Abstract
High Mobility Group Nucleosome-binding (HMGN) proteins are architectural non-histone chromosomal proteins that bind to nucleosomes and modulate the structure and function of chromatin. The interaction of HMGN proteins with nucleosomes is dynamic and the proteins compete with the linker histone H1 chromatin binding sites. HMGNs reduce the H1 mediated compaction of the chromatin fiber and facilitate the targeting of regulatory factors to chromatin. They modulate the cellular epigenetic profile, affect gene expression, and impact the biological processes such as development and the cellular response to environmental and hormonal signals. Here we review the role of HMGN in chromatin structure, the link between HMGN proteins and histone modifications, and discuss the consequence of this link on nuclear processes and cellular phenotype.
Keywords: High Mobility Group Proteins, Chromatin, Nucleosomes, Histones, Epigenesis
Introduction
The chromatin fiber stores and organizes the genetic information encoded in the sequence of the DNA and contains the epigenetic regulatory information encoded in histone variants and in the covalent chemical modifications of nucleosomes. The structure of the chromatin fiber and the accessibility of nucleosomes to various regulatory factors are key elements affecting DNA-dependent nuclear activities such as transcription, replication, recombination and repair, and the orderly progression of biological processes such as the cell cycle, development and differentiation. The ability of chromatin to affect this wide range of processes is related to its dynamic structure; chromatin compaction impedes accessibility to nucleosomes and represses genomic activity, whereas chromatin decompaction is associated with increased accessibility to the nucleosomal DNA and gene activation. Because the interaction of HMGN with chromatin affects both the structure of the chromatin fiber and the levels of histone modifications they impact numerous biological processes. In this review we focus on the role of HMGN in chromatin dynamics and in regulating the levels of histone modifications, and highlight recent findings on their role in determining the cellular phenotype. Additional information of the properties of these and other HMG proteins can be found in previous reviews [1-5]. In addition, reviews by Mahadevan, Hansen, Hock, Gerlitz, Rochman and Furusawa in this issue present more information and additional insights into various aspects of HMGN structure and function.
HMGNs: non-histone proteins that bind to nucleosomes
The HMGN protein family consists of 5 members encoded by 5 specific genes with a similar intron-exon organization. HMGN proteins share a common domain structure: a bipartite nucleosome localization signal, a conserved 30-amino acid long nucleosome-binding domain (NBD), and a less conserved C-terminal that is enriched in negatively charged residues (Fig 1). The most recently HMGN discovered, HMGN5, contains a long C terminal region that contains 13 highly negative repeated sequences motifs (see Rochman this issue and [6, 7]). Embedded in the NBD of all HMGNs is an absolutely conserved octapeptide, RRSARLSA, which is encoded by a specific exon, and is considered to be the signature of this protein family. This “NBD core” acts as a module that anchors the HMGN proteins to nucleosome core particles. Detailed analyses of numerous deletion and point mutants of HMGNs revealed that while several regions of the protein affect the chromatin-binding affinity of the HMGNs, the conserved NBD core is the sole determinant of the specific interaction of HMGN with nucleosome core particles. In vivo and in vitro studies demonstrated that even a single mutation in the R-S-RL motif contained in the NBD core will abolish the specific interaction of HMGNs with nucleosome cores. Any of these point mutants will bind to DNA better than to isolated core particles [8].
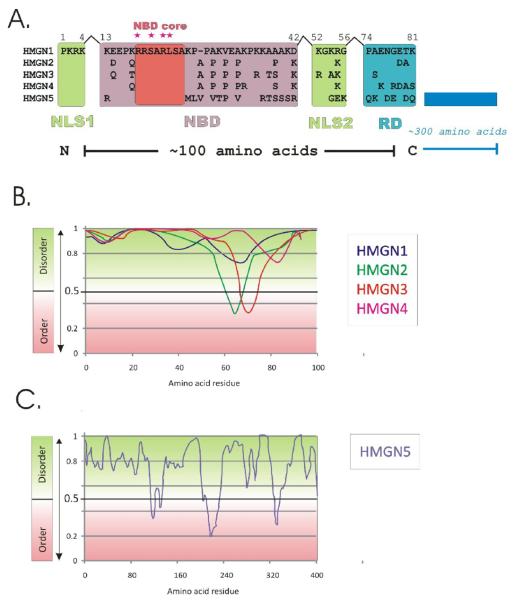
Figure 1. Structural features of the High Mobility Group N (HMGN) proteins.
A. Functional domains of the High Mobility Group N (HMGN) proteins. Sequences of the human HMGN proteins are aligned. All HMGN proteins contain four functional domains (shadowed): two Nuclear Localization Signal domains (NLS1 and NLS2, light green), a Nucleosomal Binding Domain (NBD, light purple) and a Regulatory Domain (RD, cyan). The invariant amino-acid residues within NBD domain are named as NBD core (shadowed by brick red) and four residues essential for specific binding to nucleosomes are marked by magenta stars above the core NBD. The RD domain is less conserved and has a net negative charge. The C-terminus of HMGN5 is ~300 amino acids longer than that of the other HMGNs.
B. Intrinsically disordered regions (IDRs) in HMGN family proteins. The sequences of human HMGN proteins has been analyzed by PONDR (Predictors of Natural Disordered Regions) - a series of neural network predictors (NNPs) that use amino acid sequence data to predict disorder in a given region [72].Values greater than 0.5 represent intrinsically disorder regions in the protein.
