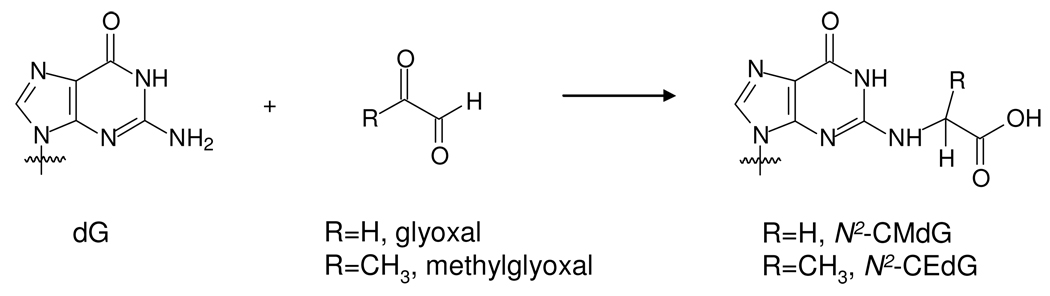
Abstract
Glyoxal is generated endogenously from the degradation of glucose and the oxidation of carbohydrates, lipids as well as the 2-deoxyribose moieties of DNA. Glyoxal is also widely used in industry and is present cigarette smoke and food. Glyoxal can conjugate with nucleobases and proteins to give advanced glycation end products. N2-carboxymethyl-2’-deoxyguanosine (N2-CMdG) and the cyclic 1,N2-glyoxal-dG are the major glyoxal adducts formed in DNA. In this study, we first assessed the stabilities of these two adducts. It turned out that 1,N2-glyoxal-dG was very unstable, with more than 70% of the adduct being decomposed to dG upon a 24-hr incubation at 37°C in phosphate buffered saline. However, N2-CMdG was very stable, less than 0.5% of the lesion was degraded to dG after a 7-day incubation under the same conditions. We further developed a sensitive capillary LC-ESI-MS/MS/MS coupled with stable isotope dilution method and quantified the formation of N2-CMdG in calf thymus DNA and 293T human kidney epithelial cells that were exposed to glyoxal and in calf thymus DNA treated with D-glucose. Our results showed that N2-CMdG was produced at 2–134 lesions per 106 nucleosides in calf thymus DNA when the surrounding glyoxal concentration was increased from 10 to 500 µM and approximately 3–27 lesions per 107 nucleosides while the D-glucose concentration changed from 2 to 50 mM. Furthermore, N2-CMdG was induced endogenously in 293T human kidney epithelial cells and exposure to glyoxal further stimulated the formation of this lesion; the level of this adduct ranged from 7 to 15 lesions per 108 nucleosides while the glyoxal concentration increased from 10 µM to 1.25 mM. Collectively, our results suggested that N2-CMdG might serve as a biomarker for glyoxal exposure.
Introduction
DNA is intrinsically chemically unstable, its nucleobases can be oxidized or alkylated during normal metabolic processes and its glycosidic linkage can be spontaneously hydrolyzed (1). In addition, UV light, ionizing radiation and various environmental agents can modify DNA directly or after metabolic activation (1). Although a certain level of genomic instability may be necessary for fostering genetic diversity, the damaged genome can lead to an enhanced rate of mutagenesis, resulting in compromised cellular function and tumor formation (1).
Humans are exposed to glyoxal from both endogenous and exogenous sources. Glyoxal can arise endogenously from a multitude of pathways, which encompass the oxidation of lipids, unsaturated fatty acids and carbohydrates (2–5). As a degradation product of glucose under physiological conditions, glyoxal was observed to be present at elevated levels in plasma of patients with diabetes, peritoneal dialysis and uremia (6–8). In addition, Awada and Dedon (9) demonstrated that the oxidation of the 2-deoxyribose moiety in DNA resulted in the formation of 3’-phosphoglycolaldehyde, which was subsequently converted to glyoxal to modify nucleobases in DNA. On the other hand, glyoxal is widely used in industry. For instance, it is utilized to serve as an intermediate for the dyestuffs and pharmaceuticals, to crosslink cellulose in paper industry, and to enhance resistance to shrinkage of cotton, fiber and rayon in textiles industry (10). Glyoxal is also used as a disinfectant in health care and dentistry work (10). Moreover, glyoxal is present in polluted air in urban regions, food, beverages, and cigarette smoke (11, 12).
The reactions of glyoxal with 2’-deoxyribonucleosides have been studied (13–16). Under physiological conditions, glyoxal reacts preferentially with 2’-deoxyguanosine (dG) to form 3-(2'-deoxy-β-D-erythro-pentofuranosyl)-5,6,7-trihydro-6,7-dihydroxyimidazo[1,2-a]purine-9-o ne (1,N2-glyoxal-dG) in DNA; this adduct can further couple with dA, dC and dG to render dA-glyoxal-dG, dC-glyoxal-dG and dG-glyoxal-dG cross-links (17, 18). The latter crosslinks were recently quantified in calf thymus and human placental DNA by using capillary liquid chromatography nano-electrospray ionization tandem mass spectrometry (19).
Pluskota-Karwatka et al. (20) also reported two adducts in addition to the known 1,N2-glyoxal-dG adduct in the reactions of glyoxal with dG and calf thymus DNA under physiological conditions. The identities of the two new adducts were characterized as 5-carboxymethyl-3-(2'-deoxy-β-D-erythro-pentofuranosyl)-5,6,7-trihydro-6,7-dihydroxyimidazo [1,2-a]purine-9-one (Gx2-dG) and N2-carboxymethyl-2’-deoxyguanosine (N2-CMdG, Scheme 1) (20). In the same study, 1,N2-glyoxal-dG and Gx2-dG were found to be unstable and transformed partially to N2-CMdG. The same transformation was also observed in single-stranded DNA after prolonged incubation. Therefore, N2-CMdG is the only stable adduct formed in calf thymus DNA upon treatment with glyoxal. Olsen and co-workers (21) quantified 1,N2-glyoxal-dG adduct in calf thymus DNA and mouse hepatoma cells upon treatment with high concentrations of glyoxal by using column switching capillary LC-ESI-MS method. However, 1,N2-glyoxal-dG adduct was only detected in mouse hepatoma cells exposed to 5 and 7 mM of glyoxal for 48 h, and the adduct was not detectable in cells treated with 1 or 3 mM of glyoxal (21).
Scheme 1.
Formation of N2-CMdG and N2-CEdG.
Glyoxal is mutagenic in both bacteria and mammalian cells. In bacteria, the majority of glyoxal-induced single-base substitutions was G:C➔ A:T transitions; in mammalian cells, the predominant the glyoxal-induced single-base substitution was G:C➔ T:A transversion (22, 23). In addition, DNA strand breaks were observed in human skin cells administered with glyoxal (24).
We reason that, as the only stable adduct formed from the reactions of glyoxal with dG or DNA, N2-CMdG may serve as a reliable biomarker for glyoxal exposure. At this point of view, we developed a sensitive capillary LC-ESI-MS3 coupled with isotope-dilution technique and quantified the formation of N2-CMdG in calf thymus DNA exposed to D-glucose and glyoxal as well as in human kidney epithelial cells treated with glyoxal.
