Abstract
The regulation of apoptosis (programmed cell death) has been the subject of a vast body of research because of its implication in normal development, tissue homeostasis and a wide range of diseases. The ubiquitin-proteasome system (UPS) plays a prominent role in the control of apoptosis by targeting key cell death proteins, including caspases, the central executioners of apoptosis. Here we summarize the major findings on the function of the UPS in both pro- and anti- apoptotic regulation.
Introduction
Ubiquitination, the conjugation of the protein ubiquitin to target proteins, has emerged as a prominent means of regulating cellular processes. Discovered more than three decades ago for its role in the degradation of unwanted proteins by the proteasome [1,2], it is now known to have additional roles in signaling, transcription, DNA repair, endosomal trafficking and cell viability. [3] [4] Over the past few years it has become increasingly clear that the ubiquitin-proteasome system (UPS) plays a central and complex role in regulating apoptosis by directly targeting key cell death proteins, including caspases, the key executioners of apoptosis.
Apoptosis is a tightly controlled form of active cell death that is necessary for development and organismal homeostasis [5] [6]. Death is achieved by the activation of a family of highly potent and specific proteases, termed caspases (for cysteine-aspartate protease) [7] [8] [9]. Given the potentially fatal consequence of their activity, these enzymes are tightly regulated; the cell maintains several “checkpoints” before it enables them to act. The first level of regulation is intrinsic to caspases themselves. Caspases are initially transcribed as weakly active zymogens, which upon proper stimulation are cleaved to form the active enzyme. This step is brought forth by either internal signals that initiate the formation of the apoptosome [10], or by external cues through receptors that make up the Death Inducing Signaling Complex (DISC) [11]. The second level of caspase regulation is achieved by inhibitors, namely by a family of proteins called IAPs (Inhibitor of Apoptosis Protein) [12] [13].[14] [3] IAPs harbor between one to three copies of a baculovirus IAP repeat (BIR) domain that enable interaction with activated caspases. The BIR domains of certain IAPs, in particular XIAP, have the ability to directly inhibit caspase activity in vitro [15] [16]. Some IAPs also contain a Really Interesting New Gene (RING) domain, which mediates binding to E2 ubiquitin-conjugating enzymes and enables these IAPs to act an E3 ubiquitin ligases [17]. E3 ligases serve as the substrate-binding module and thus convey substrate specificity. Studies in both mammalian systems and Drosophila revealed that the RING domain catalyzes many of the ubiquitination events associated with regulating apoptosis.
Role and Regulation of IAPs in apoptosis
In cells that are destined to die, IAPs are inactivated by pro-apoptotic IAP-antagonists which bind to BIR-domains with higher affinity than caspases [18] [14]. IAP-antagonists, such as Reaper, Hid and Grim, were initially identified in Drosophila based on their essential role for the initiation of apoptosis ([19] and reviewed in [14]). Reaper-family proteins contain a short N-terminal motif, termed IBM (IAP-Binding-Motif), which is required for IAP-binding and cell killing [14]. In mammals, IBM-domain proteins like Smac/DIABLO and Omi/HtrA2 have been identified as well (reviewed in [20]; [21]). Like in Drosophila, these proteins use their N-terminal IBM for IAP-binding and inhibition. However, targeted gene disruption of either Smac/DIABLO, Omi/HtrA2, or both together in double-mutant mice did not cause increased resistance towards apoptosis [22–24]. Therefore, a physiological role of these proteins for regulating IAPs remains to be established, and it is likely that additional IAP-regulators remain to be discovered in mammals. One example is ARTS, which binds to mammalian IAPs, such as XIAP, and inhibits their anti-apoptotic activity [25]. However, ARTS contains no detectable IBM motif and appears to use a distinct mechanism for IAP-binding and inhibition. Finally, cleaved caspases also contain an IBM, although it has been shown that the monomeric caspase-9 and its Drosophila homologue Dronc are also able to bind IAPs [26] [27,28] [3].