C. A graph of intrinsically disordered regions in mouse HMGN5.
All HMGNs have a high content of charged amino acids and a disordered secondary structure (Fig 1). Disordered proteins can form multiple complexes and interact with many proteins [9-11]. Indeed, it has been demonstrated that both HMGN1 and HMGN2 form multiple metastable protein complexes, and that the chromatin interaction of HMGN in the context of a complex is different from that of purified HMGN [12]. The potential ability of HMGN proteins to participate in numerous multiprotein complexes may have significant implications for their biological functions. Conceivably, HMGN proteins facilitates the interaction of protein complexes, or of specific proteins, with chromatin [12-16]
HMGNs are the only nuclear proteins known to specifically recognize the generic structural features of the 147 base pair nucleosome core particles, the building block of the chromatin fiber. These proteins bind to nucleosome particles better than to either purified DNA or to histones. In vitro analyses demonstrated that the binding of HMGNs to chromatin is highly dependent on ionic strength. At low ionic strength nucleosome core particles (CP) can bind either one or two molecules of HMGN with very high affinity [17-19]. Under these conditions, the CP complexes formed can contain different HMGN variants (i.e. one HMGN1 and one HMGN2). However at higher ionic strengths, which are close to physiological, the interaction of HMGNs with CPs is highly specific. Under these conditions the association constant of HMGN with CP is significantly lower and the only complexes detected are CPs associated with two molecules of one type of HMGN variant. Thus, addition of CPs to a mixture of HMGN1 and HMGN2, or HMGN2 and HMGN5, results in complexes containing two molecules of either HMGN1, or HMGN2, or HMGN5, per core particle. CPs associated with two different type of HMGN variants (i.e. an HMGN1 and an HMGN2) were not detected [7, 19]. Significantly, experiments with chromatin isolated from nuclei suggested that also in living cells the CPs contain only one type of HMGN variant [20].
Hydroxyl radical footprinting indicated that the path of HMGN1 on the surface of the nucleosomal DNA is indistinguishable from that of HMGN2 [21]. The bound HMGNs proteins protect the DNA from hydroxyl radical cleavage in each of the two major grooves of the DNA flanking the nucleosomal dyad axis and 25 base pairs from the ends. At the entry-exit points of the nucleosomal DNA the proteins bridge two adjacent DNA helices on the surface of the particle, resulting in stabilization of the nucleosomal DNA flanks [21]. Significantly, the hydroxyl radical footprinting indicated that the nucleosomal binding sites of HMGNs partially overlap with those of the linker histone H1 suggesting that the protein could affect each other's chromatin interaction. As elaborated below, more recent studies in living cells revealed that indeed, HMGNs and H1 mutually affect their binding to chromatin [6, 22].
Since HMGNs bind specifically to CPs and not to isolated DNA, it can be expected that that they also form specific contacts with the histone octamer. Indeed, early cross-linking studies indicated that HMGN1 and HMGN2 for multiple contacts with all the core histones [23-25]. Subsequent studies with site specific cross-linkers revealed that the negatively charged RD domain of HMGN1 contacts the N-terminal tail of histone H3 while the N-terminal region of HMGN1 contacts a restricted region it the N-terminal of H2B [26]. Elucidation of specific contacts between HMGN variants and nucleosomal histones could provide insights into the mechanisms underlying variant-specific biological functions. Such experiments are now in progress.
Recent studies revealed that the interactions of HMGN proteins with chromatin are cell cycle dependent [27]. While in interphase the proteins form specific complexes with nucleosomes, their binding to mitotic chromatin is less specific. HMGN-GFP proteins seem to be associated with the highly condensed mitotic chromosomes however this association is not dependent on an intact NBD, since HMGN point mutants that do not bind to nucleosomes still localize to mitotic chromosomes. FRET analyses supported the notion that the binding of HMGNs to mitotic chromosomes is different from their binding to interphase nucleosomes. The altered binding of HMGNs to mitotic chromosomes can be linked to the mitotic phosphorylation of the two serines located in the NBD cores. These two serines are phosphorylated during mitosis thereby abolishing the specific interaction of the proteins with CPs [27, 28].
With the exception of HMGN5, the HMGNs interact with all the nucleosomes in chromatin however they seem to be enriched in heterochromatin perhaps because the concentration of nucleosomes is higher in this chromatin domain. Mouse HMGN5 however is excluded from constitutive heterochromatin and binds preferentially to nucleosomes in euchromatin [6]. The exclusion is due to its highly acidic C-terminal domain. The HMGN5 variant is not absolutely conserved among species therefore it is not yet clear whether also in other species HMGN5 is excluded from heterochromatin. A major question that remains to be resolved is whether HMGNs bind preferentially to specific regions in chromatin. Early experiments suggested that these proteins generate, or maintain, the structure of DNaseI hypersensitive sites in chromatin [29, 30]. The advent of ChIP- Seq and whole genome sequence techniques make it possible to address this important question.
While the NBD is the major determinant of the specific interaction of HMGNs with chromatin, their negatively charged C-terminal domains (RD) seem to play an important role in their “regulatory” functions. Thus, the C-terminal domain determines the specificity of the effects of HMGN1 and HMGN2 on various histone modifications [31]. The C-terminal of mouse HMGN5 targets the protein to euchromatin. In fact, grafting the mouse HMGN5 to the C-terminal of other HMGNs also relocates them to euchromatin [6]. It is not clear whether this acidic region excludes the binding of HMGNs to heterochromatin or facilitates their interaction with nucleosomes in euchromatin. The negatively charged C-terminal domain of HMGN1 facilitates transcription from histone H1-containing chromatin [32, 33]. Likewise, the C-terminal of HMGN5 was shown to interact directly with the positively charged region of linker H5 histone and alleviate the H5 induced compaction of a chromatin template [6]. Taken together, the data suggest that interplay between HMGNs and the linker histone H1 affect the structural and functional plasticity of the chromatin fiber.
Dynamic Binding of HMGNs to Chromatin
Analysis of images obtained from fluorescence recovery after photobleaching (FRAP) experiments revealed that that the chromatin interaction of most nuclear proteins including HMGNs and H1 in living cells are dynamic (for a review see this issue, Hock and Catez). HMGNs are mobile molecules and the interaction of any HMGN specific molecule with a specific nucleosome, or with a specific site is transient. HMGN continuously move throughout the nucleus in a “stop and go” process in which a molecule stays associated with a binding site for a limited time and then dissociates and moves rapidly to another site. For HMGNs, this process is dependent on the integrity of the “NBD core”. Mutation that abolish the specific in vitro interaction of HMGNs with nucleosomes also greatly decrease the chromatin residence time of HMGNs in living cell, a clear indication that the mobility of the protein in living cells is regulated by their binding to chromatin.