Experimental Procedures
Regents and Methods
All reagents, unless otherwise specified, were from Sigma-Aldrich (St. Louis, MO). [U-15N5]-2’-deoxyguanosine was purchased from Cambridge Isotope Laboratories (Andover, MA). Proteinase K was obtained from New England Biolabs (Ipswich, WA). Fetal bovine serum, 293T human kidney cells, penicillin, and streptomycin were purchased from ATCC (Manassas, VA, USA). Electrospray ionization-mass spectrometry (ESI-MS) and tandem MS (MS/MS) experiments were carried out on an LCQ Deca XP ion-trap mass spectrometer (Thermo Fisher Scientific, San Jose, CA). A mixture of acetonitrile and water (50:50, v/v) was used as solvent for electrospray. The spray voltage was 3.0 kV, and the temperature for the ion transport tube was maintained at 275 °C. High-resolution mass spectra (HRMS) were acquired on an Agilent 6510 Q-TOF LC/MS instrument (Agilent Technologies, Santa Clara, CA) coupled with an electrospray ionization (ESI) source and an Agilent HPLC-Chip Cube MS interface. 1H NMR spectra were recorded at 500 MHz on a Varian Inova 500 NMR spectrometer (Varian Inc. Palo Alto, CA).
Synthesis and Characterization of [U-15N5]-N2-CMdG (25)
To a closed vial were added 7.5-µL glyoxal solution (4%), 300 µg [U-15N5]-2’-deoxyguanosine and 0.5 mL of 1 M sodium phosphate buffer. The reaction mixture was heated at 100°C for 16 h. The desired [U-15N5]-N2-CMdG was purified by HPLC on a Beckman system with pump module 125 and a UV detector (module 126). A 4.6×50 mm Luna C18 column (5 µm in particle size and 100 Å in pore size, Phenomenex Inc., Torrance, CA) was used. A solution of 10 mM ammonium formate (pH 6.3, solution A) and a mixture of 10 mM ammonium formate and acetonitrile (70:30, v/v, solution B) were employed as mobile phases. The flow rate was 0.30 mL/min, and a gradient of 5 min 0–4% B, 45 min 4–13% B and 2 min 13-100% B was used. After purification, the identity of [U-15N5]-N2-CMdG was confirmed by both high-resolution ESI-QTOF MS and ion trap LC-MS/MS analyses (see Results).
Synthesis and Characterization of [U-15N5]-1,N2-glyoxal-dG
[U-15N5]-1,N2-glyoxal-dG was synthesized following a previously described method (15). Briefly, a mixture of 544 µg of [U-15N5]-2’-deoxyguanosine and 4.54-µL glyoxal solution (40% in water) at a molar ratio of 1:20 was incubated in 0.5 mL of 50 mM sodium phosphate buffer (pH 7.4) at 37 °C for 2 h. The desired [U-15N5]-1,N2-glyoxal-dG was purified by the above-described HPLC method. The structure of [U-15N5]-1,N2-glyoxal-dG was confirmed by both high-resolution ESI-QTOF MS and ion trap LC-MS/MS analyses (see Results).
Measurement of Extinction Coefficients for N2-CMdG and 1,N2-glyoxal-dG
Unlabeled N2-CMdG and 1,N2-glyoxal-dG were synthesized and purified following the same methods as described above except that unlabeled 2’-deoxyguanosine was employed for the reaction. The extinction coefficients of N2-CMdG and 1,N2-glyoxal-dG at 260 nm were determined to be 1.03*104 and 1.07*104 L·mol−1·cm−1, respectively, by using a previously reported 1H NMR method (26).
Stability Studies of N2-CMdG and 1,N2-glyoxal-dG
A 50 µM solution of N2-CMdG in 1 mL phosphate-buffered saline (PBS, pH 7.4) was incubated at 37°C, and aliquots (200 µL each) were taken out at 0, 1, 2, 4, and 7 days. Similarly, a 100-µM solution of 1,N2-glyoxal-dG was incubated under the same conditions and aliquots (100 µL each) were removed at 0, 2, 4, 8, and 16 hrs as well as at 1, 2, 3, 6 and 8 days. The aliquots were immediately stored in a −20°C freezer prior to being analyzed with HPLC. The HPLC analysis was carried out by using a 4.6×250 mm Apollo C18 column (5 µm in particle size and 300 Å in pore size, Grace Inc., Deerfield, IL) on an Agilent 1100 capillary pump (Agilent Technologies) with a UV detector monitoring at 260 nm and a Peak Simple Chromatography Data System (SRI Instruments Inc., Las Vegas, NV, USA). A solution of 10 mM ammonium formate (pH 6.3, solution A) and a mixture of 10 mM ammonium formate and acetonitrile (70:30, v/v, solution B) were used as mobile phases. A gradient of 5 min 0–12% B, 40 min 10–35% B and 5 min 30–100% B was employed and the flow rate was 0.80 mL/min.
Glyoxal and Glucose Treatment of Calf Thymus DNA
Calf thymus DNA was incubated separately with 0, 10, 25, 50, 100, 250 and 500 µM of glyoxal or 0, 2, 5, 10, 25 and 50 mM of D-glucose in a 1.0-mL phosphate buffer (0.1 M, pH 7.4) at 37°C for 48 hrs. DNA was subsequently precipitated from the reaction mixture by the addition of 0.1 mL of 10 M ammonium acetate and 2.2 mL of cold ethanol and cooled at −20°C. The mixture was centrifuged at 5000 g for 15 min and the supernatant was removed. The recovered DNA was washed with 0.5 mL of cold 70% ethanol, centrifuged as described above, and the supernatant was removed. The same washing step was repeated. The DNA was then dried under vacuum, dissolved in doubly distilled water and quantified by UV spectrophotometry at 260 nm.
Cell Culture, Glyoxal Treatment and DNA Isolation
The 293T human kidney epithelial cells were cultured at 37°C in 5% CO2 atmosphere and in a custom-prepared medium with the same ingredients as Dulbecco’s Modified Eagle’s Medium except that the glucose concentration was 5 mM. The medium was supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA), 100 IU/mL penicillin, and 100 µg/mL streptomycin. After growing to 80% confluence, cells were detached by trypsin-EDTA treatment and harvested by centrifugation to remove the medium. The cell pellets were subsequently washed twice with PBS and resuspended in 20 mL of PBS buffer (at a density of 106 cells/mL) containing 0, 10, 50, 250 or 1250 µM of glyoxal and incubated at room temperature for 3 hrs with occasional shaking.
After treatment, cells (~ 2 × 107 cells) were harvested by centrifugation, and the cell pellets were resuspended in a lysis buffer containing 10 mM Tris-HCl (pH 8.0), 0.1 M EDTA, and 0.5% SDS. The cell lysates were then treated with 20 µg/mL of RNase A at 37°C for 1 hr and subsequently with 100 µg/mL of proteinase K at 50°C for 3 hrs. Genomic DNA was isolated by extraction with phenol/chloroform/isoamyl alcohol (25:24:1, v/v) and desalted by ethanol precipitation. The DNA pellet was redissolved in water and its concentration was measured by UV absorbance at 260 nm.
