Abstract
Autism susceptibility candidate 2 (Auts2) is a gene associated with autism and mental retardation, whose function is unknown. Expression of Auts2 mRNA and protein were studied in the developing mouse brain by in situ hybridization, immunohistochemistry, and western blotting. Auts2 mRNA was highly expressed in the developing cerebral cortex and cerebellum, regions often affected by neuropathological changes in autism, and a few other brain regions. On embryonic day (E) 12, Auts2 mRNA was expressed in the cortical preplate, where it colocalized with Tbr1, a transcription factor specific for postmitotic projection neurons. From E16 to postnatal day 21, Auts2 was expressed most abundantly in frontal cortex, hippocampus and cerebellum, including Purkinje cells and deep nuclei. High levels of Auts2 were also detected in developing dorsal thalamus, olfactory bulb, inferior colliculus and substantia nigra. Auts2 protein showed similar regional expression patterns as the mRNA. At the cellular level, Auts2 protein was localized in the nuclei of neurons and some neuronal progenitors.
Keywords: Auts2, Gene Expression, Patterning, Neurodevelopment, Autism
1. Results and Discussion
Autism Spectrum Disorders (ASD) are early childhood developmental abnormalities leading to impaired social interaction, limited communication skills and restricted range of interests. Despite the high impact of such disorders, the etiology of ASD is poorly understood. Hypotheses regarding the etiology of autism have suggested that exposure to toxins, prenatal infection, perinatal insults, cerebral cortex microstructural anomalies, and axonal pathway disturbances could all be important (Pardo, 2007; Dieter, 2008; Courchesne, 2005; Ciaranello, 1982; Conturo, 2008). Several molecules, ranging from cell migration cues such as reelin (Fatemi, 2005) to DNA methylation enzymes such as MeCP2 (Swanberg, 2008) and cell adhesion factors such as Nrxn1 (Kim, 2008), have been associated with ASD.
Interestingly, many of the genes associated with ASD are expressed at early stages of development. Reelin, for example, guides the radial migration of newly born neurons in the developing cerebral cortex and cerebellum (Jossin, 2004). In rodents, defective reelin expression or signaling causes a severe neurological disorder with roughly inverted layering of the cerebral cortex. Other molecules, such as MeCP2 are expressed from development to adulthood, with mutant mice showing a reduction in brain size and defects in dendritic and axonal morphology (Ballas, 2009). Genes linked to the development of glutamatergic neurons (such as cortical pyramidal neurons) are also important in autism (Autism Project Consortium, 2007)
A novel human gene of unknown function, designated autism susceptibility candidate 2 (AUTS2), was associated with ASD in a study describing a chromosomal translocation, t(7;20) (q11.2;p11.2), disrupting the AUTS2 locus in monozygotic twins with autism (Sultana, 2002). More recently, AUTS2 was also linked to mental retardation (Kalscheuer, 2007). However, the expression pattern of AUTS2, its subcellular localization, and its role in brain development are currently unknown. In the present study, we analyzed the expression pattern of the mouse ortholog, Auts2, in multiple brain regions from early embryonic to postnatal development.
1.1 Auts2 protein cellular localization and pattern of expression
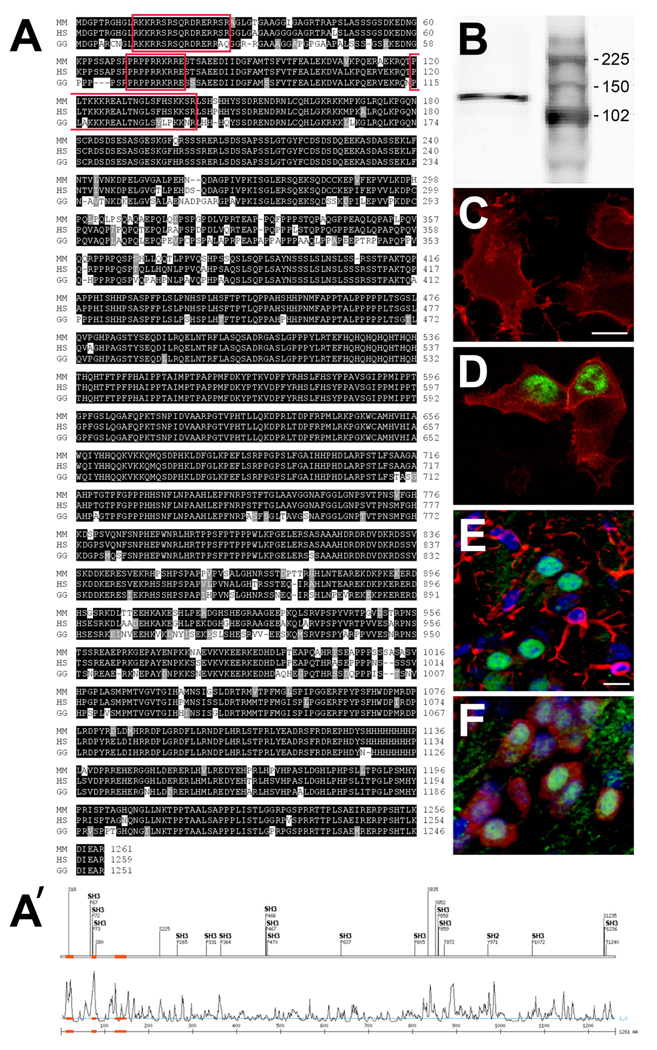
Alignment of the amino acid sequence of Auts2 from multiple species using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2) indicated that Auts2 was highly evolutionarily conserved across the species analyzed (Fig. 1A). Using Scansite software (http://scansite.mit.edu) we discovered several predicted protein-protein interaction domains (SH2 and SH3), but further analysis will be necessary to clarify the role of these sites. Using SignalP V2.0 and SignalP-HMM we were not able to find any signal peptide in the sequence of Auts2, suggesting that Auts2 is not secreted nor exposed on the cellular membrane (http://www.cbs.dtu.dk). Moreover, using WoLF PSORT we discovered several nuclear localization sequences (NLS) at the N-terminus of the protein (http://wolfpsort.org); these NLS were highly conserved between the species analyzed, suggesting that Auts2 might be a nuclear protein.
Figure 1.