The dynamic behavior of HMGNs, and other molecules, in the nucleus has important consequences. First, it ensures that all nucleosomes in chromatin will be associated with HMGNs, at some point. The amount of HMGN in the nucleus is sufficient to bind only about 1% of the nucleosomes. However, FRAP analyses calculation indicate that any given HMGN molecule can rapidly hop from one nucleosome to another and traverse the entire nucleus in less than one minute. Second, it facilitates potential functional redundancy among HMGN variants. Thus, if one variant is missing or altered, another variant can rapidly reach a “variant-preferred” binding site and functionally compensate for the missing variant. Third, it facilitates competitive chromatin binding of HMGN with H1 or with other nuclear proteins. H1 binds to chromatin significantly stronger than HMGN. The transient displacement of H1 from chromatin provides a window of opportunity for an HMGN to bind to the temporarily vacated space. Competition chromatin binding proteins for nucleosome binding sites is part of the general mechanism that provides functional and structural plasticity to the chromatin fiber ([34, 35] and also see Hock and Catez, this issue).
The transient interaction of HMGNs with chromatin does not preclude the possibility that the proteins are preferentially located at specific sites in chromatin. It is well known that dynamic components can establish stable structures [36]. In such as a scenario, at specific chromatin sites HMGN molecules would continuously exchange and their binding at these sites would be significantly higher than their random binding to nucleosomes throughout the entire chromatin. Genome wide analyses of ChIP-Seq experiments will provide information on this important aspect of the interaction of HMGNs with chromatin.
HMGNs remodel chromatin
A. HMGNs reduce chromatin compaction
Early studies by Weisbrod and Weintraub indicating that HMGNs increase the DNase I sensitivity of transcriptionally active genes were consistent with the possibility that the proteins reduce the compaction of the chromatin fiber [29]. Indeed subsequent electron microscope studies [37], nuclease restriction digestion [6, 33, 38, 39] and sedimentation studies [6, 33, 39] provide strong evidence that the proteins do indeed “decompact” chromatin. More recent immunofluorescence analysis of cells harboring condensed chromatin arrays, and of mouse cells expressing exogenous HMGN5 molecules provided further evidence that HMGNs can reduce the compaction of a condensed chromatin domain [6] (see also article by Rochman in this issue). These findings are the basis for considering HMGNs as architectural elements which function to alter the structure of the chromatin fiber.
Several mechanisms could account for the ability of HMGNs to reduce the compaction of chromatin. The most likely possibility is that HMGNs antagonizes the chromatin condensing activity of linker histone H1, a possibility suggested some time ago [40]. Indeed, the nucleosomal binding sites of H1 partially overlap with those of H1 [21] and FRAP experiments indicated that in living cells HMGNs and H1 compete for chromatin binding sites [35]. In vitro sedimentation [6] and cross linking experiments [6] provide additional direct evidence that HMGNs counteract the condensing activity of H1. Different members of the HMGN family are capable of counteracting histone H1-mediated chromatin condensation to various degrees [6]. Cross-linking experiments revealed that the negatively charged C-terminal domain of HMGN5 directly interacts with the positively charged C-terminal domain of histone H5, the linker histone domain known to stabilize chromatin compaction [6].
Sedimentation [39] and electron microscopy experiments [37] indicated that HMGNs also reduce the compaction of circular chromatin templates that are devoid of histone H1. Conceivably, HMGNs could interfere with chromatin condensation that is mediated by the histone tails or alternatively, that HMGN-mediated changes in the structure of the core particle itself lead to chromatin decondensation.
B. HMGNs affect the activity of ATP-dependent chromatin remodeling complexes?
ATP-dependent chromatin remodeling complexes play a key role in chromatin dynamics [41, 42]. Given that HMGNs bind directly to, and affect the stability of the nucleosome, they could affect the action of the chromatin remodeling complexes. Two studies addressing directly the role of HMGNs in this process gave conflicting results. Initially, it was shown that HMGN1 does not affect the rate of SWI/SNF-dependent nucleosome remodeling when assembled into mononucleosomes or nucleosomal arrays [43]. However, in a later study both HMGN1 and HMGN2 were found to suppress ATP-dependent nucleosome remodeling by ACF and BRG1 [44]. The reasons for the discrepancies in the results obtained in the two laboratories are not yet clear. Given the function of ATP-dependent nucleosome remodeling complexes in chromatin dynamics it is important to unequivocally determine whether HMGNs do affect the activities of these chromatin modifiers.
C. HMGNs affect the levels of posttranslational modification in core histones
Chemical modifications in the tails of histones are major epigenetic marks that play an important regulatory role in nuclear processes occurring in the context of chromatin such as gene expression, replication, and DNA repair. As such, they affect most biological processes including development and differentiation and impact the cellular response to various external and internal biological signals [45-47]. The nucleosomal core histones harbor more than 100 different post-translational modifications [48]. Significantly, the levels and patterns of histones modifications are not fixed; they are in a constant state of flux resulting from the equilibrium between the activities of numerous enzymes that continuously modify, and those that continuously de-modify specific histone residues. An extensive list of histone-modifying enzymes can be found elsewhere [48].
Since HMGN proteins bind specifically to nucleosomes, it could be expected that they would affect the levels of histone modifications. Indeed, analysis of cells derived from Hmgn1−/− mice and from their wild type littermates revealed that loss of HMGN1 affects the global levels of H3K14ac, H3K9ac, H3K9me, H3S10ph, H3S28ph, H4S1ph and H2AS1ph [49-51]. These findings, together with earlier observation that histone H1 represses H3 acetylation [52] establish the general principle that structural nucleosome binding proteins do affect the levels of histone modification. Likely, HMGNs affect additional histone modifications; however, it remains to be determined which of the many modifications are affected.
More detailed analysis indicated that HMGN1 increases the levels of H3K14a by enhancing the action of HATs rather than inhibiting HDACs [50]. Significantly, HMGN1 mutants defective in binding to nucleosomes, did not affect the levels of this acetylation. Furthermore, HMGN1 enhanced the PCAF-mediated acetylation of nucleosomal, but not free H3. Thus, HMGN1 modulates the levels of H3K14ac only in the context of chromatin [50]. Similar results were obtained in an expanded analysis of the mechanisms whereby HMGN1 affects the levels of H3S10ph and H3S28ph. HMGN1 affected these modifications by binding to chromatin, the modification of free histones is not affected by HMGNs [49].