Enzymatic Digestion and HPLC Enrichment
For the enzymatic digestion of DNA, nuclease P1 (4 units) was added to a mixture containing 100 µg calf thymus or cellular DNA, 200 fmol [U-15N5]-N2-CMdG, 200 fmol [U-15N5]-1,N2-glyoxal-dG, 30 mM sodium acetate (pH 5.5) and 1 mM zinc acetate, and the mixture was incubated at 37 °C for 4 hrs. Nuclease P1 is a single-stranded endonuclease; genomic DNA obtained by the above phenol/chloroform extraction and ethanol precipitation procedures might be partially denatured, rendering it to be cleaved by nuclease P1. To the digestion mixture were then added 30 units of alkaline phosphatase in a 50 mM Tris-HCl buffer (pH 8.6), the digestion was continued at 37°C for 2.5 hrs, and the enzymes were removed by chloroform extraction. The aqueous DNA layer was dried by using a Speed-vac and the dried residues were reconstituted in water. The amount of nucleosides in the mixture was quantified by UV absorbance measurements.
A previous study indicated that the addition of carbonyl scavengers could minimize the artificial formation of N2-CEdG from the involved methylglyoxal during DNA extraction and hydrolysis (27). We carried out the DNA extraction and enzymatic digestion with or without the addition of a carbonyl scavenger, D-penicillamine. The quantification results revealed that, when the enzymatic digestion was performed at 37 °C, the addition of D-penicillamine did not influence appreciably the yield of N2-CMdG. Thus, D-penicillamine was not added in subsequent experiments.
The HPLC removal of unmodified nucleosides from the digestion mixture of isolated or cellular DNA was carried out using the same HPLC system as employed for the stability studies. A solution of 10 mM ammonium formate (pH 6.3, solution A) and a mixture of 10 mM ammonium formate and acetonitrile (70:30, v/v, solution B) were used as mobile phases. A gradient of 5 min 0–8% B, 40 min 10–30% B and 5 min 30–100% B was employed and the flow rate was 0.80 mL/min. The fraction containing N2-CMdG was collected, dried in a Speedvac, reconstituted in distilled water and subjected to LC-MS/MS analysis.
LC-MS/MS Analysis
Quantitative analysis of N2-CMdG in the above DNA hydrolysates was performed by online capillary HPLC-ESI-MS/MS using an Agilent 1200 capillary HPLC pump (Agilent Technologies) interfaced with an LTQ linear ion trap mass spectrometer (Thermo Fisher Scientific, San Jose, CA). A 0.5×150 mm Zorbax SB-C18 column (5 µm in particle size, Agilent Technologies) was used for the separation of the DNA hydrolysis mixture and the flow rate was 6.0 µL/min. A gradient of 0–20% methanol (in 5-min) followed by 20–45% methanol (in 35-min) in water with 0.1% formic acid was employed. The effluent from the LC column was directed to MS/MS/MS analysis, where the LTQ mass spectrometer was set up for monitoring the further fragmentation of the protonated nucleobase portions of unlabeled and labeled N2-CMdG.
Results
Glyoxal can be generated from a variety of endogenous sources (2–8), and it is also widely used in industry and present in the environment, food and cigarette smoke (11, 12). Glyoxal can conjugate with nucleobases to give DNA adducts. Previous studies by Kronberg et al. (20) demonstrated that N2-CMdG was the only stable adduct in calf thymus DNA induced by glyoxal. Based on these previous studies, we reason that N2-CMdG has the potential to be a reliable molecular biomarker for glyoxal and D-glucose exposure. To test this, we developed LC-ESI-MS3 coupled with stable isotope-dilution technique to quantify accurately N2-CMdG formed in calf thymus DNA and 293T human kidney epithelial cells treated with glyoxal and/or D-glucose.
Synthesis and Characterization of Isotope-labeled N2-CMdG and 1,N2-glyoxal-dG
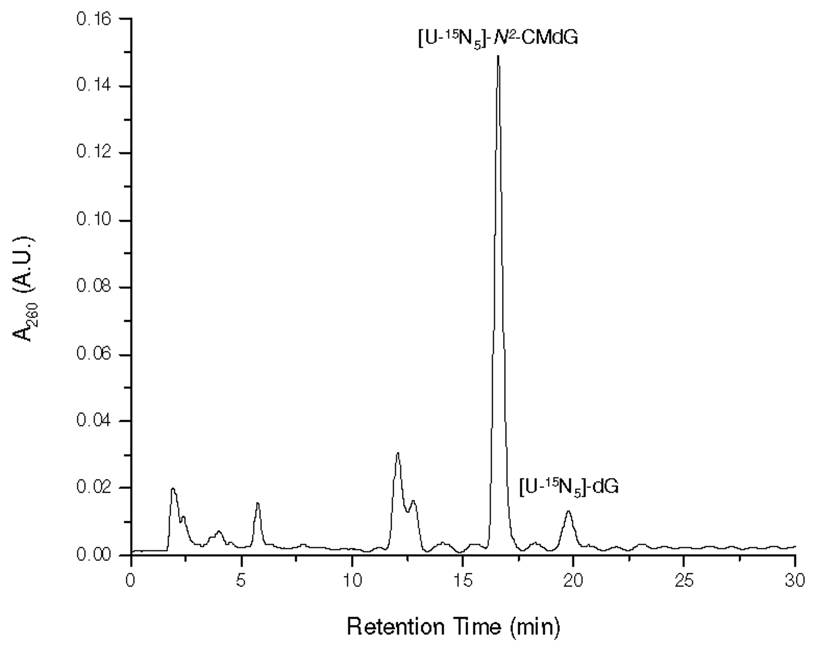
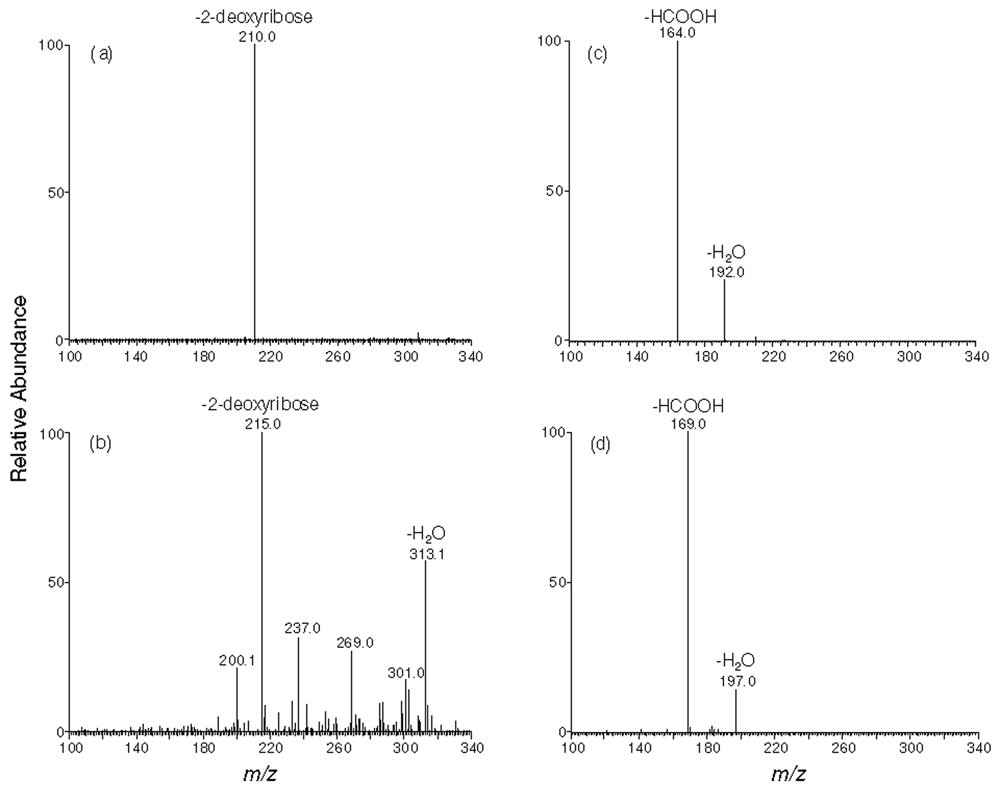
[U-15N5]-N2-CMdG was synthesized from the reaction of [U-15N5]-dG and glyoxal at 100 °C and purified from the reaction mixture (Figure 1 shows the HPLC trace for the separation of the reaction mixture). The formation of [U-15N5]-N2-CMdG increases the mass of [U-15N5]-dG by 58 Da, which gives the [M+H]+ ion at m/z 331. Collision-induced dissociation of the ion of m/z 331 led to the formation of fragment ions of m/z 215 and 313, which are attributed to the elimination of a 2-deoxyribose moiety and a H2O molecule, respectively (Figure S1a). Further fragmentation of the m/z-215 ion gave the most abundant product ion of m/z 169, which originated from the loss of a neutral HCOOH molecule and another product ion of m/z 197 for the loss of a H2O molecule from the [U-15N5]-N2-carboxymethylguanine (Figure S1b). High-resolution ESI MS gives m/z 331.0958 for the [M+H]+ ion of [U-15N5]-N2-CMdG, which is consistent with the calculated m/z of 331.0947 with a deviation of 3.4 ppm.
