Abstract
The low concentration and complex sample matrix of many clinical and environmental viral samples presents a significant challenge in the development of low cost, point-of-care viral assays. To address this problem, we investigated the use of a microfluidic passive magnetic separator combined with on-chip mixer to both purify and concentrate whole particle HIV-1 virions. Virus-containing plasma samples are first mixed to allow specific binding of the viral particles with antibody-conjugated superparamagnetic nanoparticles, and several passive mixer geometries were assessed for their mixing efficiencies. The virus-nanoparticle complexes are then separated from the plasma in a novel magnetic separation chamber, where packed micron-sized ferromagnetic particles serve as high magnetic gradient concentrators for an externally applied magnetic field. Thereafter, a viral lysis buffer was flowed through the chip and the released HIV proteins were assayed off-chip. Viral protein extraction efficiencies of 62% and 45% were achieved at 10uL/min and 30uL/min throughputs respectively. More importantly, an 80-fold concentration was observed for an initial sample volume of 1mL, and a 44-fold concentration for an initial sample volume of 0.5mL. The system is broadly applicable to microscale sample preparation of any viral sample and can be used for nucleic acid extraction as well as 40–80 fold enrichment of target viruses.
Introduction
The detection of viruses in clinical and environmental samples is of great interest for a range of diagnostic and biosafety applications. As microscale systems become increasingly applied to viral detection 1, the need to separate viruses from complex sample matrices, and the need to concentrate viruses present at extremely low concentrations, have become important challenges. As one important example, the human immunodeficiency virus (HIV), which infects 33 million people worldwide,2 has a clinically relevant concentration of 102–106 virions per milliliter of plasma. Microscale detection systems need to reliably detect only tens of virions in 100 micoliters of plasma. Currently, the only widely used commercial method for measuring such low concentrations of virus requires nucleic acid amplification (e.g., qRT-PCR, quantitative reverse transcriptase polymerase chain reaction). These systems require high-end laboratory equipment, skilled technicians, and infrastructure for transportation of samples and communication of results.3–5 Due to a lack of some or all of these components, the viral load test is currently available to fewer than 10% of HIV patients worldwide. Reducing the viral load test to a low cost lab-on-a-chip platform would have wide reaching impact in HIV care.
However, the low concentration of virus in clinical samples presents a major challenge to existing protein and RNA detection platforms in terms of sensitivity and throughput. The complex matrix of human plasma must also be addressed for many such detection systems to function. One approach is to miniaturize nucleic acid amplification and detection on microchips 6–8; to date, the lack of reliable microscale methods for extracting nucleic acid from whole virions remains a major obstacle to viable point-of-care viral load testing through PCR-on-a-chip.
An alternative approach is to concentrate virions and then use detection methods that do not require PCR. Magnetic bead-based separation is widely used in biology for the concentration and purification of a variety of analytes from complex samples.9–11 Two types of beads are popularly used: micron-sized beads (1–10μm) with a paramagnetic core, and nanometer-sized beads (50–100nm) with a superparamagnetic core. The magnetic cores are typically coated with silica or polymers that can then be functionalized with capture agents such as antibodies to target the biomolecule of choice. Both types of particles are widely available commercially with a variety of surface chemistries.
Superparamagnetic nanoparticles are particularly suitable for viral concentration, since they are on the same size scale as the viral particles themselves, and provide a larger overall surface area of interaction for the same bead volume. In addition, they do not settle out of solution, a problem experienced by micron-sized particles. As an additional advantage, some groups have used nanoparticles as a signal enhancer in impedance, surface plasmon resonance (SPR), and nuclear magnetic resonance (NMR)-based detection methods.12–14
The basic steps in magnetic separation are: 1) mixing of sample with functionalized magnetic beads targeted to the desired analyte; 2) magnetic retention of beads and any bound analytes; 3) rinsing of the beads under magnetic retention to wash off unbound contaminants; 4) elution of the desired analyte into a final volume. In all of the available commercial sample preparation kits, these steps are performed on the macro-scale, either manually or with the aid of robotic systems.