An additional significant observation is that HMGN affect histone modification in a variant specific manner, implying that the proteins are not fully functionally redundant, at least with respect to this histone modification [31]. Thus, while HMGN1 affects the phosphorylation of both H3S10 and H3S28, HMGN2 does not. Likewise, HMGN2 enhances H3K14 acetylation more robustly than HMGN1 [31]. Detailed analysis of HMGN1 and HMGN2 swap mutants identified the domains involved in regulating these histone modifications. At least two distinct HMGN domains are necessary for modulating chromatin modifications: an intact NBD which is a non-modification-specific domain necessary for chromatin binding, and a modification-specific domain localized in the C terminus of the HMGNs [31].
HMGN protein could affect the modifications levels of core histones by several mechanisms. Kinetic analysis of the effect of HMGN1 on PCAF-mediated acetylation of H3K14 indicated that HMGN affects both the apparent Vmax and the apparent Km of the reaction, suggesting that the binding of HMGN1 to nucleosomes induces changes in the nucleosome itself, which ultimately modulate the ability of the enzyme to modify the tail of core histones [50]. Extrapolating from these data, it can be hypothesized that HMGN proteins affect mostly the turnover rate of modifying and de-modifying enzymes by modulating the accessibility of the substrates (amino acid residues within histone tails) to corresponding enzymes, or the ability of enzymes to bind to nucleosomes. Additionally, by competing with the linker histone H1 for chromatin-binding sites, or by changing the entry–exit angle of the nucleosomal DNA, HMGN proteins could induce changes in the architecture of the chromatin fiber which ultimately affect the turnover balance between modifications and de-modifications of specific residues in core histones. Additional possible mechanisms include HMGN effects on the activity of ATP-driven nucleosome remodeling enzymes [43, 44], or the targeting of modifying enzymes to specific sites. The latest possibility is supported by studies indicating that HMGN proteins are organized into metastable macromolecular complexes [12]. Most likely HMGNs affect the levels of modification by more than one mechanism.
Given the importance of histone posttranslational modification in chromatin function it is likely that the biological effects of HMGNs are mainly due to their effect on the levels of histone modifications. So far two examples do support directly this possibility. The first example is the finding that the heat shock mediated induction of Hsp70 gene in Hmgn1−/− mice embryonic fibroblasts (MEF) is impaired as compared to MEF isolated from wild type littermate mice. Heat shock also elevates the levels of H3K14 acetylation in the Hsp70 chromatin of wild type cells more efficiently than in the chromatin of Hmgn1−/− cells, whereas treatment with histone deacetylase inhibitors abrogates the effects of HMGN1 on the heat shock response. The results indicated that HMGN1 enhances the rate of heat shock-induced H3K14 acetylation in the Hsp70 promoter, thereby enhancing the rate of chromatin remodeling and the activation of Hsp70 [53]. The second example is the observation that HMGN1 enhances the activation of the protein kinase ataxia telangiectesia (ATM), a key transducer of the cellular response to double stranded DNA damage. The HMGN1 mediated activation of ATM was linked to increase acetylation of H3K14 and treatments with histone acetylase inhibitors abrogated the requirement of HMGN1 for efficient ATM activation [54]. In addition, loss of HMGN1 is also linked to alteration of H3S10 phosphorylation in response to stress (see Figure 2 modified from [49]), and loss of HMGN3 leads to decreased acetylation and decreased expression of GLUT2 in the pancreatic beta cells [15].
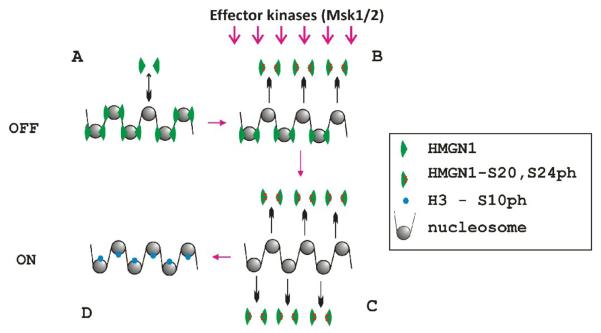
Figure 2. A model of HMGN1 modulation of the rate of stress-activated phosphorylation of H3S10 in chromatin of FosB immediate early genes (modified from [49]).
A. In quiescent cells, non-phosphorylated HMGN1 is bound to nucleosomes
B. Anisomycin or similar stress signals activate MSKs through MAPK signaling pathway. The kinases phosphorylate nuclear targets, including HMGN1. Phosphorylation of HMGN1 at Ser20 and Ser24 abolishes the binding of the protein to nucleosomes, thus shifting the equilibrium towards non-bound phosphorylated HMGN1
C. Phosphorylated HMGN1 leaves nucleosomes, removing the inhibiting activity of HMGN1 upon H3S10 phosphorylation
D. Absence of HMGN1 enhances the rate of nucleosomal H3 phosphorylation, and alleviates the rounds of transcription.
HMGNs affect transcription from chromatin templates
It has been long postulated that HMGNs play a significant role in the regulation of transcriptional processes [1, 2, 29]. This important aspect of HMGN function is the subject of a special review in this issue (see review by Hansen) and therefore is only briefly summarized below.
Transcription profiling of Hmgn1−/− [49, 55] and Hmgn3−/− [15, 56] MEFs and of cells with altered HMGN5 expression [6] indicated that loss of an HMGN alters the expression of numerous genes; however, only a few genes were significantly altered. In a few examples ChIP analysis revealed that HMGN are associated with the chromatin of the genes affected [15, 53]. Several distinct mechanisms whereby HMGNs could affect transcription from chromatin can be envisioned.