Figure 1.
HPLC trace for the separation of the reaction mixture of the [U-15N5]-dG with glyoxal.
[U-15N5]-1,N2-glyoxal-dG was synthesized by reacting [U-15N5]-dG with glyoxal at 37 °C for 2 h and purified from the reaction mixture by HPLC (15). High-resolution ESI MS gives m/z 331.0953 for the [M+H]+ ion of [U-15N5]-1,N2-glyoxal-dG, which is consistent with the calculated m/z of 331.0947 with a deviation of 1.9 ppm. Tandem MS analysis of the ion of m/z 331 shows the predominant fragment ion of m/z 215, which again arises from the loss of a 2-deoxyribose moiety (Figure S2a). Further fragmentation of the m/z-215 ion gives the most abundant product ion of m/z 197, which is attributed to the loss of a H2O molecule and another product ion of m/z 169 for the loss of a HCOOH molecule from the [U-15N5]-1,N2-glyoxal-dG (Figure S2b). In addition, we observed the protonated ion of guanine (m/z 157, Figure S2b), which was not observed in the corresponding spectrum of [U-15N5]-N2-CMdG (Figure S1b)
Stability Studies of N2-CMdG and 1,N2-glyoxal-dG
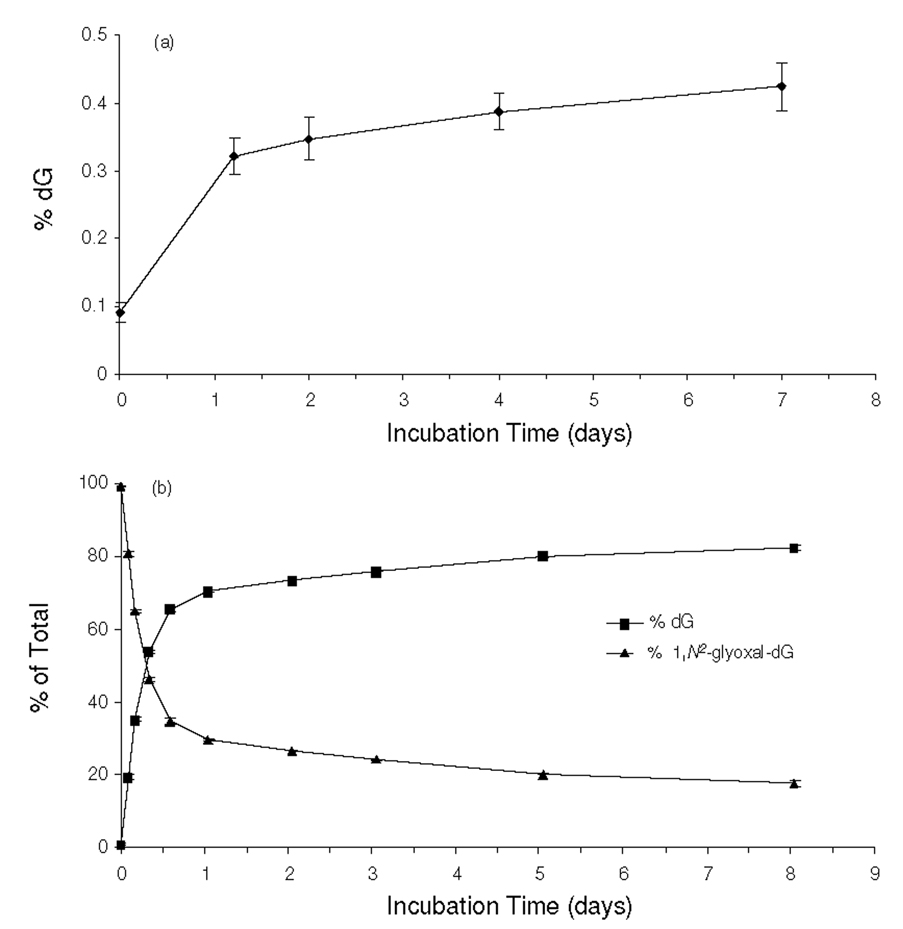
Prior to assessing quantitatively the formation of N2-CMdG and 1,N2-glyoxal-dG, we first examined the chemical stabilities of these two modified nucleosides. To this end, we synthesized the unlabeled N2-CMdG and 1,N2-glyoxal-dG (1H-NMR spectra shown in Figures S3 and S4), which were incubated separately under physiological conditions (PBS buffer, pH 7.4 at 37 °C). Aliquots were removed at different time points and analyzed by HPLC. The HPLC analysis revealed that N2-CMdG was a stable product with less than 0.5 % being decomposed to dG after a 7-day incubation and no other degradation products could be found (Figure 2a&S5). In contrast, 1,N2-glyoxal-dG was found to be very unstable; approximately 70% of 1,N2-glyoxal-dG was decomposed to dG during the first day of incubation (Figure 2b&S6). It is worth noting that, under the above conditions, the conversion from 1,N2-glyoxal-dG to N2-CMdG was too subtle to be detected.
Figure 2.