Analysis of Auts2 protein sequence and cellular localization. A: alignment of Auts2 amino acid sequence from Mus Musculus (MM), Homo Sapiens (HS) and Gallus Gallus (GG). Conserved residues are represented in black, similar residues in gray, and differently charged residues are not marked. Nuclear localization sequences (NLS) are highlighted in red. A’: Upper trace shows putative SH2 and SH3 domains. Lower trace shows surface accessibility plot with putative NLS (red). B: The specificity of Auts2 antibody was tested by Western blotting on E14 protein mouse cortical extract, revealing a band at 139.8 kDa. C–D: Auts2 protein was evaluated by transfection of membrane tomato (red) alone (C) or with Auts2 (green, D) in 293T cells. Auts2 was detected in the nuclei of cells transfected with the Auts2 expression plasmid. E–F: Expression of Auts2 (green) in postnatal day (P) 21 cortex showed no obvious colocalization with GFAP (red, E). However, Auts2 was detected in neurons, as shown by colocalization with NeuN (red, P21, F). Scale bars: C–D–E–F: 10 µm.
To test this idea, we studied the cellular distribution of Auts2 protein. The specificity of the antibody used in our analysis was first checked by calculating the apparent size of the band obtained by western blotting a protein extract purified from E14 mouse cortex (Fig. 1B). The protein migrated with an apparent molecular weight (Mr) of approximately 139.8 kDa, matching the expected mouse Auts2 size (139 KDa). Secondly, we tested Auts2 antibody specificity by transfecting a pSport6 plasmid expressing Auts2 (Open Biosystem) in 293T cells. Our staining showed that Auts2 was specifically localized to the nuclei, in agreement with the presence of NLS and the absence of any signal peptide (Fig. 1C–D). In brain tissue sections, Auts2 was found in neurons only, as no obvious colocalization was found with glial fibrillary acidic protein (GFAP), while extensive colocalization was found with NeuN, a marker of mature neurons (Fig. 1 E–F).
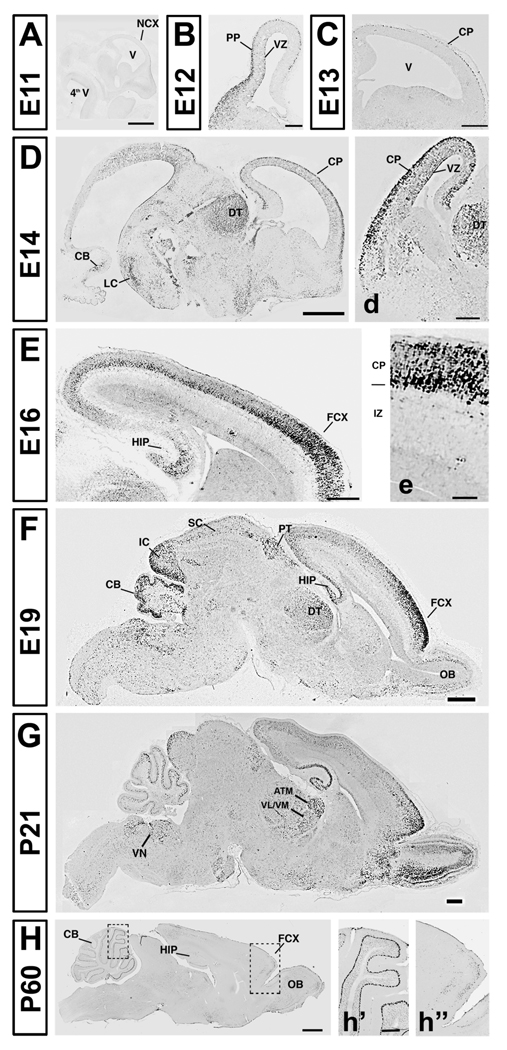
To examine Auts2 mRNA expression throughout the developing and adult brain, we used in situ hybridization. During early stages of brain neurogenesis, Auts2 mRNA was expressed very weakly; at E11 its expression was barely or not detectable at all (Fig.2 A). Between E12 and E14 several brain regions, including the developing cortex (B–D), thalamus, and cerebellum (Fig.1 D), began expressing Auts2 at a higher level. These same brain regions expressed the gene throughout embryonic development, but expression levels fluctuated with time. For example, the cerebellum showed strong expression of Auts2 at early stages of development in multiple cell types, including precursors of Purkinje cells, deep nuclei, and granule neurons (Fig.2 D). At later embryonic ages, expression became progressively more localized to the Purkinje cell layer (Fig.2 F). During adulthood, Purkinje cells were the only cell type expressing Auts2 (Fig.2 H, h’; our results matched those shown in the Allen Brain Institute Atlas of Gene Expression at http://www.brain-map.org/).
Figure 2.
Auts2 mRNA expression patterns at different embryonic and postnatal ages revealed by in situ hybridization (ISH). NCX: neocortex; V: ventricle; PP: preplate; VZ: ventricular zone; CP: cortical plate; DT: dorsal thalamus; CB: cerebellum; LC: locus ceruleus; OB: olfactory bulbs; FCX: frontal cortex; HIP: hippocampus; PT: pretectum; SC: superior colliculus; IC: inferior colliculus; ATN: anterior thalamic nuclei; VL/VM: ventrolateral/ventromedial nuclei; VN: vestibular nuclei. Scale bars: A: 500 µm; B: 200 µm; C–D–F–G: 1mm; d: 100 µm; E: 200 µm, e: 75 µm, H: 500 µm, h’ (for h’–h”) 100 µm. Ages: A: E11, B: E12, C: E13, D: E14, E: E16, F: E19, G: P21.
Refinement of Auts2 expression in relationship to ongoing regional specialization was also evident in the cerebral cortex. As depicted in Fig. 2C, expression of Auts2 was strong by E13, when a thin layer of Auts2-positive cells populated the preplate (PP). Expression was homogeneous along the rostro-caudal axis at this time. However, by E16, the Auts2 expression pattern changed as marked arealization became apparent. A strong gradient of expression was detected, with frontal cortical regions exhibiting high expression in deep and superficial layers (Fig. 2 E). In caudal cortical regions, only the deeper cortical layers exhibited Auts2 expression, which was nevertheless weaker as compared to rostral areas. At later stages of development, only the rostral region of the cortex expressed high levels of Auts2 (Fig.2 F–G–H–h”). A dynamic pattern of expression was likewise detected in dorsal thalamus, where high levels of Auts2 were evident on E14 (Fig.2 D). At later embryonic ages, the developing thalamic areas expressed Auts2 in distinct regional patterns. High levels were detected at P21 in the anterior thalamic nuclei (ATN), and in ventrolateral / ventromedial nuclei (VL/VM, Fig. 2G). By adulthood, thalamic expression of Auts2 was markedly decreased.