There have been several previous attempts to miniaturize magnetic particle separation using microfluidics. Deng et al. patterned 15μm nickel posts inside a micro-channel, and used them to enhance the field from an external permanent magnet to separate 4.5μm paramagnetic beads from flow.15 Smistrup et al. positioned permalloy elements 20μm away from microchannel side walls and used these to trap 1um beads under an external magnetic field.16 In contrast to these passive approaches, active traps use electrical power to generate magnetic fields through on-chip electromagnets. The most widely used design is that of a patterned planar coil at the base of the channel.17–20
The majority of these previous systems were developed for micron-sized paramagnetic particles, which require a much smaller field gradient for retention than superparamagnetic nanoparticles. For instance, Lien et al. used micron-sized beads to separate dengue virus particles via a magnetic field gradient generated from a planar coil 20. They were able to demonstrate an 87% particle separation efficiency, and a 1-fold sample concentration. Only a few efforts have attempted to separate and concentrate analytes using nanometer-sized particles. Xia et al. developed an embedded microcomb as a high magnetic field gradient generator, and then used 130nm beads to separate E. coli; however, they could only obtain a flow rate of 25μL/hr.21 Inglis et al. used angled nickel strips inside a microfluidic channel to magnetically deflect cells bound to nanometer-sized superparamagnetic beads at a flow rate of 110μL/min.22 This system required a minimum of 4,000 magnetic nanoparticles to be attached to each cell to generate enough magnetic force for the deflection. Both of these systems were based on continuous flow separation, which has the advantage of simplified operation and unlimited sample processing capacity, but the achievable degree of concentration of dilute virus-containing samples is low.
In this study, we demonstrate a novel passive magnetic separator which uses superparamagnetic nanoparticles to bind nanometer-size particles such as virions, as well as micron-sized ferromagnetic particles as on-chip high magnetic gradient concentrators. The separator is simple, low-cost to fabricate, and optimizes all three of the critical parameters for handling viruses at the microscale: high separation efficiencies, high throughput, and significant sample concentration. Overall, we show an 80-fold concentration of HIV from an initial plasma sample of 1mL in 40 minutes, and a 44-fold concentration from an initial sample of 0.5mL in 20 minutes, demonstrating a new, broadly applicable approach to viral separation and concentration.
Device Design
Magnetic Separator
The effective trapping of superparamagnetic nanoparticles under flow requires a combination of two major design strategies. The first is to maximize the magnetic field gradient, in order to increase the magnetic force acting on the nanoparticles. Since the force on a particle due to a magnetic field is proportional to the volume of a particle, but the hydrodynamic drag force is dependent on the radius 23, 24, this means that nanoparticles are much more difficult to separate than their microparticle counterparts. To create sufficiently high gradients, a popular strategy is to make use of high-gradient magnetic concentrators (HGMC). These are magnetic materials with geometries that contain uneven or angular features. When placed inside a magnetic field, the sharp features will locally concentrate the field and produce a high gradient in close range.15, 21, 23
The second strategy is to reduce the separation distance between individual particles and the trapping surface, which minimizes the time required for separation. Furthermore, since our ultimate goal is for concentration as well as separation, we would like to achieve this in a small device volume.
Our design uses a random close-packed column of polydisperse iron particles (25μm–75μm in diameter) physically trapped inside a microfluidic chamber. The magnetic field is supplied by a NdFeB permanent magnet positioned outside the chamber. This configuration addresses all of our design challenges: the irregular shape of the iron particles allow them to act as high magnetic field gradient concentrators; the packed bed of variable size particles creates a tortuous path for the nanoparticles to navigate through, and thus ensures proximity between the two types of particles; and random close-packing of the clusters inside a microfluidic chamber reduces its internal volume. For our separator, the capture chamber has dimensions of 5mm long × 4mm wide × 120um height, and a total volume of 2.5μL. Previous studies have estimated the filling factor of random close-packed polydisperse spherical particles to be around 0.63.25 In our case, the filling factor was found by liquid filling measurements to be around 0.5. The lower filling factor is likely to be due to the fact that the particles are irregularly shaped instead of spherical, and that the particle sizes are significant compared to the chamber height. At this filling factor, the bead filled chamber has an active internal volume of ~1.25μL.