Time-dependant decomposition of N2-CMdG (a) and 1,N2-glyoxal-dG (b) in PBS buffer (pH 7.4) at 37 °C. The data were obtained from HPLC analyses of the aliquots removed from the N2-CMdG and 1,N2-glyoxal-dG solutions after incubation in PBS buffer for the indicated periods of time. dG was found to be the predominant product arising from the decomposition of the N2-CMdG and 1,N2-glyoxal-dG, and the quantification was based on peak areas observed in the chromatograms with the consideration of the extinction coefficients of dG, N2-CMdG (1.03*104 L·mol−1 · cm−1) and 1,N2-glyoxal-dG (1.07*104 L·mol−1 · cm−1) at 260 nm.
LC-MS Quantification of N2-CMdG in Calf Thymus DNA
LC-MS/MS with the isotope dilution method constitutes a reliable quantification method. In this study, we added isotope-labeled [U-15N5]-N2-CMdG and [U-15N5]-1,N2-glyoxal-dG to the samples prior to the enzymatic hydrolysis of genomic DNA, which corrected for the potential analyte loss during various stages of sample preparation process. Owing to the presence of the carboxylic moiety in N2-CMdG and a better sensitivity afforded by the positive- than the negative-ion mode, we quantified this adduct by LC-MS in the positive-ion mode where 0.1% formic acid was added to the mobile phases to facilitate the protonation of the analyte. In this context, DNA was first digested with nuclease P1 to yield nucleoside 5’-monophosphates, which were dephosphorylated with alkaline phosphatase. The nucleoside mixture was subsequently separated by HPLC and detected by ESI-MS3.
Prior to quantifying the formation of N2-CMdG in DNA samples, we assessed the limit of detection (LOD) for our LC-MS/MS method. It turned out that, when the pure standard was analyzed by LC-MS/MS, the LOD for N2-CMdG at an S/N of 3 was 1.8 fmol. The LOD was very similar (1.9 fmol with an S/N of 3) when the adduct was enriched from the mixture of 100 µg of DNA hydrolysate, which was doped with the [U-15N5]-N2-CMdG, and analyzed by LC-MS/MS under the same conditions. The latter LOQ corresponds to the capability of detecting N2-CMdG at a frequency of 2.8 lesions per 108 normal nucleosides when 100 µg of DNA is used.
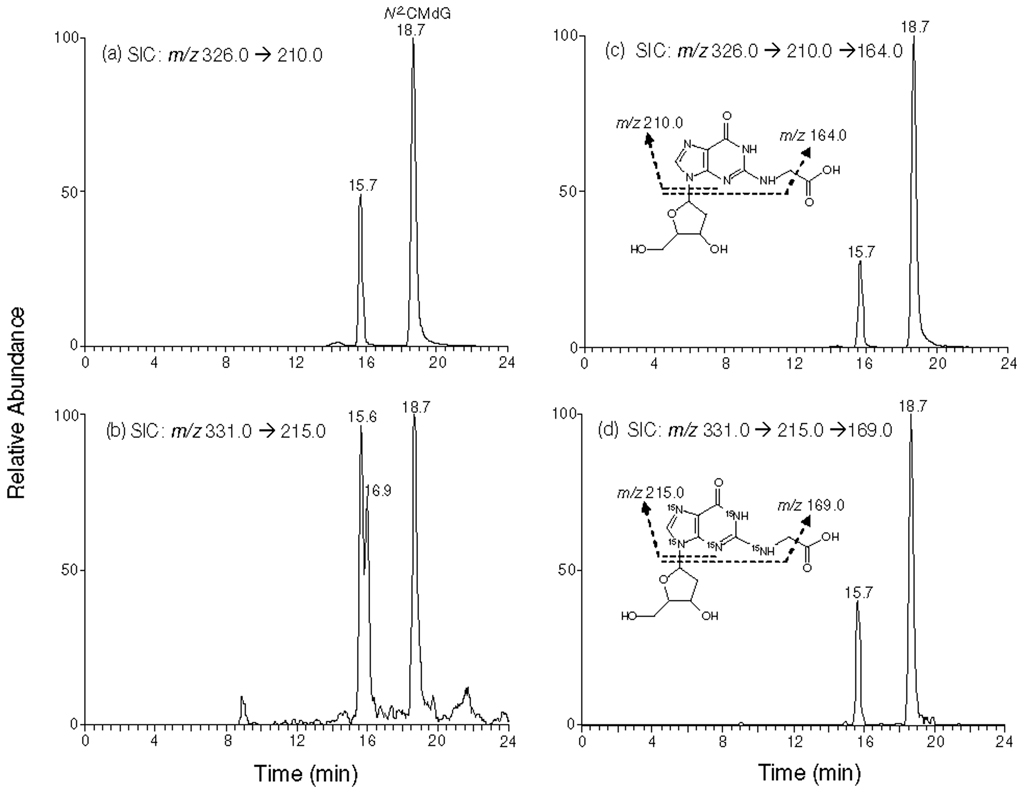
We monitored the m/z 326➔210 and m/z 331➔215 transitions for N2-CMdG and its uniformly 15N-labeled counterpart, respectively. The identity of the component eluting at 18.7 min in the SIC of Figure 3a was determined to be N2-CMdG based on the same retention time and similar tandem mass spectra as those observed for [U-15N5]-N2-CMdG (MS/MS shown in Figure4a & 4b). In this context, aside from the most abundant ion at m/z 215, there were abundant interference ions present in Figure 4b. MS3 analysis monitoring the further cleavage of the ion of m/z 210 was subsequently carried out. When compared to LC-MS/MS, LC-MS3 provided better specificity for identifying and quantifying N2-CMdG (Figure 4c & 4d). The component eluting at 15.7 min in Figure 3 was identified as 1,N2-glyoxal-dG adduct based on similar retention time and tandem MS spectra compared to the added [U-15N5]-1,N2-glyoxal-dG internal standard (MS/MS and MS3 spectra shown in Figure S7). However, due to the unstable nature of 1,N2-glyoxal-dG, we did not quantify this adduct. The origin of the peak eluting at 16.9 min in Figure 3b remains unclear. On the grounds that LC-MS/MS of [U-15N5]-1,N2-glyoxal-dG or [U-15N5]-N2-CMdG under the same conditions gives a single peak in the corresponding selected-ion chromatograms (Figures S8 and S9), it is unlikely that the 16.9-min fraction arises from the conversion of either internal standard to a chemical entity with the same molecular weight. We speculate that it might be due to some isobaric interference present in the sample.
Figure 3.
Selected-ion chromatograms (SICs) for monitoring m/z 326➔210 (a) and m/z 326➔210➔ 164 (c) (for unlabeled N2-CMdG) as well as the m/z 331➔ 215 (b) and m/z 331➔ 215➔ 169 (d) (for [U-15N5]-N2-CMdG) transitions of the digestion mixtures of calf thymus DNA which was treated with 250 µM of glyoxal. The peak at 15.7 min in Figure 3b was identified as1,N2-glyoxal-dG based on its co-elution with the added [U-15N5]-1,N2-glyoxal-dG, and the one at 16.9 min in Figure 3b might be due to the presence of isobaric interferences, as evidenced by the absence of the corresponding peak in the SIC obtained from MS/MS/MS analysis (Figure 3d and discussion in text).