Several other areas exhibited transient Auts2 expression during development. For example, we observed strong expression on E14 in the locus ceruleus (LC, Fig. 2D) and in the vestibular nuclei, but mRNA levels subsequently declined. Similarly, Auts2 expression in the inferior and superior colliculi, and in the pretectum, were highest on E19 (Fig.2 F) and then decreased. By adulthood, only a few scattered Auts2 positive cells populated these areas (Fig.2 H; results matched those shown in Allen Brain Atlas at www.mouse.brain-map.org).
1.2 Auts2 expression in cortical progenitors and projection neurons
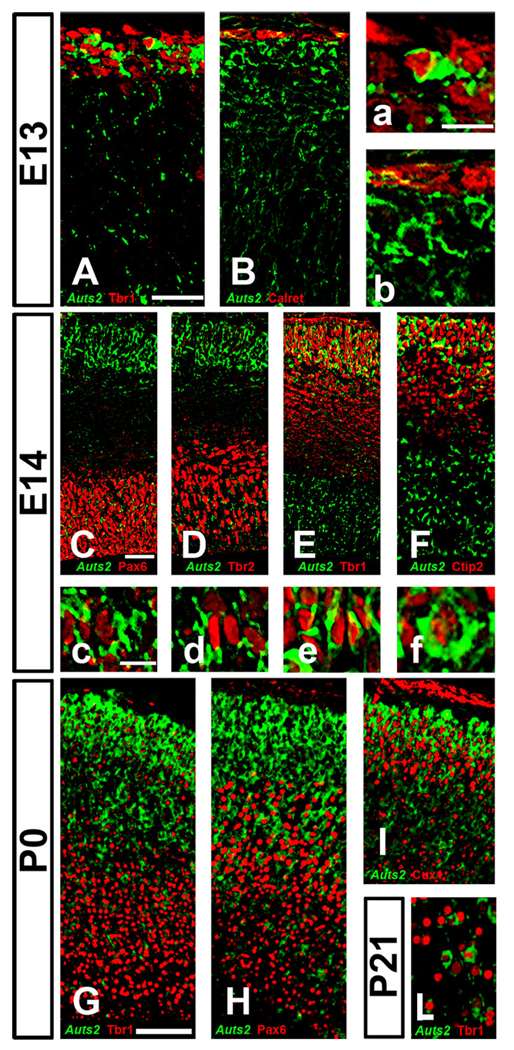
Early in development (E13), Auts2 colocalized with Tbr1, a transcription factor expressed by postmitotic neurons (Fig.3 A–a). Interestingly, no overlap was detected between Auts2 and Calretinin, a marker of Cajal-Retzius cells (Fig.3 B–b). As previously shown, at E14 cells expressing Auts2 at a high level populated the entire cortical mantle (Fig. 1D). To identify the cell types expressing Auts2 in cortex, we used antibodies against Pax6, a marker of radial glial progenitors, and Tbr2, a marker of intermediate neuronal progenitors (Englund et al., 2005). Colocalization of Auts2 with Pax6 and Tbr2 was found in the ventricular zone and subventricular zone (Fig.3 C–c; D–d). However, only a subset of all Pax6 or Tbr2 positive cells expressed Auts2, suggesting that Auts2 might be expressed in the transition between radial glia and intermediate progenitors. In the cortical plate, coexpression of Tbr1 and Auts2 was apparent in the majority of neurons consistent with the strong expression of Auts2 observed in the cortical plate at this age (Fig. 3E). While Auts2 levels were particularly high in the CP, weak or no staining at all was detected in the intermediate zone (IZ), an area where low levels of Tbr1 are expressed by migrating cells. Also on E14, Auts2 colocalized with Ctip2 (Fig.3 F–f), a protein highly expressed in the CP early during embryonic development, and in layer 5 at later ages (Arlotta et al., 2005), suggesting that Auts2 is expressed in multiple neuronal subtypes in the developing cortex.
Figure 3.
Auts2 mRNA expression in the developing cortex, revealed by ISH (green, pseudocolor). The pattern of expression is studied in association with other markers detected by immunohistochemistry (red) in the same section. A, a, E, e, G: Tbr1; B, b: calretinin; C, c: Pax6; D, d: Tbr2; F, f, H: Ctip2; I: Cux1. CP: cortical plate; IZ: intermediate zone. Scale bars: A: 50 µm (applies to A, B); a: 20 µm (applies to a, b); C: 50 µm (applies to C–F); c: 20 µm (applies to c–f); G: 100 µm (applies to G–I). Ages: A–B, a–b: E13, C–F, c–f: E14, G–I, g–i: P0.
At later stages of cortical development (P0), the pattern of Auts2 expression became progressively more superficial in frontal cortex, and coexpression with Tbr1 became more rare as Tbr1 became more selectively localized in layer 6 (Fig.3 G). Stronger coexpression was apparent with Ctip2, which is highly expressed in layer 5 (Fig. 3H), and coexpression with Cux1, an upper layer marker, was very pronounced (Fig. 3I). By P21, Auts2 expression was restricted to superficial layers in frontal cortex (Fig. 2G) and coexpression with Tbr1 was restricted to a small subset of cortical neurons (Fig. 3L). Auts2 expression in the cortex declined thereafter, and by adulthood was only weakly apparent in very superficial layers (Fig. 2H; see also Allen Brain Institute at www.mouse.brain-map.org).