Microfluidic mixer
Due to the lack of turbulence at low Reynolds numbers, mixing in microfluidic channels is a challenging problem that has been widely studied 26–30. Mixers fall under two classes: active mixers require an external energy source to perturb the fluid from their laminar flow paths, whilst passive mixers rely on the geometry of the channels to create diffusive or advective mixing, with energy supplied by the same sources as for fluid actuation. We have focused on passive mixers due to their ease of implementation, scalability, and lower cost. The established method for assessing mixer efficiency is based on optical quantification of the dilution of a tracer dye, and many mixers are optimized for the shortest length required to completely mix two different colored solutions.26 Our objective is to maximize interaction between the nanoparticles and the viral sample so that affinity binding may occur between the two. This is likely to require continued mixing beyond the point where two solutions are considered ‘mixed’ in previously described terms. For this reason, we selected two chaotic advection-based mixers which have simple modular designs and have been reported to operate well within our Reynolds number range of interest of ~5. The first was first proposed by Hong et al 31, and consists of a 2D modified Tesla structure that creates chaotic advection based on the Coanda effect. The second is the staggered herringbone structure originally described by Stroock et al.32 Both mixers have been shown to mix colored dyes within 10–15 cycles. We created a third design that combines the two by patterning herringbone structures in the straight channel sections of the modified Tesla device. Finally, a straight channel with the same dimensions as the other mixers was also tested for comparison.
Experimental
Materials
For microfluidic chamber fabrication, SU-8 photoresist and developer were obtained from MicroChem (Newton, MA); silicone elastomer and curing agent were purchased from Dow Corning (Midland, MI). Phosphate buffered saline (PBS) was obtained from Mediatech (Herndon, VA). Lyophilized bovine serum albumin (BSA) was purchased from Aldrich Chemical Co. (Milwaukee, WI). Superparamagnetic nanoparticles conjugated to anti-CD44 was obtained from Miltenyi Biotech (Auburn, CA). Polydisperse iron particles between 200 and 350 mesh was obtained from ChemicalStore.com (Clifton, NJ). NdFeB magnets were purchased from McMaster-Carr (Princeton. NJ) Purified HIV-1 culture was provided by Partners AIDS Research Center (Charlestown, MA). Human plasma was obtained from healthy donors at the BioMEMS Resource Center (Charlestown, MA). HIV-1 p24 ELISA kit was purchased from Perkin Elmer (Waltham, MA).
Device Fabrication
Multi-layer microfluidic devices were fabricated by soft lithography 33. SU-8 photoresist was patterned on a silicon wafer by standard photolithography to form a negative mold. A 10:1 mixture of PDMS prepolymer and curing agent was poured onto the mold and baked at 75°C until cured. The PDMS chambers were then bonded to microscope cover slips after oxygen plasma treatment. Polydisperse iron particles suspended in water were injected into the 125μm high chamber using a syringe until the full length of the separation chamber was filled. The flow compacts the beads into a random close-packing configuration. The device was flushed with 3% BSA in PBS solution to reduce non-specific binding on the channel surfaces. Before operation, a 1″ × 1/4″ × 1/4″ NdFeB permanent magnet was positioned under the separation chamber to provide the magnetic field.
Fluid handling
Separator only experiments
Plasma samples spiked with ~106 HIV virions/ml were mixed with a 5:1 ratio of anti-CD44 superparamagnetic beads, which attach to HIV virions, and incubated on a rocker at room temperature for 60 minutes. 200μL of the mixed sample was then injected into the microchip separator using a syringe pump at controlled flow rates ranging between 30μL/min and 100μL/min. This was followed by a wash step using 25μL of PBS, flowing at 25μL/min for 1 minute. We found that no free-floating viral particles remained in the device after a single 25μL wash. The flowthrough and the wash fluid were collected together, and 25μL of Triton X (5%) was added to lyse the viral envelope in preparation for analysis by HIV p24 protein ELISA. To determine the amount of virus captured, 250μL of 0.5% Triton X in PBS was flowed through the device at 50μL/min, and the lysate was collected for analysis. Comparison experiments with the macro-scale commercial system were performed using the Miltenyi ?-MACS separation column and magnet stand, following vendor protocol.
