA. 2000;97:11014–11019. doi: 10.1073/pnas.97.20.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CB, Schmidt KC, Qin M, Burlin TV, Cook MP, Kang J, Saunders RC, Bacher JD, Carson RE, Channing MA, Eckelman WC, Herscovitch P, Laverman P, Vuong BK. Measurement of regional rates of cerebral protein synthesis with L-[1–11C]leucine and PET with correction for recycling of tissue amino acids: II. Validation in rhesus monkeys. J Cereb Blood Flow Metab. 2005;25:629–640. doi: 10.1038/sj.jcbfm.9600066. [DOI] [PubMed] [Google Scholar]
- Sugiura N. Further analysis of the data by Akaike’s information criterion and the finite corrections. Commun Stat Theory Meth. 1978;A7:13–26. [Google Scholar]
- Sun Y, Deibler GE, Jehle J, Macedonia J, Dumont I, Dang T, Smith CB. Rates of local cerebral protein synthesis in the rat during normal postnatal development. Am J Physiol. 1995;268:R549–R561. doi: 10.1152/ajpregu.1995.268.2.R549. [DOI] [PubMed] [Google Scholar]
- Sundaram SK, Muzik O, Chugani DC, Mu F, Mangner TJ, Chugani HT. Quantification of protein synthesis in the human brain using L-[1–11C]-leucine PET: incorporation of factors for large neutral amino acids in plasma and for amino acids recycled from tissue. J Nucl Med. 2006;47:1787–1795. [PubMed] [Google Scholar]
- Tomasi G, Bertoldo A, Schmidt K, Turkheimer FE, Smith CB, Cobelli C. Residual-based comparison of different weighting schemes for pixel-wise kinetic analysis at the pixel level in PET. Neuroimage. 2008;41:T84. [Google Scholar]
- Widmann R, Kuroiwa T, Bonnekoh P, Hossmann KA. [14C]leucine incorporation into brain proteins in gerbils after transient ischemia: relationship to selective vulnerability of hippocampus. J Neurochem. 1991;56:789–796. doi: 10.1111/j.1471-4159.1991.tb01993.x. [DOI] [PubMed] [Google Scholar]
- Widmann R, Kocher M, Ernestus RI, Hossmann KA. Biochemical and autoradiographical determination of protein synthesis in experimental brain tumors of rats. J Neurochem. 1992;59:18–25. doi: 10.1111/j.1471-4159.1992.tb08870.x. [DOI] [PubMed] [Google Scholar]
- Wienhard K, Schmand M, Casey ME, Baker K, Bao J, Eriksson L, Jones WF, Knoess C, Lenox M, Lercher M, Luk P, Michel C, Reed JH, Richerzhagen N, Treffert J, Vollmar S, Young JW, Heiss WD, Nutt The ECAT HRRT: Performance and first clinical application of the new high resolution research tomograph. IEEE Trans Nucl Sci. 2002;49:104–10. [Google Scholar]
- Wu Y, Carson R. Noise reduction in the simplified reference tissue model for neuroreceptor functional imaging. J Cereb Blood Flow Metab. 2002;22 :1440–1452. doi: 10.1097/01.WCB.0000033967.83623.34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.