1.3 Auts2 expression in the developing and adult hippocampus
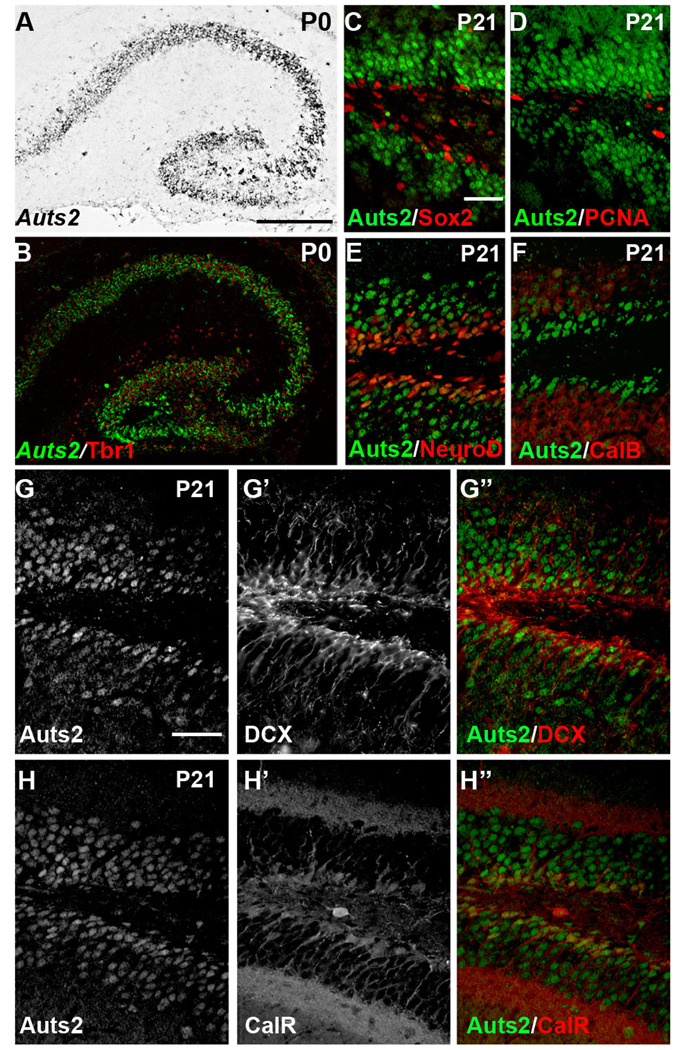
Auts2 was expressed in the hippocampal primordium from early in development (E14, Fig. 2D, d). Auts2 was not detected in the cortical hem or fimbria, directly adjacent to the developing dentate gyrus (DG) of hippocampus. After E14, the hippocampal fields showed increasing Auts2 expression, in the DG as well as CA1 and CA3. On E19 (Fig. 2F), strong Auts2 expression was apparent in the dentate migratory stream, in both the suprapyramidal and infrapyramidal blades of the dentate gyrus, throughout the hilus, and in the CA fields as well. By P0, Auts2 was expressed in the CA fields and DG, and colocalization with Tbr1 was apparent in the majority of cells in these regions (Fig. 4A, B)
Figure 4.
Auts2 mRNA and protein expression in the developing hippocampus. A–B: At P0, Auts2 expression, as detected by ISH (gray or green), was apparent throughout the developing hippocampus where it colocalized with Tbr1 immunofluorescence (red). C–H: Auts2 protein, detected by immunofluorescence at P21, was apparent throughout the granule cell layer (GCL) of the dentate gyrus. Coexpression with specific markers of progenitor cells and differentiating neurons was assessed by double immunofluorescent labeling with C) Sox2; D) PCNA; E) NeuroD1; F) calbindin (CalB); G–G”) DCX; H–H”) calretinin (CalR). Scale bars: A 100 µm (applies to A, B); C: 50 µm (applies to C–F) G: 50 µm (applies to G–H”).
By P21, a time at which development of the hippocampus is largely complete and the subgranular zone (SGZ) neurogenic niche has been established, Auts2 was expressed throughout the SGZ and the granule cell layer (GCL), suggesting that Auts2 exhibits an expression pattern similar to the GCL marker Prox1 (Fig. 4) (Elliott et al., 2001). In the DG, Auts2 also colocalized with markers of immature neurons, including NeuroD, DCX, and calretinin, suggesting that it is upregulated in newborn granule neurons (Fig. 4E, G, H). Generally, Auts2 did not colocalize with markers of progenitor cells such as Sox2 (Fig. 4C). Accordingly, coexpression with markers of dividing cells such as PCNA (Fig. 4D) was not apparent in the SGZ. Auts2 expression persisted into mature granule neurons in the DG, as evidenced by colocalization with calbindin in the outer margins of the GCL (Fig. 4F). Auts2 continued to be expressed in both the CA fields and the GCL of the DG into adulthood (Fig. 2H; results identical to Allen Brain Map of adult gene expression).
1.4 Comparison of Auts2 and Tbr1 outside the cortex
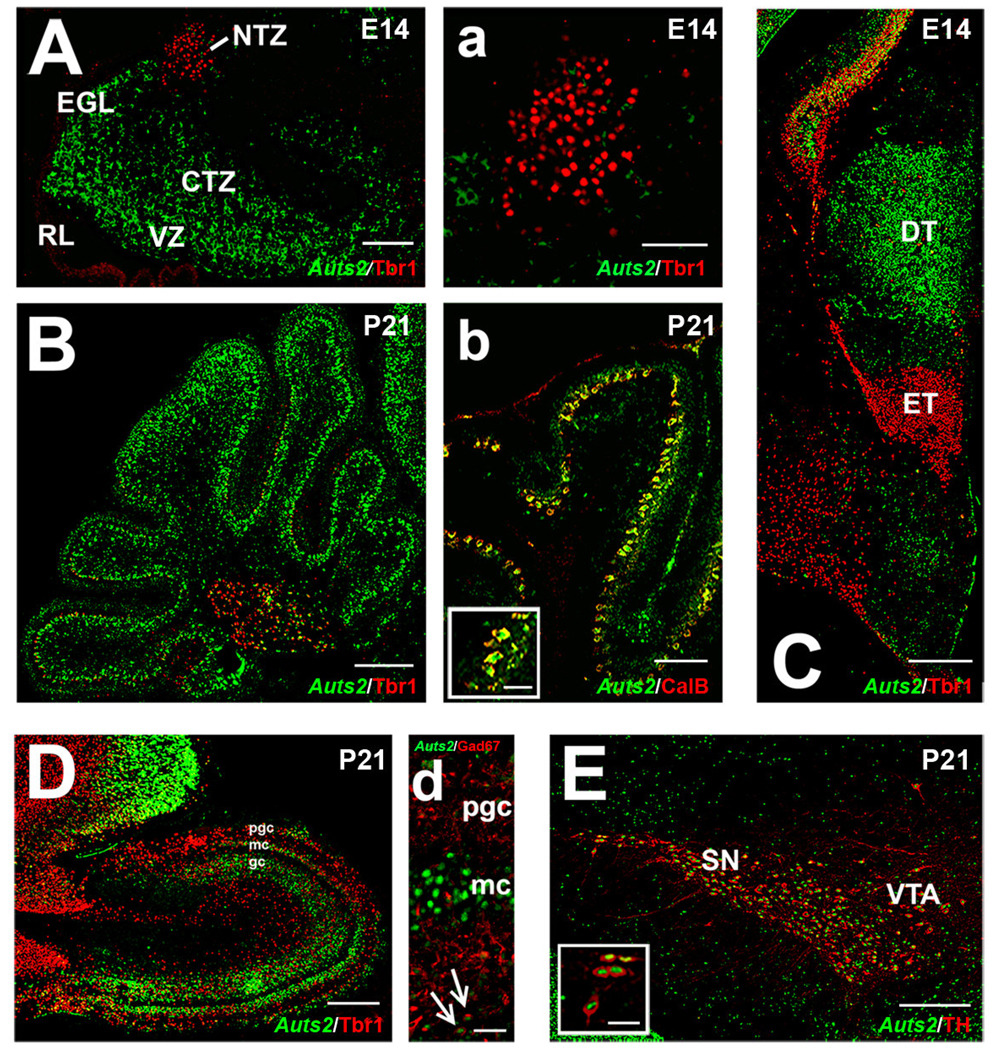
Given the diversity of areas expressing Auts2 and the notable overlap with Tbr1 in cortex, we analyzed the possibility that this relationship might exist in other regions expressing Auts2. We first focused our attention on the cerebellum (Fig. 5A–B), an area known to express Tbr1 in a transcriptional cascade similar to that of the cortex (Fink et al., 2006). On E14 (Fig.5 A), the ventricular zone (VZ), an area that gives birth to Purkinje cells and most of the GABAergic and inhibitory interneurons, and the cortical transitory zone (CTZ) containing immature Purkinje cell precursors, both expressed high levels of Auts2. Conversely, the rhombic lip (RL), the external granular layer and the nuclear transitory zone (NTZ), areas committed to give birth to cell types such as unipolar brush cells and deep cerebellar nuclei, showed only weak expression of Auts2 (Fig.5 A). As previously demonstrated (Fink et al., 2006), these latter areas express Pax6, Tbr2 and Tbr1 in a temporal fashion that begins in the RL and culminates in the nuclear transitory zone (NTZ), where Tbr1 and Tbr2 are highly expressed. The low level of Auts2 expression in these areas indicated that Auts2 and Tbr1 are not strictly coexpressed in all brain regions, and suggested that Auts2 is expressed in a much wider range of neuronal subtypes than Tbr1. Lack of Auts2 and Tbr1 coexpression in the cerebellum was even more noticeable at P21 (Fig.5 B), when Auts2 was localized to calbindin-positive Purkinje cells (Fig.5 b), while Tbr1 was expressed only in the deep nuclei, where scattered Auts2-positive/Tbr1-negative cells (presumably GABAergic interneurons) were found.