Integrated mixer and separator experiments
Spiked viral samples were injected into one of the device inlets at various flow rates using a syringe pump. Anti-CD44 superparamagnetic beads were injected into a second inlet at 20% of the viral sample flow rate. Flow through from the integrated device was collected together with 25μL wash fluid. Finally, lysis of captured virions was performed with 0.5% Triton X and the lysate was collected for analysis by p24 ELISA. For capture efficiency experiments, 250μL of lysate was continuously collected from the device. For viral concentration experiments, only the first 5μL of lysate was collected by flowing the Triton solution at 5μL/min for 1 minute.
Quantification of separation efficiency
p24 is a 24kDa protein found specifically in HIV-1 and comprises the viral capsid. There are approximately 2,000 units equivalent to 8×10−17g of p24 protein per HIV virion 34; thus the p24 concentration directly relates to the viral concentration in a spiked sample. The Perkin Elmer HIV-1 p24 ELISA kit was used to determine the viral concentration in all samples. The kit uses 200μL of sample volume and has a dynamic range between 10 and 400pg/ml, corresponding to 1.25×105 and 5×106 virions/ml concentrations. For our experiments, the viral separation efficiency was found by calculating the ratio between p24 values of the post-wash lysis solution and the flow through plus wash solution. In addition, the p24 concentration of the original sample was used to check for mass balance.
Statistics and data analysis
The experiments were repeated using at least 3 different devices at each condition. Data shown in Figs. 3, 4 and 5 on viral capture efficiency represent ELISA measurement results averaged over these devices, and each error bar represents the standard deviation of the mean.
Fig. 3.
Separation efficiencies of the magnetic separator unit alone at varying flow rates (open circles). Comparison with the Miltenyi MACS μ-column and separator (filled circle). Control with no iron particle gradient concentrators (open triangle).
Fig. 4.
(a) Viral capture efficiency of different mixers combined with the magnetic separator. 250 cycles of each mixer at 30μL/min. (b) Herringbone mixers of different lengths (c) Herringbone mixer (250 cycles) at different flow rates.
Fig. 5.
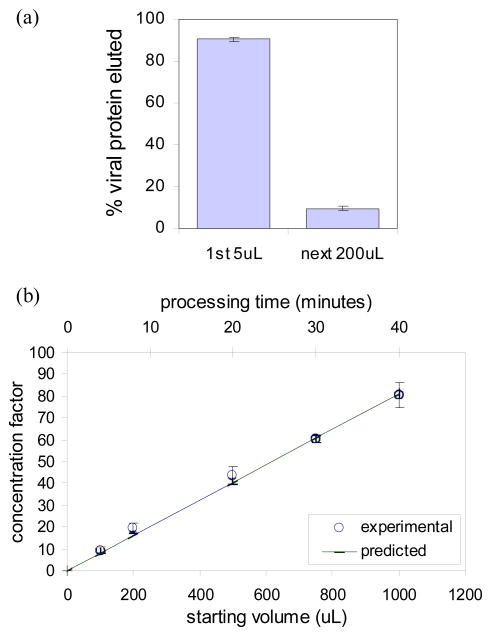
(a) Viral protein recovered in first 5μL of eluted fluid. (b) Concentration factor versus initial sample volume and processing time, predicted and actual values for 30μL/min flow rate.
Results
Magnetic separator unit
The performance of the magnetic separator unit alone was assessed by first mixing the sample with the superparamagnetic beads in a tube off-chip. The mixture was then injected into the microseparator column at flow rates between 30uL/min to 100uL/min and the percentage of viruses trapped inside the micro-separator column was characterized. These correspond to flow velocities of 2.8mm/s to 9.4mm/s. Results in Figure 3 show that the trapping efficiency of the on-chip system is comparable to that of the commercial macro-scale system at flow rates up to 5μL/min. The efficiency at this point averages 78%, and the flow speed is 4.7mm/s. A set of control chips were tested in which an external permanent magnet was placed next to the separation chamber, but the iron micro-particle gradient concentrators were not injected. These obtained almost no viral capture, showing that field gradient concentration from the iron particles are necessary for magnetic viral capture.