The E3 ubiquitin ligase activity of IAPs has been implicated in both promoting and inhibiting apoptosis. Reaper, Hid and Grim (RHG) can induce IAP auto-ubiquitination and degradation, thus removing caspase inhibition [29] [30]. Furthermore, this degradation is mediated by other ubiquitination machinery proteins, such as the E1 UB-activating enzyme UBA1, [31] and the E2 UB-conjugase Effete (UBCD1) [29], which are needed for the efficient removal of DIAP1. Apoptosis has also been shown to be stimulated by the de-ubiquitinases (DUBs) fat-facet, which enhances RHG-induced cell death phenotypes [29] [32], and emperor’s thumb [33], which causes cell-death when over-expressed. In mammals, the IAP antagonist ARTS has also been shown to bind and stimulate XIAP ubiquitination [25], and the SMAC/DIABLO peptide can induce cIAP1/2 ubiquitination and degradation [34] [35]. Furthermore, caspase-8 can be directly activated by cullin3-mediated ubiquitination, which enables the binding of proteins that facilitate caspase oligomerization and auto-activation [36]. In all these contexts, ubiquitination promotes caspase activation and apoptosis. However, ubiquitination can also play an anti-apoptotic role, and this appears to be the dominant function in most cases studied. The physical interaction between IAPs and caspases is insufficient to block apoptosis, and mutations in DIAP1 that eliminate or disrupt the RING motif behave as loss-of-function alleles and die as embryos due to massive cell death [37], [30]. Furthermore, loss of DIAP1 RING function in mutant clones leads to rapid caspase activation and apoptosis, but mutant cells can be permanently rescued by expressing the caspase inhibitor protein p35 [38]. Significantly, these “undead cells” survive for extended periods of time and differentiate into adult structures. This indicates that the primary function of the DIAP1 RING is to inhibit caspases, and that DIAP1 RING function is not essential for many basic cellular functions as long as caspases are kept inactive. Furthermore, it was shown that IAPs can directly ubiquitinate and inactivate caspases. For instance, the Drosophila caspase-9 homologue DRONC is inactivated by DIAP1 ubiquitination, and caspase-3 has been reported to be both poly- and mono-ubiquitinated by both XIAP1 and cIAP1 respectively [3]. However, much remains to be learned about the precise mechanism by which caspase inactivation occurs. In particular, in many cases it is not clear to what extent the ubiquitination of caspases leads to their degradation, or their inhibition by other mechanisms.
A recent study aimed at elucidating the effect of caspase ubiquitination showed that the Drosophila caspase DRICE is ubiquitinated, and that this event is important for its inhibition by DIAP1 [39]. In this case ubiquitination does not target DRICE for degradation, because the addition of a proteasome inhibitor did not change its steady-state levels. In addition, over-expressing DIAP1 in tissue clones did not alter DRICE stability.
Another indicator that caspases are controlled by IAP-mediated ubiquitination came from targeted disruption of the XIAP gene in the mouse [40]. Targeted deletion of the XIAP RING (“ΔRING”) mutant mice led, as expected, to increases stability of the truncated XIAP protein. However, contrary to expectations based on biochemical studies suggesting that caspase inhibition is only mediated by the BIR domains, RING MEFs display elevated caspase-3 activity in response to different apoptotic stimuli. This elevated activity was associated with less caspase-3 ubiquitination, but like in the case of DRICE, this ubiquitination did not influence steady-state protein levels. ΔRING mice also displayed more apoptosis and suppressed E -myc-induced lymphomas. Taken together, these results reveal a physiological requirement of XIAP ubiquitin-ligase activity for the inhibition of mammalian caspases and for tumor suppression in vivo. The general picture that emerges is that while IAP-RING function serves to opposing roles in cell death, the net effect of eliminating RING function for most cases studied so far appears to favor caspase activation and apoptosis. Based on biochemical and structural studies it was suggested that XIAP is the only bona fide mammalian caspase inhibitor [41]. However, these conclusions were based on based on simplified in vitro assays with individual domains of IAPs that ignore their E3-ligase activity. Furthermore, because different IAPs can form heteromeric complexes, it is possible that IAPs that do not have the ability to inhibit caspases in vitro may regulate caspases in vivo.