Figure 5.
Expression of Auts2 mRNA in brain regions outside the cortex at different stages of development. A–a) Double labeling for Auts2 mRNA revealed by in situ hybridization (green) and Tbr1 protein by immunofluorescence (red) in the E14 cerebellum. NTZ: nuclear transitory zone; EGL: external granular layer; CTZ: cortical transitory zone; RL: rhombic lip; VZ: ventricular zone. B) Postnatal day (P) 21 cerebellum labeled with Auts2 (green) in relationship to Tbr1 (red); b) expression of Auts2 protein (green, immunofluorescence) in Purkinje cells identified by calbindin immunofluorescence (red). C) Auts2 mRNA (green) was expressed in the dorsal thalamus (DT), in a complementary fashion compared to the eminentia thalami (ET), stained by Tbr1 (red, immunofluorescence). D) Auts2 mRNA (green) was localized in the P21 olfactory bulb mitral cells (MC) and the periglomerular cells (PGC); overlap with Tbr1 (red) was found in some MC. Also, some overlap of Auts2 mRNA (green) with GAD67 (red, d), a marker of GABAergic neurons, was detected in the GC (arrows). E) Strong coexpression of Auts2 protein (green) and TH (red) was found in the substantia nigra (SN) and the ventral tegmental area (VTA). Scale bars: A: 100 µm; a: 50 µm; B: 100 µm; b: 50 µm; inset b: 20 µm; C: 200 µm; D: 100 µm; d: 30 µm; E: 100 µm; inset E: 20 µm. Ages: A–a: E14, B–b: P21, C: E14, D–d: P21, E: P21.
The pattern of Auts2 expression was particularly interesting around the eminentia thalami (ET), an area of high Tbr1 expression. Tbr1 and Auts2 were expressed in a very complementary fashion around this region (Fig. 5C), with Tbr1 confined to the ET and Auts2 specifically localized to the dorsal thalamus.
In the olfactory bulbs, high levels of Tbr1 were detected from early ages (E14) to adulthood. This structure also exhibited high expression of Auts2. On P21 (Fig. 5D), mitral cells and periglomerular cells expressed both Auts2 and Tbr1, although coexpression was apparent only within a subset of these cells (Fig. 5D). In rare examples, Auts2 was detected in the granular layer (Fig. 5D, arrows), possibly in glutamatergic precursors (Brill et al., in press). These results, along with those from the cerebral cortex and hippocampus, suggest that Auts2 and Tbr1 might be coexpressed mainly in glutamatergic neuron populations in the forebrain, while other transcriptional regulators likely influence the expression of Auts2 in other regions.
Besides glutamatergic neurons (cortex, olfactory bulbs, hippocampus) and GABAergic neurons (Purkinje cells), we also detected high levels of Auts2 in tyrosine hydroxylase (TH) positive dopaminergic neurons of the substantia nigra (SN) and the ventral tegmental area (VTA) on P21 (Fig. 5E). Interestingly, Auts2 was expressed in virtually all of the dopaminergic TH positive neurons. Similar to the other brain regions analyzed, Auts2 expression tended to decrease with age in these neurons, such that in adulthood, expression in the VTA and the SN was very low.
1.5 Different brain areas and neuronal types involved in autism express Auts2
Our results indicate that Auts2 is a nuclear protein that is highly expressed in developing neurons of certain brain regions, notably the frontal cortex and cerebellum, which have been closely associated with the neuropathology of autism (Fatemi, 2002; Palmen, 2004; Amaral, 2008; Casanova, 2002; Geschwind, 2007; Bauman, 2005). Our findings suggest that Auts2 could be important for the migration, axon guidance, dendrite elaboration, synaptogenesis, or related aspects of neuronal development in these key brain regions. However, the precise function of Auts2 remains unknown and will require further studies using genetic perturbation approaches, such as targeted knockout.