Combined mixer and separator devices
The four different mixer designs were integrated with the separator and tested for their overall viral capture efficiency at a flow rate of 30μL/min (25μL/min sample flow rate and 5μL/min bead flow rate). The mixing efficiency was derived by dividing by the 78% efficiency of the separator. This assumes that 60 minutes of off-chip mixing in a test tube provides 100% binding between viruses and the superparamagnetic nanoparticles.
We found that the herringbone mixer narrowly outperformed the 2D Tesla and the combined Tesla and herringbone devices, and all three were considerably more efficient than the straight channel mixer. We then selected the herringbone mixer and further assessed its performance at different flow rates and different mixing lengths. Lowering the number of mixing cycles from 250 decreased the mixing efficiency, but lowering the flow rate increased it. The highest mixing efficiency of 79% was achieved at a flow rate of 10μL/min in a 250 cycle mixer.
Viral concentration
In order to achieve concentration of viral proteins for downstream detection, the volume of viral lysate collected from the device must be less than the initial sample volume. We show in figure 5(a) that more than 90% of the viral proteins elute out in the first 5μL. Figure 5(b) shows the predicted and measured values of the concentration factors given different starting sample volumes, and the time taken to process each sample at a 30μL/min flow rate. Predictions are based on previous measurements of 45% viral capture efficiency from the combined 250 cycle herringbone mixer and separator device, and a 90% viral recovery rate in the first 5uL of eluted fluid. The concentration factor is greater for larger starting volumes, with a highest measured concentration factor was 80x for 1mL of initial sample. The tradeoff is that longer processing time is required.
Discussion
Bringing viral detection from the centralized laboratory into the field has important implications for global public health, biodefense, and environmental safety. Recent advances in micro- and nanotechnology has seen the emergence of numerous micro-fabricated platforms that can sense viral nucleic acids, proteins, or intact virions 1, 35, 36. A major barrier in the practical adoptability of these platforms lies in the difficulty of processing large volumes of low concentration samples that contain many other contaminants besides the virus of interest. Our work attempts to bridge this gap by providing a simple and cost-effective method for concentrating and purifying viruses from a plasma sample.
The major considerations for assessing the performance of a viral extraction and enrichment unit are throughput, extraction efficiency, and concentration factor. Our system consists of three distinct steps: mixing of the sample with the immunomagnetic nanoparticles, magnetic separation of the virus-nanoparticle complexes from flow, and extraction of viral markers by membrane lysis. The overall viral product extraction efficiency is the multiplication product of the efficiencies at each stage, whereas the overall throughput is limited by the lowest throughput stage. The degree of viral product concentration is determined by the amount of sample processed, the overall extraction efficiency, and the final resuspension volume.
Our magnetic separator unit demonstrates a maximum efficiency of 78% for viral extraction, which is comparable to that of the commercial Miltenyi MACS system designed to be used with the 50nm superparamagnetic beads. In our design, we can maintain this up to flow rates of 50μL/min, maintaining the possibility of high throughput; increasing loss of capture occurs at higher flow rates. This throughput is more than 100 times greater than previous designs described by Xia et al. for the separation of 130nm superparamagnetic beads, which ran at 25μL/hour (0.42μL/min) 21, and is equivalent to the flow rate demonstrated with micron-sized beads17, 18, 19.
In this study we also assessed several passive mixer designs for their ability to mix nanoparticles with viral samples for efficient separation. Our assessment of chaotic advection microfluidic mixers revealed that although these devices have been reported to efficiently mix colored dyes, they are inefficient for achieving specific binding between nanoparticles and viral particles. Of the four different device geometries tested, we found that the staggered herringbone mixer described by Stroock et al32 narrowly outperformed the others.
A 250 cycle version of this mixer with a 30μL/min flow rate had an efficiency of 57%, but reducing flow to 10μL/min increased the efficiency to 79%. This suggests that both diffusive and advective effects play important roles in the overall nanoparticle-virus mixing process. It may be possible to improve the mixing efficiency by utilizing an active mixer. rather than the passive mixers investigated here. However, this is likely to increase the system complexity and the cost of the end device, which is an important consideration in devices designed for detecting viruses like HIV and dengue, prevalent in impoverished settings.