One possible mechanism is that HMGNs alleviate the H1 induced inhibition of transcription from chromatin. Conceivably, the chromatin condensing activity of histone H1 must be antagonized in order to convert chromatin to a less condensed, more transcriptionally competent template. Direct support for this possibility comes from experiments using Simian Virus 40 minichromosomes as a model system, which demonstrated that HMGN1 reduces the histone H1-mediated inhibition of transcription by RNA polymerase II [33]. The effect was linked to an HMGN1-mediated disruption of histone H1-dependent compaction of chromatin. Both the transcriptional enhancement and chromatin-unfolding activities of HMGN1 are mediated through its acidic, C-terminal region [33, 57]. These results are in full agreement with more recent studies indicating that in vivo HMGNs and H1 compete for chromatin binding sites ([22, 35], see also Hock and Catez in this issue) and that the negatively charged C-terminal of HMGN5 targets the C-terminal of histone H5 and reduces the linker -mediated chromatin compaction. Earlier studies with chromatin templates reconstituted in Xenopus extracts also indicated that deletion of the C-terminal amino acids of either HMGN2 or HMGN1 significantly reduces the ability of the proteins to enhance transcription [32].
A second possible mechanism, that was already discussed above is that HMGNs do affect the activity of ATP dependent and /or the ATP-independent chromatin remodeling complexes [43, 44]. Changes in nucleosome position or in the levels of histone modifications at gene regulatory sites will have major effects on transcription in the context of chromatin. In this respect, genome wide analysis of the organization of HMGNs in chromatin may provide important insights into the involvement of HMGNs in transcription.
A third possibility is that HMGNs facilitate the binding of regulators to chromatin. Support for this possibility comes from the observation that HMGN is found in multiprotein complexes [12]. It is noteworthy that the structure of HMGN is highly disordered (Fig 1) and that proteins with a disordered conformation have the potential to complex with multiple partners [9, 12, 58]. Furthermore, the high mobility of HMGN proteins within the nucleus increases the likelihood of collisions between HMGNs and potential partners. Several experiments provide direct evidence that HMGNs interact with specific protein partners in chromatin. Thus, incorporation of HMGN2 into chromatin templates facilitated the ability of GAL4-VP16 to activate transcription [59]. More recent studies revealed that Cockayne syndrome protein B recruits HMGN1 to the RNA polymerase II –containing complex stalled at UV-induced chromatin lesions [13, 14]. Likewise, HMGN1 affects the induction of several estrogen-regulated genes by interacting with estrogen receptor alpha or with serum response factor [16], while HMGN3 and the transcription initiation factor PDX1 mutually affect each other interaction in the promoter region of the Glut2 gene chromatin [15].
Obviously, these 3 possible mechanisms can be interrelated and act in concert. In one such possible scenario, HMGN alters the interaction of H1 with chromatin thereby facilitating the binding of transcription factors which in turn recruit histone modification complexes to distinct regulatory sites. For a more detailed discussion of the role of HMGNs in transcription please see the review by Hansen in this issue.
Biological functions of HMGNs
Most of the studies on the biological function of HMGNs were done in cell free system, or in tissue culture. An understanding of the biological function of HMGNs in the context of an entire organism will provide important insights into mechanisms whereby events occurring in chromatin affect the cellular phenotype of a complex system. So far HMGNs were detected only in vertebrates. Analysis of their expression during mouse [60-63] and Xenopus development [64] and during the differentiation of blood [65] and muscle [66] indicated that the expression of HMGN is linked to differentiation. The developmental expression of HMGN1 was studied in some detail in the hair follicle [60], in the developing limb bud [61] and in the eye [62, 67]. Loss of HMGN1 affected the development and maturation of the corneal epithelium but had no obvious effects on the skin differentiation or hair follicle cycling. The role of HMGNs in differentiation is the subject of a special review in this issue (Furusawa and Cherukuri, see also [5]) and is only briefly summarized here.
In Xenopus laevis, the expression and cellular location of the HMGN proteins are developmentally regulated, and either up- or down-regulation of both HMGN1 and HMGN2, but not of HMGN1 or HMGN2 alone, leads to gross developmental defects in post-blastula embryos, perhaps because HMGN proteins are involved in the regulation of mesoderm-specific genes [64]. Likewise, down regulation of both HMGN1 and HMGN2 affected the development of mouse pre-implantation embryos [68]. The results suggested functional redundancy among the proteins. However, analysis of genetically altered mice lacking a single HMGN provides evidence that the proteins are not fully functionally redundant. In fact preliminary studies indicate that loss of HMGN2 is embryonic lethal (Cherukuri and Bustin, unpublished).
Studies with mice and cells lacking HMGN1 revealed that loss of the protein impairs the proper response to stress, especially the repair of damaged DNA (for a comprehensive review on the role of HMGNs in DNA Repair and Cancer see review by G. Gerlitz, this issue). Significantly, loss of HMGN1 affects the repair of DNA damaged by either UV or gamma irradiation [55, 69]. The cellular process that repairs DNA damaged by UV is completely different from the process that repairs DNA damaged by gamma irradiation. Thus, HMGN1 is not a bona fide DNA repair factor. HMGN1 affects DNA repair because these processes occur in the context of chromatin. Indeed, the effect of HMGN1 on the repair of DNA damaged by gamma irradiation is linked to its ability to enhance the acetylation of H3K14, which in turn affects the activation of ATM, a key molecule in the cellular response to double stranded DNA breaks [54, 70]. Similarly, it has been proposed but not experimentally proven, that the involvement of HMGN1 in the repair of UV damaged DNA is also linked to histone modification [71].
The expression of HMGN3 and HMGN5 variant seem to be tissue specific. HMGN3 is highly expressed in pancreatic islet cells and Hmgn3−/− are mildly diabetic most likely because the transcriptional network of the insulin secreting beta cells is disrupted [15]. Interestingly, ectopic expression of HMGN3 in Hepa-1 cells alters the expression of approximately 0.8% of the genes [56]. HMGN5, the most recently discovered member of this family is highly expressed in the placenta and modulates the expression of specific differentiation markers in placental RCHO-1 cells [7]. The phenotype of mice lacking HMGN5 has not yet been determined.