Notably, we observed heterogeneity of neuron types expressing Auts2, ranging from glutamatergic neurons in the cortex, to GABAergic Purkinje cells in the cerebellum, to TH positive neurons in the substantia nigra (SN) and the VTA, the sites of most dopamine biosynthesis in the brain. This wider expression of Auts2 in the developing brain may also link with autism, as ASD frequently results from dysfunction of several neuronal types in different brain areas (Pardo, 2007; Polleux, 2005; Muhle, 2004).
The frontal expression of Auts2 in cerebral cortex was particularly interesting. On E14, Auts2 mRNA was homogeneous along the rostro-caudal axis. Thereafter, a gradient of high rostral to low caudal expression of Auts2 was detectable by E16. The arealization of Auts2 expression appeared complete on P0, when expression was almost entirely confined to the frontal cortex. Interestingly, Auts2 expression was also associated with the ongoing stratification of cortical layers. Tbr1 or Ctip2 positive cells tended to express high levels of Auts2 on E14 but lower levels on P0, when the upper layer marker Cux1 strongly colocalized with Auts2. These findings correlated with the well known inside-first, outside-last layering mechanism of cortical development (Rakic, 1995). The dynamic expression of Auts2 coupled to different layer specific markers suggests that Auts2 is expressed by newly born neurons migrating into their final cortical location, and that Auts2 expression declines as these neurons become fully mature. Taken together, these results suggest that in the cortex, Auts2 may be expressed in a transient phase of neuronal maturation or differentiation.
Outside the cortex, Auts2 was strongly expressed in the hippocampus from very early in embryonic development to adulthood. Unlike the cerebral cortex, Auts2 in hippocampus was present both in immature neurons and in developmentally older cells throughout the GCL, suggesting that Auts2 expression is maintained in mature granule neurons. In adulthood, Auts2 expression decreased in the CA fields and became progressively restricted to the DG, where it was apparent throughout the GCL. These observations suggest a dual role for Auts2 in the hippocampus, where it may be important for both the maturation and the maintenance, or perhaps the plasticity, of granule neurons.
Our findings regarding Auts2 expression suggest interesting links to the neurobiology of ASD (Palmen, 2004). Defects in the structural organization of the cortex are well described in ASD: cortical minicolumns, basic units formed by chains of neurons vertically oriented across the layers (Rakic, 1995), appear more numerous, smaller and less dense than normal, especially in frontal cortex (Pardo, 2007; Casanova, 2002). Reelin, a molecule playing a pivotal role in the migration of neurons from the cortical VZ/SVZ to their final destination in the CP (Rakic, 1995), has been associated with ASD (Fatemi, 2005). Moreover, several studies found an association between autism and several molecules associated with glutamatergic and GABAergic transmission and the acquisition of fully mature neuronal subtypes (Polleux, 2005). The overall picture of the cortical defects displayed in ASD suggests that glutamatergic and GABAergic tones play a pivotal role. Current hypotheses suggest that autism may be a hypoglutamatergic disorder (Purcell, 2001; Jamain, 2002; Serajee, 2003); conversely, a decreased level of inhibition has also been hypothesized (Rubenstein, 2003; Belmonte, 2004). These observations leave open the possibility for a more complex hypothesis in which different brain regions display unbalanced glutamatergic and GABAergic activity ratios (Polleux, 2005). Accordingly, perturbed Auts2 expression might, at least in part, contribute to the wide heterogeneity of the phenotypes displayed in autism. Future studies will focus on examining the functional role of Auts2 in neuronal development.
2. Experimental Procedures
2.1 Animals and tissue preparation
Wild-type CD1 mice were purchased from Charles River Laboratories and maintained as a breeding colony. Animals were housed under standard conditions (12 h light/dark cycle). All experimental procedures were approved by Institutional Animal Care and Use Committees at the University of Washington and Seattle Children's Hospital Research Institute. Timed-pregnant embryos were identified by vaginal plugs, with noon on the day of plug appearance designated embryonic day (E) 0.5. Animals were killed by cervical dislocation under Avertin anesthesia (pregnant females) or cryoanesthesia (neonatal mice), as described previously (Englund, 2005). Embryos were rapidly removed and perfused with 4% paraformaldehyde diluted in PBS. Embryonic brains were post-fixed by immersion in cold (4 °C) 4% paraformaldehyde for 4–6 h (for immunofluorescence) or overnight for in-situ hybridization. Brains were cryoprotected in cold-buffered sucrose (30% in 0.1 M sodium phosphate, pH 7.0), frozen in optimum cutting temperature compound (Sakura Finetek, Torrance, CA), cryosectioned at 12 µm, and mounted on Superfrost Plus slides (Thermo Fisher Scientific, Waltham, MA). Slides were stored at −80 °C until needed. The figures shown are representative of at least three separate experiments run on three different animals. For western blotting analysis tissues from E14 embryos were surgically dissected and rapidly frozen on dry ice, then stored at −80°C.
2.2 Western Blotting
Cortexes from E14 embryos were mechanically triturated in a hypotonic lysis buffer [150 mM Tris, 150 mM NaCl, 5 mM EDTA, 1% protease inhibitor cocktail (Sigma-Aldrich), pH 7.6]. Aliquots of 20 µg of protein were combined with β-Mercaptoethanol and SDS reducing buffer, electrophoretically separated and transferred to an Immobilon-FL PVDF membrane (Millipore) for 1 hour at 100V in the cold. Membranes were blocked in Odyssey Blocking Buffer (LI-COR Biosciences) for 1 hour and incubated over night in the cold in a 1:250 dilution of Auts2 rabbit antibody in blocking solution. After washing, the membranes were incubated into a dilution 1:5000 of goat anti-rabbit infrared dye-labeled secondary antibody (Alexa Fluor 680, Molecular Probes, Inc.) for 2 hour at room temperature protected from light then washed thoroughly. Infrared fluorescence was used for signal detection and quantitation (Odyssey Infrared Imaging System, LI-COR Biosciences).