The difference in viral extraction efficiency at different flow rates presents a tradeoff between speed and efficiency when selecting the operation mode for the device. In our experiments with large volume blood plasma samples, we chose to favor speed in the interest of completing the processing at realistic time frames suitable for point-of-care settings. Setting the flow rate at 30μL/min, we were able to achieve viral protein enrichments of 44-fold for 0.5mL of starting sample in 20min, and 80-fold for 1mL of starting sample in 40min. If lower flow rates were used, higher concentration factors would have been achieved for the same sample volume, but the assay would take longer to complete. If the application is for low initial sample volumes of <100μL, then a flow rate of 10μL/min is recommended, since more viral products would be extracted, whilst still having a processing time of less than 10min.
Magnetic particles for viral extraction have been extensively explored using macro scale systems for a range of viruses and sample media.37–39 On the micro-scale, Lien et al. 20 previously described a microfluidic system for capturing dengue virus using micron-sized magnetic beads, a rotary mixer, and current carrying coils acting as electromagnets. The system acted as a sample purifier for downstream PCR detection, and the study presented produced 25uL of viral extracts from 25μL of initial sample, so no enrichment was achieved. The authors postulated a potential 20-fold enrichment with 1mL of starting sample, assuming 100% extraction efficiency.
The viral purification system developed in this work shows a significant increase in the degree of viral concentration compared to other on-chip viral purification systems, as well as a significant improvement in throughput over previous microfluidic systems for superparamagnetic nanoparticle separation. Practical processing times of tens of minutes for sample volumes up to 1ml is realistic for point-of-care applications, and may be further improved upon by multiplexing of the mixer units, which form the current bottleneck in throughput.
Conclusions
We demonstrated a simple, low-cost device for the concentration and purification of HIV viral products from human plasma samples. The three-step process of bead mixing, magnetic separation and viral product elution can be completed in 20 minutes for a 0.5ml sample, and can generate a highly enriched solution of viral products 40–80 fold more concentrated than the initial sample, and free from plasma contaminants. The approach could ultimately be integrated with downstream systems for rapid, point-of-care detection of pathogenic viruses.
Fig. 1.
a) Schematic of the device. Viral plasma sample is mixed with functionalized magnetic nanoparticles, then trapped in the magnetic separation chamber. b) Schematic of the magnetic separator, The microfluidic trapping chamber is 5mm long × 4mm wide × 120μm high. 25–75μm iron particles are physically trapped before the 20μm high outlet channel and are compacted into random close packing by the force of the flow. c) Suspension of polydisperse iron particles
Fig. 2.
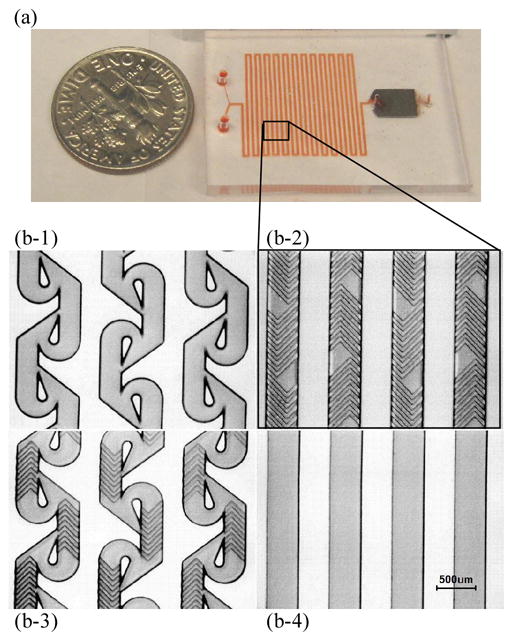
(a) Integrated mixer and separator device, with mixing channel on the left, and magnetic separation chamber on the right. (b) Microscope pictures of SU8 channel molds for the different mixer designs. (b-1) modified 2D Tesla structure. Channels are 100um high and 200um wide in the straight sections. (b-2) Staggered herringbone device. Main channels are 100um high and 300um wide. Herringbone features are 20um high and 50um wide. (b-3) Combined tesla + herringbone. Channels are 100um high and 200um widein the straight sections. Herringbone features are 20um high and 50um wide. (b-4) Straight channel device. Channels are 100um high and 300um wide.