Protein stability controlled by ubiquitination can also be used to regulate pre-activated caspases, as a way of maintaining continuous low levels of the apoptotic machinery and creating a negative feedback loop for preventing unwanted death. A study by Shapiro et. al [42] showed that Drosophila apoptosome components Apaf-1 and DRONC reciprocally control each other’s protein level. Inactive full-length DRONC levels are reduced by over-expressing Apaf-1, and are elevated in mutant Apaf-1 tissue-clones. Similarly, Apaf-1 levels were increased in DRONC mutant clones. This interaction is mediated by active DRONC, since Apaf-1 cleavage-resistant constructs were no longer able to change full-length DRONC levels. The DIAP1 RING domain also plays a role in this system, by possibly mediating the ubiquitin-dependant degradation of both DRONC and Apaf-1. These findings support the idea that low levels of caspase activation can induce the ubiquitination and degradation of pro-apoptotic factors, so as to create a barrier against a “run-away” proteolytic cascade.
IAP antagonists are also subject to ubiquitination, presumably as another means of preventing unwanted death. Drosophila reaper is ubiquitinated by DIAP1, which promotes its degradation, while constructs of reaper that cannot be ubiquitinated are more potent killers [43]. Likewise, the mammalian IAP-antagonist ARTS is kept in check by ubiquitin-dependent degradation, which is overcome when apoptosis is induced [44]. Taken together, these examples of the anti-apoptotic role of IAP-induced ubiquitination demonstrate ways for a cell to maintain viability while keeping a “finger on the trigger” to enable apoptosis when appropriate. Further work in this area is likely to advance our understanding of both the regulation of normal cell death, and also provide insight into the mechanisms of de-regulated cell death in human diseases. In particular, IAPs are commonly over-expressed in human tumors and have become prominent targets for the development of novel anti-cancer drugs [13]. Initially, small IBM-motif peptides and small molecule derivatives that mimic the physical interaction between IAPs and their antagonist were designed to compete with IAP-caspase binding, thus removing their inhibition and allowing death to occur. However, it turns out that these molecules can also target certain IAPs for ubiquitination and degradation [35] [34]. Therefore, the ability to modulate IAP-mediated ubiqutination is expected to have profound implications for the development of novel anti-cancer therapies.
A large body of work in the last several years has focused on the role of IAPs in signaling, most prominently the Nf-kappa-B pathway, as is discussed in recent reviews [13] [3]. It is worth noting that this regulation affects apoptosis somewhat indirectly, and usually involves non-degradative ubiquitination. Nonetheless, the discoveries that the RING domain of several IAPs can mediate K-63 linked ubiquitination [45] [46] [47], which promotes protein-protein interactions, and contain ubiquitin-binding domains [48], are another indicator that IAPs control cell viability in many ways through complex interactions.
Ubiquitination in non-apoptotic caspase activation
An added level of complexity to caspase regulation is their non-apoptotic role in certain contexts. Caspases are involved in diverse pathways, including processing of pro-inflammatory cytokines, cellular signaling, differentiation and remodeling [49] [50]. At this point, very little is known about how the potentially lethal activity of caspases is restricted in time and space to specific cellular compartments. However, several studies indicate that the ubiquitination machinery is used also in these cases to permit spatially limited proteolysis.