2.3 Cell Transfection
293T cells were cotransfected with expression plasmids containing the sequence for a membrane localized tdTomato fluorescent protein (Shaner, 2004) and full length Auts2 (pSport6-Auts2) using Fugene 6 transfection reagent (Roche). Transfections were carried out for 12 hours in serum free medium (DMEM, Mediatech), after which time, cells were transferred to growth medium (DMEM plus 10% fetal bovine serum, Mediatech) and cultured for an additional 18 hours. Cells were then fixed in 4% paraformaldehyde for 30 minutes and processed for immunofluorescent detection of Auts2 as described.
2.4 Immunofluorescence
Slide-mounted cryostat sections were thawed and washed in PBS. Single- and double-label immunofluorescence were done routinely as described (Hodge, 2008). Blocking solution, consisting of PBS with 0.1% Triton X-100 (PBS-T), 10% normal horse serum was applied to slides for 30 minutes. Slides were then incubated overnight at 4 °C with primary antibodies (listed below), rinsed 3 times in PBS for 30 min, and then incubated for 1 h at room temperature with fluorescent secondary antibodies (Alexa Fluor, Invitrogen). Sections were rinsed 3 times in PBS for 10 min and counterstained with DAPI (0.01%, Invitrogen). The following primary antibodies were used: rabbit polyclonal Auts2 (1:200, HPA000390, Sigma Aldrich); mouse monoclonal anti-glial fibrillary acidic protein (GFAP) (1:1000, MAB360, Millipore/Chemicon); mouse monoclonal anti-NeuN (1:1000, MAB337, Chemicon); rabbit polyclonal anti-Tbr1 (1:1500, R.F.H. laboratory); mouse anti-Calretinin (1:1000, MAB1568, Chemicon); rabbit polyclonal anti-Pax6 (1:1000, Covance); anti rabbit polyclonal anti-Tbr2 (1:2000, R.F.H. laboratory); rat anti Ctip2 (1:1000, AB18465, AbCam); rabbit anti-Cux1 (1:1000, sc-13024, Santa Cruz Biotechnology); goat Doublecortin (1:250, sc-8066, Santa Cruz Biotechnology); goat NeuroD (1:400, sc-1084, Santa Cruz Biotechnology); mouse Calbindin (1:1000, CB-955, Sigma-Aldrich); mouse anti-GAD67 (1:1000, MAB5406, Chemicon); mouse monoclonal anti-TH (1:5000, T1299, Sigma Aldrich). Secondary antibodies were conjugates of Alexa Fluors 488, 568, and 647 (1:500, Invitrogen).
2.5 In Situ Hybridization
Auts2 full-length mouse cDNA in a pSport6 expression vector was purchased from Open Biosystem (cat. Number EMM1002-5823733). A BamHI and EcoRV 1243bp fragment (from nucleotide 47 to 1290) was excised and cloned into a BamHI/EcoRV linearized pBluescript II SK. DIG-labeled probes were synthesized by T3 polymerase transcription on a HindIII linearized template. As a control a T7 transcribed sense probe was obtained using a SpeI linearized template. In situ hybridizations were run on slide-mounted cryostat sections using a protocol based on a previously published paper (Tongiorgi, 1998). Double labeling by in situ hybridization and immunofluorescence in the same section was also done as described previously (Fink, 2006). For figures, the in situ hybridization color reaction was photographed in bright field gray scale, inverted using Adobe Photoshop CS2, and pasted in the red color channel for dark field pseudocolor in combination with Pax6, Tbr2, or Tbr1 immunofluorescence.
Acknowledgments
We thank Laura Jansen and William Roden for their assistance with western blotting. This work was supported by National Institutes of Health Grant to R.F.H. (R01 NS050248). F.B. was the recipient of a research fellowship from the Universita’ degli Studi di Milano and R.D.H. was supported by a research fellowship from the Heart & Stroke Foundation of Canada.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amaral D, Schumann C, Nordahl C. Neuroanatomy of autism. Trends. Neurosci. 2008;31:137–145. doi: 10.1016/j.tins.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Autism Genome Project Consortium. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat. Genet. 2007;39(3):319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N, Lioy D, Grunseich C, Mandel G. Non–cell autonomous influence of MeCP2-deficient glia on neuronal dendritic morphology. Nat. Neurosci. 2009;12:311–317. doi: 10.1038/nn.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman ML, Kemper TL. Neuroanatomic observations of the brain in autism: a review and future directions. Int. J. Dev. Neurosci. 2005;23(2–3):183–187. doi: 10.1016/j.ijdevneu.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Belmonte M, Cook E, Anderson G, Rubenstein J, Greenough W, Beckel-Mitchener A, Courchesne E, Boulanger L, Powell S, Levitt P, Perry E, Jiang Y, Delorey T, Tierney E. Autism as a disorder of neural information processing: directions for research and targets for therapy. Mol. Psychiatry. 2004;9:646–663. doi: 10.1038/sj.mp.4001499. [DOI] [PubMed] [Google Scholar]
- Belmonte MK, Allen G, Beckel-Mitchener A, Boulanger LM, Carper RA, Webb SJ. Autism and abnormal development of brain connectivity. J. Neurosci. 2004;24:9228–9231. doi: 10.1523/JNEUROSCI.3340-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill MS, Ninkovic J, Winpenny E, Hodge RD, Ozen I, Yang R, Erdelyi F, Szabo G, Parras C, Guillemot F, Frotscher M, Berninger B, Hevner RF, Raineteau O, Götz M. Adult generation of glutamatergic olfactory bulb interneurons. Nat. Neurosci. doi: 10.1038/nn.2416. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova MF, Buxhoeveden DP, Switala AE, Roy E. Minicolumnar pathology in autism. Neurology. 2002;58:428–432. doi: 10.1212/wnl.58.3.428. [DOI] [PubMed] [Google Scholar]
- Ciaranello RD, VandenBerg SR, Anders TF. Intrinsic and extrinsic determinants of neuronal development: relation to infantile autism. J. Autism Dev. Disord. 1982;12(2):115–145. doi: 10.1007/BF01531304. [DOI] [PubMed] [Google Scholar]