Acknowledgments
We thank Mr Octavio Hurtado for technical support with microfabrication procedures, and Ms Alicja Trocha for assistance with the biosafety of HIV handling. We thank Partners AIDS Research Center for provision of HIV samples and use of tissue culture facilities. This work was supported by the Center for Integration of Medicine and Innovative Technology under Grant No. 09-441 and the National Institute of Biomedical Imaging and Bioengineering under Grant No. P41 EB002503 (BioMEMS Resource Center).
References
- 1.Cheng X, Chen G, Rodriguez WR. Anal Bioanal Chem. 2009;393:487. doi: 10.1007/s00216-008-2514-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Global summary of the AIDS epidemic 2008. World Health Oraganization; 2008. [accessed 7th April 2009]. available at http://www.who.int/hiv/data/2008_global_summary_AIDS_ep.png. [Google Scholar]
- 3.Calmy A, Ford N, Hirschel B, Reynolds SJ, Lynen L, Goemaere E, Garcia de la Vega F, Perrin L, Rodriguez W. Clin Infect Dis. 2007;44:128. doi: 10.1086/510073. [DOI] [PubMed] [Google Scholar]
- 4.Rouet F, Rouzioux C. Clinical Laboratory. 2007;53:135. [PubMed] [Google Scholar]
- 5.Fiscus SA, Cheng B, Crowe SM, Demeter L, Jennings C, Miller V, Respess R, Stevens W. Plos Medicine. 2006;3:1743. doi: 10.1371/journal.pmed.0030417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Auroux PA, Koc Y, deMello A, Manz A, Day PJR. Lab on a Chip. 2004;4:534. doi: 10.1039/b408850f. [DOI] [PubMed] [Google Scholar]
- 7.Espy MJ, Uhl JR, Sloan LM, Buckwalter SP, Jones MF, Vetter EA, Yao JDC, Wengenack NL, Rosenblatt JE, Cockerill FR, Smith TF. Clinical Microbiology Reviews. 2006;19:165. doi: 10.1128/CMR.19.1.165-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu YJ, Rauch CB, Stevens RL, Lenigk R, Yang JN, Rhine DB, Grodzinski P. Analytical Chemistry. 2002;74:3063. doi: 10.1021/ac020094q. [DOI] [PubMed] [Google Scholar]
- 9.Haukanes BI, Kvam C. Bio-Technology. 1993;11:60. doi: 10.1038/nbt0193-60. [DOI] [PubMed] [Google Scholar]
- 10.Miltenyi S, Muller W, Weichel W, Radbruch A. Cytometry. 1990;11:231. doi: 10.1002/cyto.990110203. [DOI] [PubMed] [Google Scholar]
- 11.Safarik I, Safarikova M. J Chromatogr B Biomed Sci Appl. 1999;722:33. [PubMed] [Google Scholar]
- 12.Varshney M, Li Y. Biosens Bioelectron. 2007;22:2408. doi: 10.1016/j.bios.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 13.Soelberg SD, Stevens RC, Limaye AP, Furlong CE. Anal Chem. 2009 doi: 10.1021/ac900007c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez JM, Simeone FJ, Saeki Y, Josephson L, Weissleder R. Journal of the American Chemical Society. 2003;125:10192. doi: 10.1021/ja036409g. [DOI] [PubMed] [Google Scholar]
- 15.Deng T, Prentiss M, Whitesides GM. Applied Physics Letters. 2002;80:461. [Google Scholar]
- 16.Smistrup K, Kjeldsen BG, Reimers JL, Dufva M, Petersen J, Hansen MF. Lab on a Chip. 2005;5:1315. doi: 10.1039/b510995g. [DOI] [PubMed] [Google Scholar]