With the possible exception of the HMGN2 variant, loss of HMGN do not result in major developmental abnormalities. It is possible that partial redundancy among HMGN variants partially compensates for loss of a specific variant. On the other hand, loss of any HMGN variant does have significant effects on the ability of a cell to withstand stress. The emerging picture suggests that HMGNs optimize the fidelity of gene expression and thereby optimize the function and survival of an organism, especially under stress. The availability of genetically altered mice that either lack, or over-express a single, or a combination of HMGN variants, will provide additional insights into the biological function of the HMGN protein family. This information, together with detailed studies on the interaction of HMGN with their chromatin targets, and on the chromatin remodeling effects of HMGN will provide information on the mechanisms whereby events occurring in the chromatin fiber ultimately shape the cellular phenotype.
Acknowledgements
We acknowledge NIH Fellows Editorial Board for help with editing the manuscript. The research of the laboratory is supported by CCR, the intramural research program of NCI, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 2.Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol. 1999;19:5237–5246. doi: 10.1128/mcb.19.8.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bustin M. Chromatin unfolding and activation by HMGN(*) chromosomal proteins. Trends Biochem Sci. 2001;26:431–437. doi: 10.1016/s0968-0004(01)01855-2. [DOI] [PubMed] [Google Scholar]
- 4.West KL, Postnikov YV, Birger Y, Bustin M. Chromatin decompaction method by HMGN proteins. Methods Enzymol. 2003;371:521–536. doi: 10.1016/S0076-6879(03)71039-8. [DOI] [PubMed] [Google Scholar]
- 5.Hock R, Furusawa T, Ueda T, Bustin M. HMG chromosomal proteins in development and disease. Trends Cell Biol. 2007;17:72–79. doi: 10.1016/j.tcb.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rochman M, Postnikov Y, Correll S, Malicet C, Wincovitch S, Karpova TS, McNally JG, Wu X, Bubunenko NA, Grigoryev S, Bustin M. The Interaction Of NSBP1 With Nucleosomes In Euchromatin Counteracts Linker Histone-Mediated Chromatin Compaction And Modulates The Fidelity Of The Cellular Transcription Profile. Mol Cell. 2009;35:642–656. doi: 10.1016/j.molcel.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shirakawa H, Landsman D, Postnikov YV, Bustin M. NBP-45, a novel nucleosomal binding protein with a tissue-specific and developmentally regulated expression. J Biol Chem. 2000;275:6368–6374. doi: 10.1074/jbc.275.9.6368. [DOI] [PubMed] [Google Scholar]
- 8.Ueda T, Catez F, Gerlitz G, Bustin M. Delineation of the protein module that anchors HMGN proteins to nucleosomes in the chromatin of living cells. Mol Cell Biol. 2008;28:2872–2883. doi: 10.1128/MCB.02181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patil A, Nakamura H. Disordered domains and high surface charge confer hubs with the ability to interact with multiple proteins in interaction networks. FEBS Lett. 2006;580:2041–2045. doi: 10.1016/j.febslet.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 10.Singh GP, Ganapathi M, Dash D. Role of intrinsic disorder in transient interactions of hub proteins. Proteins. 2007;66:761–765. doi: 10.1002/prot.21281. [DOI] [PubMed] [Google Scholar]
- 11.Uversky VN, Oldfield CJ, Midic U, Xie H, Xue B, Vucetic S, Iakoucheva LM, Obradovic Z, Dunker AK. Unfoldomics of human diseases: linking protein intrinsic disorder with diseases. BMC Genomics. 2009;10(Suppl 1):S7. doi: 10.1186/1471-2164-10-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim JH, Bustin M, Ogryzko VV, Postnikov YV. Metastable macromolecular complexes containing high mobility group nucleosome-binding chromosomal proteins in HeLa nuclei. J Biol Chem. 2002;277:20774–20782. doi: 10.1074/jbc.M200404200. [DOI] [PubMed] [Google Scholar]
- 13.Fousteri M, Mullenders LH. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 2008;18:73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- 14.Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LH. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell. 2006;23:471–482. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 15.Ueda T, Furusawa T, Kurahashi T, Tessarollo L, Bustin M. The Nucleosome Binding Protein HMGN3 Modulates The Transcription Profile Of Pancreatic {beta}-Cells And Affects Insulin Secretion. Mol Cell Biol. 2009 doi: 10.1128/MCB.00526-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu N, Hansen U. HMGN1 modulates estrogen-mediated transcriptional activation through interactions with specific DNA-binding transcription factors. Mol Cell Biol. 2007;27:8859–8873. doi: 10.1128/MCB.01724-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mardian JK, Paton AE, Bunick GJ, Olins DE. Nucleosome cores have two specific binding sites for nonhistone chromosomal proteins HMG 14 and HMG 17. Science. 1980;209:1534–1536. doi: 10.1126/science.7433974. [DOI] [PubMed] [Google Scholar]
- 18.Sandeen G, Wood WI, Felsenfeld G. The interaction of high mobility proteins HMG14 and 17 with nucleosomes. Nucleic Acids Res. 1980;8:3757–3778. doi: 10.1093/nar/8.17.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Postnikov YV, Trieschmann L, Rickers A, Bustin M. Homodimers of chromosomal proteins HMG-14 and HMG-17 in nucleosome cores. J Mol Biol. 1995;252:423–432. doi: 10.1006/jmbi.1995.0508. [DOI] [PubMed] [Google Scholar]
- 20.Postnikov YV, Herrera JE, Hock R, Scheer U, Bustin M. Clusters of nucleosomes containing chromosomal protein HMG-17 in chromatin. J Mol Biol. 1997;274:454–465. doi: 10.1006/jmbi.1997.1391. [DOI] [PubMed] [Google Scholar]
- 21.Alfonso PJ, Crippa MP, Hayes JJ, Bustin M. The footprint of chromosomal proteins HMG-14 and HMG-17 on chromatin subunits. J Mol Biol. 1994;236:189–198. doi: 10.1006/jmbi.1994.1128. [DOI] [PubMed] [Google Scholar]