Figure 4.
Product-ion spectra of the ions m/z 326 (a), m/z 331 (b) and MS3 spectra of the ions of m/z 210 (c) and m/z 215 (d) (a, c for unlabeled and b, d for the [U-15N5]-N2-CMdG in calf thymus DNA treated with 250 µM of glyoxal, respectively).
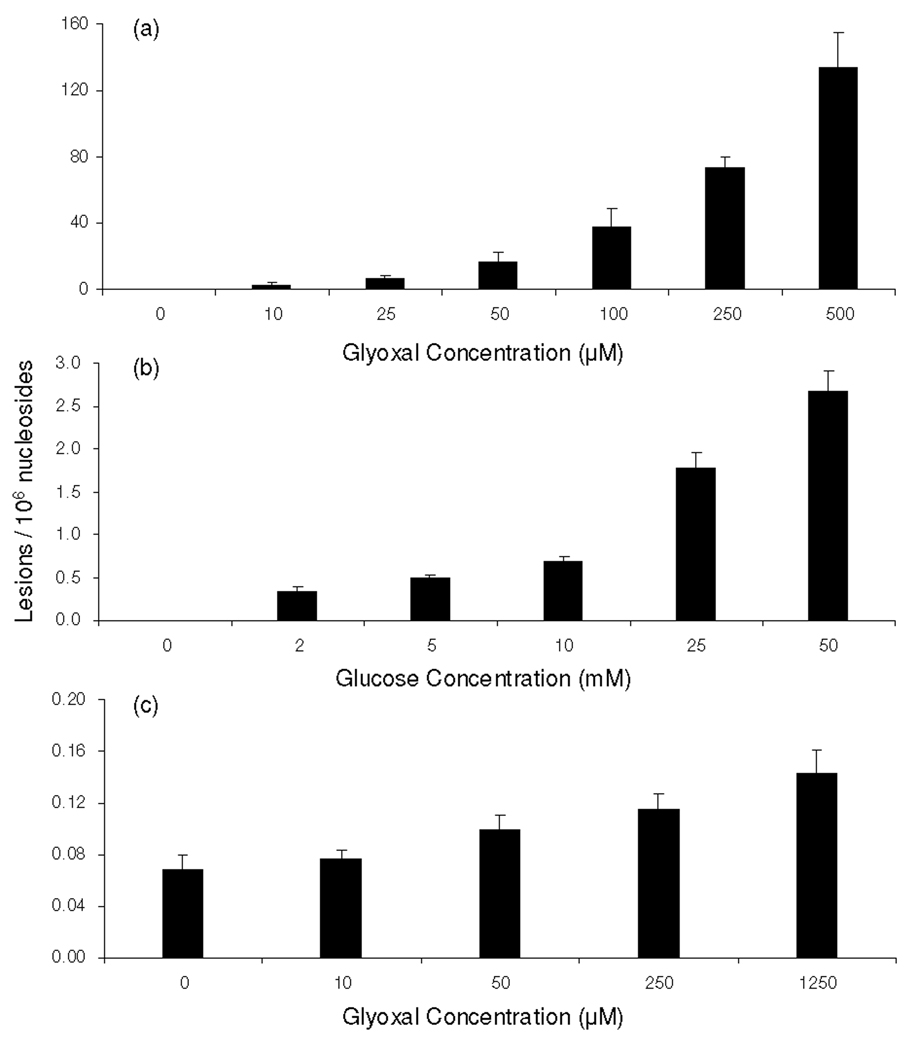
Our LC-MS/MS/MS quantification results revealed that, under physiological conditions, N2-CMdG was formed at a frequency of approximately 2 lesions per 106 nucleosides in calf thymus DNA treated with 10 µM glyoxal. The level of N2-CMdG increased from 7 to 134 adducts per 106 nucleosides when the concentration of glyoxal increased from 25 to 500 µM (Figure 5a). However, this adduct was not detectable in the control calf thymus DNA that was incubated under the same conditions but without the addition of glyoxal, suggesting that, if this adduct is present in calf thymus DNA, it should exist at a level that is lower than 2.8 lesions per 108 nucleosides.
Figure 5.
The dose-dependent formation of N2-CMdG in calf thymus DNA, either untreated or treated with glyoxal (a), glucose (b) and in DNA isolated from glyoxal-treated 293T human kidney cells treated with glyoxal(c).
We further quantified N2-CMdG produced in calf thymus DNA upon D-glucose treatment. It turned out that N2-CMdG could be detected in all DNA samples treated with D-glucose. Treatment of calf thymus DNA with 2, 5, 10, 25 and 50 mM of D-glucose resulted in the formation of N2-CMdG at levels of 0.32 ± 0.06, 0.50 ± 0.03, 0.68 ± 0.07, 1.8 ± 0.2 and 2.7 ± 0.2 lesions per 106 nucleosides, respectively (Figure 5b). Thus, the incubation of calf thymus DNA with 25 mM D-glucose induced approximately 3.5-fold more N2-CMdG, compared to the DNA treated with 5 mM D-glucose. Taken together, N2-CMdG can be observed in isolated DNA upon exposure to D-glucose or glyoxal, and the adduct formation is dependent on the concentrations of D-glucose and glyoxal.
Quantification of N2-CMdG in Cultured 293T Human Kidney Epithelial Cells
By using the same LC-ESI-MS3 coupled with isotope-dilution method, we were able to quantify the formation of N2-CMdG in genomic DNA isolated from 293T human kidney epithelial cells exposed with glyoxal. LC-MS3 again revealed unambiguously the formation of N2-CMdG in 293T cells upon treatment with glyoxal (Figures S10 & S11). The quantification results further demonstrated the dose-dependent formation of N2-CMdG (Figure 5c). The yield of N2-CMdG increased from 0.07 to 0.15 lesions per 106 nucleosides when the concentration of glyoxal rose from 10 to 1250 µM. It is of note that N2-CMdG can be detected in untreated 293T cells at a frequency of approximately 0.07 lesions per 106 nucleosides. The much less efficient formation of N2-CMdG in 293T cells than in calf thymus DNA might be attributed to several factors. First, not all glyoxal added to the medium is uptaken into cells, transported to the nucleus, and reacted with DNA. In addition, the package of DNA in nucleosomes in cells may also result in different frequency of adduct formation compared with isolated calf thymus DNA. Furthermore, DNA adducts in cells can be repaired, whereas those formed in isolated calf thymus DNA are not.