- 17.Choi JW, Ahn CH, Bhansali S, Henderson HT. Sensors and Actuators B-Chemical. 2000;68:34. [Google Scholar]
- 18.Ramadan Q, Samper V, Poenar D, Yu C. Biomed Microdevices. 2006;8:151. doi: 10.1007/s10544-006-7710-x. [DOI] [PubMed] [Google Scholar]
- 19.Smistrup K, Hansen O, Bruus H, Hansen MF. Journal of Magnetism and Magnetic Materials. 2005;293:597. [Google Scholar]
- 20.Lien KY, Lin JL, Liu CY, Lei HY, Lee GB. Lab Chip. 2007;7:868. doi: 10.1039/b700516d. [DOI] [PubMed] [Google Scholar]
- 21.Xia N, Hunt TP, Mayers BT, Alsberg E, Whitesides GM, Westervelt RM, Ingber DE. Biomed Microdevices. 2006;8:299. doi: 10.1007/s10544-006-0033-0. [DOI] [PubMed] [Google Scholar]
- 22.Inglis DW, Riehn R, Austin RH, Sturm JC. Applied Physics Letters. 2004;85:5093. [Google Scholar]
- 23.Pamme N. Lab Chip. 2006;6:24. doi: 10.1039/b513005k. [DOI] [PubMed] [Google Scholar]
- 24.Gijs MAM. Microfluidics and Nanofluidics. 2004;1:22. [Google Scholar]
- 25.Al-Raoush R, Alsaleh M. Powder Technology. 2007;176:47. [Google Scholar]
- 26.Nguyen NT, Wu ZG. Journal of Micromechanics and Microengineering. 2005;15:R1. [Google Scholar]
- 27.Mansur EA, Ye MX, Wang YD, Dai YY. Chinese Journal of Chemical Engineering. 2008;16:503. [Google Scholar]
- 28.Xia HM, Wan SYM, Shu C, Chew YT. Lab on a Chip. 2005;5:748. doi: 10.1039/b502031j. [DOI] [PubMed] [Google Scholar]
- 29.Munson MS, Yager P. Analytica Chimica Acta. 2004;507:63. [Google Scholar]
- 30.Jen CP, Wu CY, Lin YC, Wu CY. Lab on a Chip. 2003;3:77. doi: 10.1039/b211091a. [DOI] [PubMed] [Google Scholar]
- 31.Hong CC, Choi JW, Ahn CH. Lab on a Chip. 2004;4:109. doi: 10.1039/b305892a. [DOI] [PubMed] [Google Scholar]
- 32.Stroock AD, Dertinger SKW, Ajdari A, Mezic I, Stone HA, Whitesides GM. Science. 2002;295:647. doi: 10.1126/science.1066238. [DOI] [PubMed] [Google Scholar]
- 33.McDonald JC, Whitesides GM. Accounts of Chemical Research. 2002;35:491. doi: 10.1021/ar010110q. [DOI] [PubMed] [Google Scholar]
- 34.Blumel J, Burger R, Gerlich W, Gurtler L, Heiden M, Hitzler W, Jansen B, Klamm H, Lefevre H, Lower J, Ludwig WD, Montag-Lessing T, Offergeld R, Paessens A, Pauli G, Seitz R, Schlenkrich U, Werner E, Willkommen H. Transfusion Medicine and Hemotherapy. 2005;32:196. [Google Scholar]
- 35.Bentzen E, Wright DW, Crowe JE. Future Virology. 2006;1:769. [Google Scholar]
- 36.Patolsky F, Zheng GF, Hayden O, Lakadamyali M, Zhuang XW, Lieber CM. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:14017. doi: 10.1073/pnas.0406159101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myrmel M, Rimstad E, Wasteson Y. International Journal of Food Microbiology. 2000;62:17. doi: 10.1016/s0168-1605(00)00262-2. [DOI] [PubMed] [Google Scholar]
- 38.Jothikumar N, Cliver DO, Mariam TW. Applied and Environmental Microbiology. 1998;64:504. doi: 10.1128/aem.64.2.504-508.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Veyret R, Elaissari A, Marianneau P, Sall AA, Delair T. Analytical Biochemistry. 2005;346:59. doi: 10.1016/j.ab.2005.07.036. [DOI] [PubMed] [Google Scholar]