- 22.Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24:4321–4328. doi: 10.1128/MCB.24.10.4321-4328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brawley JV, Martinson HG. HMG proteins 14 and 17 become cross-linked to the globular domain of histone H3 near the nucleosome core particle dyad. Biochemistry. 1992;31:364–370. doi: 10.1021/bi00117a008. [DOI] [PubMed] [Google Scholar]
- 24.Cook GR, Yau P, Yasuda H, Traut RR, Bradbury EM. High mobility group protein 17 cross-links primarily to histone H2A in the reconstituted HMG 17-nucleosome core particle complex. J Biol Chem. 1986;261:16185–16190. [PubMed] [Google Scholar]
- 25.Espel E, Bernues J, Perez-Pons JA, Querol E. Binding of HMG14 non-histone protein to histones H2A, H2B, H1 and DNA in reconstituted chromatin. Biochem Biophys Res Commun. 1985;132:1031–1037. doi: 10.1016/0006-291x(85)91910-2. [DOI] [PubMed] [Google Scholar]
- 26.Trieschmann L, Martin B, Bustin M. The chromatin unfolding domain of chromosomal protein HMG-14 targets the N-terminal tail of histone H3 in nucleosomes. Proc Natl Acad Sci U S A. 1998;95:5468–5473. doi: 10.1073/pnas.95.10.5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cherukuri S, Hock R, Ueda T, Catez F, Rochman M, Bustin M. Cell Cycle-dependent Binding of HMGN Proteins to Chromatin. Mol Biol Cell. 2008;19:1816–1824. doi: 10.1091/mbc.E07-10-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prymakowska-Bosak M, Misteli T, Herrera JE, Shirakawa H, Birger Y, Garfield S, Bustin M. Mitotic phosphorylation prevents the binding of HMGN proteins to chromatin. Mol Cell Biol. 2001;21:5169–5178. doi: 10.1128/MCB.21.15.5169-5178.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weisbrod S, Groudine M, Weintraub H. Interaction of HMG 14 and 17 with actively transcribed genes. Cell. 1980;19:289–301. doi: 10.1016/0092-8674(80)90410-9. [DOI] [PubMed] [Google Scholar]
- 30.Weisbrod S, Weintraub H. Isolation of actively transcribed nucleosomes using immobilized HMG 14 and 17 and an analysis of alpha-globin chromatin. Cell. 1981;23:391–400. doi: 10.1016/0092-8674(81)90134-3. [DOI] [PubMed] [Google Scholar]
- 31.Ueda T, Postnikov YV, Bustin M. Distinct domains in high mobility group N variants modulate specific chromatin modifications. J Biol Chem. 2006;281:10182–10187. doi: 10.1074/jbc.M600821200. [DOI] [PubMed] [Google Scholar]
- 32.Trieschmann L, Postnikov YV, Rickers A, Bustin M. Modular structure of chromosomal proteins HMG-14 and HMG-17: definition of a transcriptional enhancement domain distinct from the nucleosomal binding domain. Mol Cell Biol. 1995;15:6663–6669. doi: 10.1128/mcb.15.12.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding HF, Bustin M, Hansen U. Alleviation of histone H1-mediated transcriptional repression and chromatin compaction by the acidic activation region in chromosomal protein HMG-14. Mol Cell Biol. 1997;17:5843–5855. doi: 10.1128/mcb.17.10.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bustin M, Catez F, Lim JH. The dynamics of histone H1 function in chromatin. Mol Cell. 2005;17:617–620. doi: 10.1016/j.molcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 35.Catez F, Ueda T, Bustin M. Determinants of histone H1 mobility and chromatin binding in living cells. Nat Struct Mol Biol. 2006;13:305–310. doi: 10.1038/nsmb1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Misteli T. The concept of self-organization in cellular architecture. J Cell Biol. 2001;155:181–185. doi: 10.1083/jcb.200108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vestner B, Bustin M, Gruss C. Stimulation of replication efficiency of a chromatin template by chromosomal protein HMG-17. J Biol Chem. 1998;273:9409–9414. doi: 10.1074/jbc.273.16.9409. [DOI] [PubMed] [Google Scholar]
- 38.Crippa MP, Trieschmann L, Alfonso PJ, Wolffe AP, Bustin M. Deposition of chromosomal protein HMG-17 during replication affects the nucleosomal ladder and transcriptional potential of nascent chromatin. EMBO J. 1993;12:3855–3864. doi: 10.1002/j.1460-2075.1993.tb06064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trieschmann L, Alfonso PJ, Crippa MP, Wolffe AP, Bustin M. Incorporation of chromosomal proteins HMG-14/HMG-17 into nascent nucleosomes induces an extended chromatin conformation and enhances the utilization of active transcription complexes. Embo J. 1995;14:1478–1489. doi: 10.1002/j.1460-2075.1995.tb07134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Postnikov YV, Shick VV, Belyavsky AV, Khrapko KR, Brodolin KL, Nikolskaya TA, Mirzabekov AD. Distribution of high mobility group proteins 1/2, E and 14/17 and linker histones H1 and H5 on transcribed and non-transcribed regions of chicken erythrocyte chromatin. Nucleic Acids Res. 1991;19:717–725. doi: 10.1093/nar/19.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horn PJ, Peterson CL. Molecular biology. Chromatin higher order folding--wrapping up transcription. Science. 2002;297:1824–1827. doi: 10.1126/science.1074200. [DOI] [PubMed] [Google Scholar]
- 42.Smith CL, Peterson CL. ATP-dependent chromatin remodeling. Curr Top Dev Biol. 2005;65:115–148. doi: 10.1016/S0070-2153(04)65004-6. [DOI] [PubMed] [Google Scholar]
- 43.Hill DA, Peterson CL, Imbalzano AN. Effects of HMGN1 on chromatin structure and SWI/SNF-mediated chromatin remodeling. J Biol Chem. 2005;280:41777–41783. doi: 10.1074/jbc.M509637200. [DOI] [PubMed] [Google Scholar]