During differentiation and development, certain cells undergo major re-structuring and remodeling. This often involves compartmentalized degradation of portions of a cell, such as branched regions and/or elimination organelles. In some cases, this degradation event involves both non-apoptotic local caspase activation and the Ubiquitin-Proteasome system (UPS), as is in the most recently explored Drosophila sperm differentiation and neuronal pruning during metamorphosis. In the former case, almost all the cytoplasm and excess organelles are removed from the elongated spermatids to create highly motile sperm [51,52]. This process was shown to require caspase activation and known apoptosis regulators, including cytochrome- c, Apaf-1, DRONC, DRICE [53] [54] [55] [56]. Interestingly, DIAP1, which is required to prevent unwanted caspase activation and death in most somatic cells, does not appear to play a dominant role for caspase regulation in this system. From a genetic screen, Arama and colleagues discovered that components of a cullin-3 based E3 ubiquitin-ligase are required for caspase activation and sperm differentiation [57]. Mutations in all three components of the complex, a testis specific cullin-3 isoform, the RING protein Roc1B and the substrate binding BTB protein KLHL10 cause male sterility and abolish active-caspase-3 staining from differentiating spermatids. Furthermore, these proteins affect global ubiquitination, as conjugated poly-ubiquitin staining is greatly reduced in these mutants. In this regard, mammalian spermatogenesis displays similarities to Drosophila spermatogenesis, in that the removal of organelles and cytoplasm involves apoptotic proteins and the cells display caspase-3 staining [50] [58]. Interestingly, it was recently reported that mutations in the human form of KLHL10 are associated with male infertility and low sperm count [59], indicating that ubiquitination may be used in a similar fashion for cell sculpting in mammals.
Neuronal pruning is the process by which excess or inaccurate projections are eliminated [60]. It is believed that this is achieved by local degeneration, characterized by cytoskeleton destabilization, fragmentation and clearance. Although an array of proteases have been implicated in different pruning systems, pruning of class IV dendritic arborization sensory neurons (C4da) in Drosophila requires local caspase activity [61]. This activity can be detected by restricted caspase-3-like staining in degenerating dendrites, and verified by the fact that mutations in DRONC and APAF-1 or DIAP1 over-expression all suppress branch removal. As in sperm maturation, the UPS is also involved in this process. Proteasome subunits, Uba1 and UBCD1 are necessary for proper dendrite severing, as well as DIAP1 ubiquitination. Since caspase-6 activity has been recently implicated in mammalian axonal-pruning [62], it will be interesting to see whether the UPS also plays a part in regulating localized degeneration in mammals as well, especially as there has been evidence of proteasome sequestration and movement along dendritic spines during excitation [63].
Localized degeneration in these systems can be viewed as “controlled atrophy”, which requires coupling of both caspases and the proteasome protease activity. The emerging duel role of these regulation pathways could represent a way of controlling “macro-proteolysis” of organelles and massive protein turnover and destruction. Caspase activation in the testis, for instance, occurs in parallel to the re-organization of the proteasome with specialized catalytic subunits, which, when mutated, affect caspase activity [64]. Changing the proteolytic profile of the cell (or sub-compartment) may be a way of controlling large-scale morphological changes. Therefore, it would be interesting to investigate whether the activity of proteasomes themselves can be regulated at the sub-cellular level to control these changes.
Conclusion
Virtually all our cells constitutively express all the components necessary to carry out apoptosis and can rapidly self-destruct in response to a wide range of cell death stimuli. At the same time, healthy cells can avoid the activation of a caspase cascade and live for many years, or even the lifetime of an organism. Therefore, very stringent yet responsive mechanisms must govern caspase activation and cell death. The UPS, with its ability to rapidly eliminate and modify regulatory proteins in a very precise manner well-suited for this purpose. A growing number of observations support a major role of ubiquitnation in the regulation of key cell death proteins, including caspases. At this point, the major family of relevant E3-ligases are members of the IAP family, which can inhibit caspases by direct binding and ubiquitination. In cells that are doomed to die, IAP-antagonists, such as Reaper, are activated and promote the self-conjugation and degradation of IAPs. In this way, the E3-ligase activity of IAPs is switched from an anti-apoptotic to a pro-apototic function upon the induction of apoptosis. These activities can be, at least partially, mimicked by small molecules that provide leads for the development of novel anti-cancer drugs that target elevated levels of IAPs that found in many human tumors. On the other hand, IAP-mediated ubiquitination does not always lead to protein degradation, but can also alter enzymatic activity, protein-protein interaction and sub-cellular targeting. Besides IAPs, other E3-ligases have also been implicated in the regulation of caspases, including cullin-based multi-subunit proteins, and it is likely that additional players will emerge. Another open question is whether proteasomes are also directly regulated during apoptosis, and how the UPS is used in coordination with non-lethal caspase activity for the targeted degradation of specific cellular structures during cellular remodeling. Further advances in this area are likely to not only provide major new insights into basic biological mechanisms, but also drive the development of novel therapeutics to treat a range of human diseases.