Discussion
Humans are exposed to glyoxal from a variety of sources. The exposure of DNA and isolated nucleosides to glyoxal facilitated the preferential formation of adducts with dG relative to other nucleosides (28). In light of the relatively high yield of 1,N2-glyoxal-dG adduct, it was proposed that this adduct could be a potential biomarker for the assessment of glyoxal exposure (15, 21). Aside from 1,N2-glyoxal-dG, two other glyoxal adducts of dG were found to be present in the reaction mixture of glyoxal with calf thymus DNA, i.e., Gx2-dG and N2-CMdG (20). Among these three adducts, 1,N2-glyoxal-dG adduct was found to undergo spontaneous decomposition to dG at pH 7.4 and 37°C (17, 20). Indeed, our results revealed that, when incubated in PBS buffer at 37°C, more than 70% of 1,N2-glyoxal-dG could be decomposed to dG within one day. Likewise, the Gx2-dG was unstable and can be partially transformed to render N2-CMdG; the same transformation was also observed in single-stranded DNA after prolonged incubation (20). N2-CMdG, however, is very stable; we found that only 0.43 % of N2-CMdG was decomposed after a 7-day incubation under the same conditions. Therefore, N2-CMdG is the only stable adduct formed in calf thymus DNA upon treatment with glyoxal and we only assessed the formation of N2-CMdG in isolated DNA and cultured human cells. Our data revealed a dose-responsive formation of N2-CMdG in calf thymus DNA upon glyoxal exposure. The yield of N2-CMdG increased from 2 to 134 lesions per 106 nucleosides when calf thymus DNA was exposed with 10–500 µM of glyoxal (Figure 5a).
Under physiological conditions, glucose can undergo degradation to form α-ketoaldehydes including glyoxal, methylglyoxal and 3-deoxyglucosone (5). Thus, hyperglycemia may also stimulate the formation of N2-CMdG in DNA. In this respect, it was observed that the incubation of guanosine with glucose under aerobic conditions at 37 °C for 3 weeks could give rise to N2-carboxymethyl-guanosine as the major product (25). We monitored the formation of N2-CMdG in the reaction mixture where calf thymus DNA was incubated with 2, 5, 10, 25 and 50 mM of D-glucose under physiological conditions for 2 days. It turned out that N2-CMdG could be detected in all DNA samples exposed to D-glucose. The level of N2-CMdG in isolated calf thymus DNA treated with 25 mM of glucose was approximately 3.5 fold higher than that in DNA exposed with 5 mM glucose (Figure 5b). In this respect, the glucose concentration in blood samples of healthy human subjects is around 3.6–5 mM; however, it can reach up to 30 mM for diabetic patients. Our experimental result, therefore, suggests that hyperglycemia may stimulate the formation of N2-CMdG in DNA.
To understand the biological implications of N2-CMdG, it is important to assess its formation in human cells. Along this line, N2-CEdG, a DNA advanced glycation endproduct induced by methylglyoxal (Scheme 1), was found to occur frequently in kidney and aorta cells of diabetic and uremic patients as well as in human breast cancer tissues (27, 29). Exposure of cultured human cells to methylglyoxal or glucose further stimulated the formation of N2-CEdG (30). Here we assessed the formation of N2-CMdG in 293T human kidney epithelial cells that were either untreated or treated with glyoxal. Our results showed that N2-CMdG was induced endogenously in 293T cells, and the treatment of these cells with glyoxal further enhanced the formation of N2-CMdG (Figure 5c). It is worth mentioning that we did not quantify the formation of N2-CMdG in 293T cells treated with D-glucose because of the much lower rate for the formation of this product in calf thymus DNA treated with D-glucose (1.8 adduct per 106 normal nucleosides when exposed with 25 mM of D-glucose) than with glyoxal (72 adducts per 106 normal nucleosides when treated with 250 µM of glyoxal) and due to its relatively low rate of formation in 293T cells upon exposure to glyoxal (Figure 5).
Glyoxal exhibits tumor promoting activity on rat glandular stomach carcinogenesis pretreated with N-methyl-N'-nitro-N-nitrosoguanidine (31). In addition, glyoxal has been shown to be a potent mutagen in mammalian cells (23). The majority of glyoxal-induced mutations was G:C➔T:A followed by G:C➔C:G transversions as well as deletions and frameshift mutations, with 83% of the single-base substitutions occurring at G:C base pairs (23). The facile formation of N2-CMdG may account, in part, for the mutations occurring at G:C site. Future studies on the cytotoxic and mutagenic properties of N2-CMdG will further illuminate the relationship between glyoxal exposure and the risk of cancer development.
Supplementary Material
Acknowledgement
This work was supported by the National Institutes of Health (R01 DK082779) and Tobacco-Related Disease Research Program (18XT-0073).
Abbreviations
- N2-CMdG
N2-carboxymethyl-2’-deoxyguanosine
- dG
2’-deoxyguanosine
- 1,N2-glyoxal-dG
3-(2'-deoxy-β-D-erythro-pentofuranosyl)-5,6,7-trihydro-6,7-dihydroxyimidazo[1,2-a]purine-9-one
- Gx2-dG
5-carboxymethyl-3-(2'-deoxy-β-D-erythro-pentofuranosyl)-5,6,7-trihydro-6,7-dihydroxyimidazo [1,2-a]purine-9-one
- ESI-MS
electrospray ionization-mass spectrometry
- MS/MS
tandem MS
Footnotes
Supporting Information Available: LC-MS results and calibration curves. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Frankel EN. Volatile lipid oxidation-products. Prog. Lipid Res. 1983;22:1–33. doi: 10.1016/0163-7827(83)90002-4. [DOI] [PubMed] [Google Scholar]
- 3.Loidlstahlhofen A, Spiteller G. Alpha-hydroxyaldehydes, products of lipid-peroxidation. Biochim. Biophys. Acta- Lipids Lipid Metabol. 1994;1211:156–160. doi: 10.1016/0005-2760(94)90264-x. [DOI] [PubMed] [Google Scholar]
- 4.Mizutari K, Ono T, Ikeda K, Kayashima K, Horiuchi S. Photo-enhanced modification of human skin elastin in actinic elastosis by N-epsilon-(carboxymethyl)lysine, one of the glycoxidation products of the Maillard reaction. J. Invest. Dermatol. 1997;108:797–802. doi: 10.1111/1523-1747.ep12292244. [DOI] [PubMed] [Google Scholar]
- 5.Thornalley PJ, Langborg A, Minhas HS. Formation of glyoxal, methylglyoxal and 3-deoxyglucosone in the glycation of proteins by glucose. Biochem. J. 1999;344:109–116. [PMC free article] [PubMed] [Google Scholar]
- 6.Lapolla A, Flamini R, Vedova AD, Senesi A, Reitano R, Fedele D, Basso E, Seraglia R, Traldi P. Glyoxal and methylglyoxal levels in diabetic patients: quantitative determination by a new GC/MS method. Clin. Chem. Lab. Med. 2003;41:1166–1173. doi: 10.1515/CCLM.2003.180. [DOI] [PubMed] [Google Scholar]
- 7.Ueda Y, Miyata T, Goffin E, Yoshino A, Inagi R, Ishibashi Y, Izuhara Y, Saito A, Kurokawa K, de Strihou CV. Effect of dwell time on carbonyl stress using icodextrin and amino acid peritoneal dialysis fluids. Kidney Int. 2000;58:2518–2524. doi: 10.1046/j.1523-1755.2000.00436.x. [DOI] [PubMed] [Google Scholar]
- 8.Raj DSC, Choudhury D, Welbourne TC, Levi M. Advanced glycation end products: a nephrologist's perspective. Am. J. Kidney Dis. 2000;35:365–380. doi: 10.1016/s0272-6386(00)70189-2. [DOI] [PubMed] [Google Scholar]
- 9.Awada M, Dedon PC. Formation of the 1,N2-glyoxal adduct of deoxyguanosine by phosphoglycolaldehyde, a product of 3 '-deoxyribose oxidation in DNA. Chem. Res. Toxicol. 2001;14:1247–1253. doi: 10.1021/tx0155092. [DOI] [PubMed] [Google Scholar]
- 10.World Health Organization. Concise International Chemical Assessment Document No 57; Glyoxal. 2004:1–47.