- 44.Rattner BP, Yusufzai T, Kadonaga JT. HMGN proteins act in opposition to ATP-dependent chromatin remodeling factors to restrict nucleosome mobility. Mol Cell. 2009;34:620–626. doi: 10.1016/j.molcel.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001;13:263–273. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- 46.Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 47.Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- 48.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 49.Lim JH, Catez F, Birger Y, West KL, Prymakowska-Bosak M, Postnikov YV, Bustin M. Chromosomal protein HMGN1 modulates histone H3 phosphorylation. Mol Cell. 2004;15:573–584. doi: 10.1016/j.molcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 50.Lim JH, West KL, Rubinstein Y, Bergel M, Postnikov YV, Bustin M. Chromosomal protein HMGN1 enhances the acetylation of lysine 14 in histone H3. Embo J. 2005;24:3038–3048. doi: 10.1038/sj.emboj.7600768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Postnikov YV, Belova GI, Lim JH, Bustin M. Chromosomal protein HMGN1 modulates the phosphorylation of serine 1 in histone H2A. Biochemistry. 2006;45:15092–15099. doi: 10.1021/bi0613271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Herrera JE, Schiltz RL, Bustin M. The accessibility of histone H3 tails in chromatin modulates their acetylation by P300/CBP-associated factor. J Biol Chem. 2000;275:12994–12999. doi: 10.1074/jbc.275.17.12994. [DOI] [PubMed] [Google Scholar]
- 53.Belova GI, Postnikov YV, Furusawa T, Birger Y, Bustin M. Chromosomal protein HMGN1 enhances the heat shock-induced remodeling of Hsp70 chromatin. J Biol Chem. 2008;283:8080–8088. doi: 10.1074/jbc.M709782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim YC, Gerlitz G, Furusawa T, Catez F, Nussenzweig A, Oh KS, Kraemer KH, Shiloh Y, Bustin M. Activation of ATM depends on chromatin interactions occurring before induction of DNA damage. Nat Cell Biol. 2009;11:92–96. doi: 10.1038/ncb1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Birger Y, West KL, Postnikov YV, Lim JH, Furusawa T, Wagner JP, Laufer CS, Kraemer KH, Bustin M. Chromosomal protein HMGN1 enhances the rate of DNA repair in chromatin. Embo J. 2003;22:1665–1675. doi: 10.1093/emboj/cdg142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.West KL, Castellini MA, Duncan MK, Bustin M. Chromosomal proteins HMGN3a and HMGN3b regulate the expression of glycine transporter 1. Mol Cell Biol. 2004;24:3747–3756. doi: 10.1128/MCB.24.9.3747-3756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ding HF, Rimsky S, Batson SC, Bustin M, Hansen U. Stimulation of RNA polymerase II elongation by chromosomal protein HMG-14. Science. 1994;265:796–799. doi: 10.1126/science.8047885. [DOI] [PubMed] [Google Scholar]
- 58.Abercrombie BD, Kneale GG, Crane-Robinson C, Bradbury EM, Goodwin GH, Walker JM, Johns EW. Studies on the conformational properties of the high-mobility-group chromosomal protein HMG 17 and its interaction with DNA. Eur J Biochem. 1978;84:173–177. doi: 10.1111/j.1432-1033.1978.tb12154.x. [DOI] [PubMed] [Google Scholar]
- 59.Paranjape SM, Krumm A, Kadonaga JT. HMG17 is a chromatin-specific transcriptional coactivator that increases the efficiency of transcription initiation. Genes Dev. 1995;9:1978–1991. doi: 10.1101/gad.9.16.1978. [DOI] [PubMed] [Google Scholar]
- 60.Furusawa T, Ko JH, Birger Y, Bustin M. Expression of nucleosomal protein HMGN1 in the cycling mouse hair follicle. Gene Expr Patterns. 2009;9:289–295. doi: 10.1016/j.gep.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Furusawa T, Lim JH, Catez F, Birger Y, Mackem S, Bustin M. Down-regulation of nucleosomal binding protein HMGN1 expression during embryogenesis modulates Sox9 expression in chondrocytes. Mol Cell Biol. 2006;26:592–604. doi: 10.1128/MCB.26.2.592-604.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Birger Y, Davis J, Furusawa T, Rand E, Piatigorsky J, Bustin M. A role for chromosomal protein HMGN1 in corneal maturation. Differentiation. 2006;74:19–29. doi: 10.1111/j.1432-0436.2006.00054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lehtonen S, Lehtonen E. HMG-17 is an early marker of inductive interactions in the developing mouse kidney. Differentiation. 2001;67:154–163. doi: 10.1046/j.1432-0436.2001.670407.x. [DOI] [PubMed] [Google Scholar]
- 64.Korner U, Bustin M, Scheer U, Hock R. Developmental role of HMGN proteins in Xenopus laevis. Mech Dev. 2003;120:1177–1192. doi: 10.1016/j.mod.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 65.Crippa MP, Nickol JM, Bustin M. Differentiation-dependent alteration in the chromatin structure of chromosomal protein HMG-17 gene during erythropoiesis. J Mol Biol. 1991;217:75–84. doi: 10.1016/0022-2836(91)90612-a. [DOI] [PubMed] [Google Scholar]
- 66.Pash JM, Alfonso PJ, Bustin M. Aberrant expression of high mobility group chromosomal protein 14 affects cellular differentiation. J Biol Chem. 1993;268:13632–13638. [PubMed] [Google Scholar]
- 67.Lucey MM, Wang Y, Bustin M, Duncan MK. Differential expression of the HMGN family of chromatin proteins during ocular development. Gene Expr Patterns. 2008;8:433–437. doi: 10.1016/j.gep.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mohamed OA, Bustin M, Clarke HJ. High-mobility group proteins 14 and 17 maintain the timing of early embryonic development in the mouse. Dev Biol. 2001;229:237–249. doi: 10.1006/dbio.2000.9942. [DOI] [PubMed] [Google Scholar]
- 69.Birger Y, Catez F, Furusawa T, Lim JH, Prymakowska-Bosak M, West KL, Postnikov YV, Haines DC, Bustin M. Increased tumorigenicity and sensitivity to ionizing radiation upon loss of chromosomal protein HMGN1. Cancer Res. 2005;65:6711–6718. doi: 10.1158/0008-5472.CAN-05-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gerlitz G, Bustin M. Nucleosome binding proteins potentiate ATM activation and DNA damage response by modifying chromatin. Cell Cycle. 2009;8:1641. doi: 10.4161/cc.8.11.8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 72.Romero P, Obradovic Z, Dunker AK. Natively disordered proteins: functions and predictions. Appl Bioinformatics. 2004;3:105–113. doi: 10.2165/00822942-200403020-00005. [DOI] [PubMed] [Google Scholar]