Fig. 1. Role of Ubiquitin in the regulation of apoptosis.
Ubiquitination plays opposing roles at different stages during the life-death decision in a cell. In cells that live, IAP-mediarted ubiquitination serves to inhibit unwanted caspase activity. IAP-dependant ubiquitination of caspases leads to inhibition of apoptosis through both proteasomal degradation and a non-degradative pathway. In addition, it can also be used to eliminate caspase activators, like apoptosome subunits [42]. Upon induction of apoptosis, IAP-RING function promotes self-conjugation and UPS-mediated degradation of IAP-proteins, thereby removing the “brakes on death” and facilitating the execution of apoptosis [29] [17] [25] [30]. Additionally, non-degradative ubiquitination can also directly activate caspases, by mediating self-cleavage through the induction of specific protein-protein interactions [36]. Besides IAPS, other E3’s, such as cullin-based E3 ligases, have also been shown to regulate caspase activation, as in sperm differentiation [57].
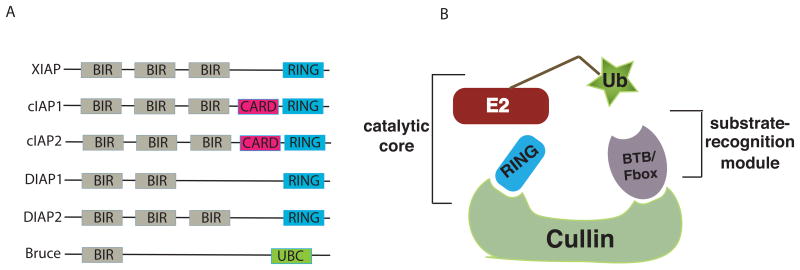
Fig. 2.
(A)Domain structure of RING-containing IAPs
IAP family members are defined by containing at least one BIR domain, which mediates their interaction with caspases. In addition, the IAPs listed here also harbor a RING domain, enabling them to direct ubiquitination of substrate proteins. The three best studied mammalian IAPs (cIAP1, cIAP2, and XIAP) are shown here. cIAP1 and cIAP2 contain a Casapse Recruitment Domain (CARD). In Drosophila, DIAP1 is strictly required to prevent unwanted caspase activation and apoptosis in somatic cells. DIAP2 has a domain arrangement very similar to mammalian IAPs; it is important for regulating the innate immune response [65,66] and also inhibits the effector caspase Drice [67]. Bruce/Apollon is a giant BIR protein that also contains a Ubiquitin Conjugating (UBC) domain [68]. In mammals, Bruce was shown to inhibit apoptosis by binding and mediating the ubiquitination of Smac and caspase-9 [69,70], while in Drosophila it suppress death induced by RHG proteins [71], and might have a role in caspase activation during sperm differentiation [57].
(B) Cullin-based E3 ligases
Cullins are scaffolding proteins that mediate the physical interaction between different subunits, like BTB and Fbox proteins, which are responsible for substrate recognition and RING domain containing proteins, necessary for E2 ligase recruitment. Cullins are known to interact with several distinct E2 ligase and numerous substrate recognition proteins, and this combinatorial arragement of proteins allows cullins to mediate both general and specific biological events (reviewed in [72]).