- Conturo TE, Williams DL, Smith CD, Gultepe E, Akbudak E, Minshew NJ. Neuronal fiber pathway abnormalities in autism: an initial MRI diffusion tensor tracking study of hippocampo-fusiform and amygdalo-fusiform pathways. J. Int. Neuropsychol. Soc. 2008;14(6):933–946. doi: 10.1017/S1355617708081381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne E, Redcay E, Morgan JT, Kennedy DP. Autism at the beginning: microstructural and growth abnormalities underlying the cognitive and behavioral phenotype of autism. Dev. Psychopathol. 2005;17(3):577–597. doi: 10.1017/S0954579405050285. [DOI] [PubMed] [Google Scholar]
- Dietert RR, Dietert JM. Potential for early-life immune insult including developmental immunotoxicity in autism and autism spectrum disorders: focus on critical windows of immune vulnerability. J. Toxicol. Environ. Health. B. Crit. Rev. 2008;11(8):660–680. doi: 10.1080/10937400802370923. [DOI] [PubMed] [Google Scholar]
- Elliott RC, Khademi S, Pleasure SJ, Parent JM, Lowenstein DH. Differential regulation of basic helix-loop-helix mRNAs in the dentate gyrus following status epilepticus. Neuroscience. 2001;106:79–88. doi: 10.1016/s0306-4522(01)00198-1. [DOI] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J. Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi S, Snow A, Stary J, Araghiniknam M, Reutiman T, Lee S, Brooks A, Pearce D. Reelin signaling is impaired in autism. Biol Psychiatry. 2005;57:777–787. doi: 10.1016/j.biopsych.2004.12.018. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Halt AR, Stary JM, Kanodia R, Schulz SC, Realmuto GR. Glutamic acid decarboxylase 65 and 67 kDa proteins are reduced in autistic parietal and cerebellar cortices. Biol. Psychiatry. 2002;52:805–810. doi: 10.1016/s0006-3223(02)01430-0. [DOI] [PubMed] [Google Scholar]
- Fink AJ, Englund C, Daza RA, Pham D, Lau C, Nivison M, Kowalczyk T, Hevner RF. Development of the deep cerebellar nuclei: transcription factors and cell migration from the rhombic lip. J. Neurosci. 2006;26:3066–3076. doi: 10.1523/JNEUROSCI.5203-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Levitt P. Autism spectrum disorders: developmental disconnection syndromes. Curr. Opin. Neurobiol. 2007;17(1):103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Hodge RD, Kowalczyk TD, Wolf SA, Encinas JM, Rippey C, Enikolopov G, Kempermann G, Hevner RF. Intermediate progenitors in adult hippocampal neurogenesis: Tbr2 expression and coordinate regulation of neuronal output. J. Neurosci. 2008;28:3707–3717. doi: 10.1523/JNEUROSCI.4280-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamain S, Betancur C, Quach H, Philippe A, Fellous M, Giros B, Gillberg C, Leboyer M, Bourgeron T Study PARISP. Linkage and association of the glutamate receptor 6 gene with autism. Mol Psychiatry. 2002;7:302–310. doi: 10.1038/sj.mp.4000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossin Y. Neuronal migration and the role of reelin during early development of the cerebral cortex. Mol. Neurobiol. 2004;30:225–251. doi: 10.1385/MN:30:3:225. [DOI] [PubMed] [Google Scholar]
- Kalscheuer VM, FitzPatrick D, Tommerup N, Bugge M, Niebuhr E, Neumann LM, Tzschach A, Shoichet SA, Menzel C, Erdogan F, Arkesteijn G, Ropers HH, Ullmann R. Mutations in autism susceptibility candidate 2 (AUTS2) in patients with mental retardation. Hum. Genet. 2007;121(3–4):501–509. doi: 10.1007/s00439-006-0284-0. [DOI] [PubMed] [Google Scholar]
- Kim H, Kishikawa S, Higgins A, Seong I, Donovan D, Shen Y, Lally E, Weiss L, Najm J, Kutsche K. Disruption of Neurexin 1 Associated with Autism Spectrum Disorder. Am. J. Human Genetics. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhle R, Trentacoste S, Rapin I. The Genetics of Autism. Pediatrics. 2004;113:472–486. doi: 10.1542/peds.113.5.e472. [DOI] [PubMed] [Google Scholar]
- Palmen S. Neuropathological findings in autism. Brain. 2004;127:2572–2583. doi: 10.1093/brain/awh287. [DOI] [PubMed] [Google Scholar]
- Pardo C, Eberhart C. The Neurobiology of Autism. Brain Pathology. 2007;17:434–447. doi: 10.1111/j.1750-3639.2007.00102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polleux F, Lauder J. Toward a developmental neurobiology of autism. Mental Retardation Dev. Dis. Res. Rev. 2005;10:303–317. doi: 10.1002/mrdd.20044. [DOI] [PubMed] [Google Scholar]
- Purcell AE, Jeon OH, Zimmerman AW, Blue ME, Pevsner J. Postmortem brain abnormalities of the glutamate neurotransmitter system in autism. Neurology. 2001;57(9):1618–1628. doi: 10.1212/wnl.57.9.1618. [DOI] [PubMed] [Google Scholar]
- Rakic P, Caviness VS. Cortical development: view from neurological mutants two decades later. Neuron. 1995;14(6):1101–1104. doi: 10.1016/0896-6273(95)90258-9. [DOI] [PubMed] [Google Scholar]
- Rubenstein J, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2003;2:255–267. doi: 10.1034/j.1601-183x.2003.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serajee F. The metabotropic glutamate receptor 8 gene at 7q31: partial duplication and possible association with autism. J. Med. Genetics. 2003;40:42–42. doi: 10.1136/jmg.40.4.e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY. Improved monomeric red, orange, and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature Biotechnology. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Sultana R, Yu C, Yu J, Munson J, Chen D, Hua W, Estes A, Cortes F, De La Barra F, Yu D. Identification of a Novel Gene on Chromosome 7q11.2 Interrupted by a Translocation Breakpoint in a Pair of Autistic Twins. Genomics. 2002;80:129–134. doi: 10.1006/geno.2002.6810. [DOI] [PubMed] [Google Scholar]
- Swanberg S, Nagarajan R, Peddada S, Yasui D, Lasalle J. Reciprocal co-regulation of EGR2 and MECP2 is disrupted in Rett syndrome and autism. Human. Mol. Genetics. 2008;18:525–534. doi: 10.1093/hmg/ddn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tongiorgi E, Righi M, Cattaneo A. A non-radioactive in situ hybridization method that does not require RNAse-free conditions. J. Neurosci. Methods. 1998;85(2):129–139. doi: 10.1016/s0165-0270(98)00123-x. [DOI] [PubMed] [Google Scholar]