- 11.Nagao M, Fujita Y, Wakabayashi K, Nukaya H, Kosuge T, Sugimura T. Mutagens in coffee and other beverages. Environ. Health Perspect. 1986;67:89–91. doi: 10.1289/ehp.866789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moreetesta P, Saintjalm Y. Determination of alpha-dicarbonyl compounds in cigarette-smoke. J. Chromatogr. 1981;217:197–208. [Google Scholar]
- 13.Shapiro R, Hachmann H. Reaction of guanine derivatives with 1,2-dicarbonyl compounds. Biochemistry. 1966;5:2799–2807. doi: 10.1021/bi00873a004. [DOI] [PubMed] [Google Scholar]
- 14.Staehelin M. Inactivation of virus nucleic acid with glyoxal derivatives. Biochim. Biophys. Acta. 1959;31:448–454. doi: 10.1016/0006-3002(59)90019-8. [DOI] [PubMed] [Google Scholar]
- 15.Olsen R, Molander P, Ovrebo S, Ellingsen DG, Thorud S, Thomassen Y, Lundanes E, Greibrokk T, Backman J, Sjoholm R, Kronberg L. Reaction of glyoxal with 2'-deoxyguanosine, 2'-deoxyadenosine, 2'-deoxycytidine, cytidine, thymidine, and calf thymus DNA: identification of DNA adducts. Chem. Res. Toxicol. 2005;18:730–739. doi: 10.1021/tx0496688. [DOI] [PubMed] [Google Scholar]
- 16.Li YY, Cohenford MA, Dutta U, Dain JA. The structural modification of DNA nucleosides by nonenzymatic glycation: an in vitro study based on the reactions of glyoxal and methylglyoxal with 2 '-deoxyguanosine. Anal. Bioanal. Chem. 2008;390:679–688. doi: 10.1007/s00216-007-1682-4. [DOI] [PubMed] [Google Scholar]
- 17.Kasai H, Iwamoto-Tanaka N, Fukada S. DNA modifications by the mutagen glyoxal: adduction to G and C, deamination of C and GC and GA cross-linking. Carcinogenesis. 1998;19:1459–1465. doi: 10.1093/carcin/19.8.1459. [DOI] [PubMed] [Google Scholar]
- 18.Brock AK, Kozekov ID, Rizzo CJ, Harris TM. Coupling products of nucleosides with the glyoxal adduct of deoxyguanosine. Chem. Res. Toxicol. 2004;17:1047–1056. doi: 10.1021/tx049906z. [DOI] [PubMed] [Google Scholar]
- 19.Chen HJC, Chen YC. Analysis of glyoxal-induced DNA cross-links by capillary liquid chromatography nanospray ionization tandem mass spectrometry. Chem. Res. Toxicol. 2009;22:1334–1341. doi: 10.1021/tx900129e. [DOI] [PubMed] [Google Scholar]
- 20.Pluskota-Karwatka D, Pawlowicz AJ, Tomas M, Kronberg L. Formation of adducts in the reaction of glyoxal with 2 '-deoxyguanosine and with calf thymus DNA. Bioorg. Chem. 2008;36:57–64. doi: 10.1016/j.bioorg.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Olsen R, Ovrebo S, Thorud S, Lundanes E, Thomassen Y, Greibrokk T, Molander P. Sensitive determination of a glyoxal-DNA adduct biomarker candidate by column switching capillary liquid chromatography electrospray ionization mass spectrometry. Analyst. 2008;133:802–809. doi: 10.1039/b719842f. [DOI] [PubMed] [Google Scholar]
- 22.MurataKamiya N, Kamiya H, Kaji H, Kasai H. Mutational specificity of glyoxal, a product of DNA oxidation, in the lacI gene of wild-type Escherichia coli W3110. Mutat. Res. 1997;377:255–262. doi: 10.1016/s0027-5107(97)00083-3. [DOI] [PubMed] [Google Scholar]
- 23.MurataKamiya N, Kamiya H, Kaji H, Kasai H. Glyoxal, a major product of DNA oxidation, induces mutations at G:C sites on a shuttle vector plasmid replicated in mammalian cells. Nucleic Acids Res. 1997;25:1897–1902. doi: 10.1093/nar/25.10.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts MJ, Wondrak GT, Laurean DC, Jacobson MK, Jacobson EL. DNA damage by carbonyl stress in human skin cells. Mutat. Res. 2003;522:45–56. doi: 10.1016/s0027-5107(02)00232-4. [DOI] [PubMed] [Google Scholar]
- 25.Seidel W, Pischetsrieder M. Reaction of guanosine with glucose under oxidative conditions. Bioorg. Med. Chem. Lett. 1998;8:2017–2022. doi: 10.1016/s0960-894x(98)00343-6. [DOI] [PubMed] [Google Scholar]
- 26.Hong HZ, Wang YS. Formation of intrastrand cross-link products between cytosine and adenine from UV irradiation of d((Br)CA) and duplex DNA containing a 5-bromocytosine. J. Am. Chem. Soc. 2005;127:13969–13977. doi: 10.1021/ja0531677. [DOI] [PubMed] [Google Scholar]
- 27.Synold T, Xi BX, Wuenschell GE, Tamae D, Figarola JL, Rahbar S, Termini J. Advanced glycation end products of DNA: quantification of N2-(1-Carboxyethyl)-2'-deoxyguanosine in biological samples by liquid chromatography electrospray ionization tandem mass spectrometry. Chem. Res. Toxicol. 2008;21:2148–2155. doi: 10.1021/tx800224y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dutta U, Cohenford MA, Dain JA. Nonenzymatic glycation of DNA nucleosides with reducing sugars. Anal. Biochem. 2005;345:171–180. doi: 10.1016/j.ab.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 29.Li H, Nakamura S, Miyazaki S, Morita T, Suzuki M, Pischetsrieder M, Niwa T. N2-carboxyethyl-2 '-deoxyguanosine, a DNA glycation marker, in kidneys and aortas of diabetic and uremic patients. Kidney Int. 2006;69:388–392. doi: 10.1038/sj.ki.5000064. [DOI] [PubMed] [Google Scholar]
- 30.Yuan B, Cao H, Jiang Y, Hong H, Wang Y. Efficient and accurate bypass of N2-(1-carboxyethyl)-2’-deoxyguanosine by DinB DNA polymerase in vitro and in vivo. Proc. Natl. Acad. Sci. USA. 2008;105:8679–8684. doi: 10.1073/pnas.0711546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi M, Okamiya H, Furukawa F, Toyoda K, Sato H, Imaida K, Hayashi Y. Effects of glyoxal and methylglyoxal administration on gastric carcinogenesis in wistar rats after initiation with N-methyl-N'-nitro-N-nitrosoguanidine. Carcinogenesis. 1989;10:1925–1927. doi: 10.1093/carcin/10.10.1925. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.