Acknowledgments
We thank Sigi Benjamin and Samara Brown for critically reading this manuscript. H Steller is an investigator of the Howard Hughes Medical Institute. This work was supported by NIH grant RO1GM60124 to HS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hershko A, Ciechanover A, Rose IA. Resolution of the ATP-dependent proteolytic system from reticulocytes: a component that interacts with ATP. Proc Natl Acad Sci U S A. 1979;76:3107–3110. doi: 10.1073/pnas.76.7.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciechanover A, Heller H, Elias S, Haas AL, Hershko A. ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc Natl Acad Sci U S A. 1980;77:1365–1368. doi: 10.1073/pnas.77.3.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broemer M, Meier P. Ubiquitin-mediated regulation of apoptosis. Trends Cell Biol. 2009;19:130–140. doi: 10.1016/j.tcb.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Thompson SJ, Loftus LT, Ashley MD, Meller R. Ubiquitin-proteasome system as a modulator of cell fate. Curr Opin Pharmacol. 2008;8:90–95. doi: 10.1016/j.coph.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steller H. Mechanisms and genes of cellular suicide. Science. 1995;267:1445–1449. doi: 10.1126/science.7878463. [DOI] [PubMed] [Google Scholar]
- 6.Abraham MC, Shaham S. Death without caspases, caspases without death. Trends Cell Biol. 2004;14:184–193. doi: 10.1016/j.tcb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Yuan J, Shaham S, Ledoux S, Ellis HM, Horvitz HR. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 8.Thornberry NA, Lazebnik Y. Caspases: enemies within. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 9.Nicholson DW, Thornberry NA. Caspases: killer proteases. Trends Biochem Sci. 1997;22:299–306. doi: 10.1016/s0968-0004(97)01085-2. [DOI] [PubMed] [Google Scholar]
- 10.Schafer ZT, Kornbluth S. The apoptosome: physiological, developmental, and pathological modes of regulation. Dev Cell. 2006;10:549–561. doi: 10.1016/j.devcel.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Song Z, Steller H. Death by design: mechanism and control of apoptosis. Trends Cell Biol. 1999;9:M49–52. [PubMed] [Google Scholar]
- 12.Clem RJ, Miller LK. Control of programmed cell death by the baculovirus genes p35 and iap. Mol Cell Biol. 1994;14:5212–5222. doi: 10.1128/mcb.14.8.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LaCasse EC, Mahoney DJ, Cheung HH, Plenchette S, Baird S, Korneluk RG. IAP-targeted therapies for cancer. Oncogene. 2008;27:6252–6275. doi: 10.1038/onc.2008.302. [DOI] [PubMed] [Google Scholar]
- 14.Steller H. Regulation of apoptosis in Drosophila. Cell Death Differ. 2008;15:1132–1138. doi: 10.1038/cdd.2008.50. [DOI] [PubMed] [Google Scholar]
- 15.Deveraux QL, Takahashi R, Salvesen GS, Reed JC. X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- 16.Deveraux QL, Roy N, Stennicke HR, Van Arsdale T, Zhou Q, Srinivasula SM, Alnemri ES, Salvesen GS, Reed JC. IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. Embo J. 1998;17:2215–2223. doi: 10.1093/emboj/17.8.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- 18.Kornbluth S, White K. Apoptosis in Drosophila: neither fish nor fowl (nor man, nor worm) J Cell Sci. 2005;118:1779–1787. doi: 10.1242/jcs.02377. [DOI] [PubMed] [Google Scholar]
- 19.White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- 20.Salvesen GS, Duckett CS. IAP proteins: blocking the road to death’s door. Nat Rev Mol Cell Biol. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- 21.Vaux DL, Silke J. IAPs, RINGs and ubiquitylation. Nat Rev Mol Cell Biol. 2005;6:287–297. doi: 10.1038/nrm1621. [DOI] [PubMed] [Google Scholar]
- 22.Jones JM, Datta P, Srinivasula SM, Ji W, Gupta S, Zhang Z, Davies E, Hajnoczky G, Saunders TL, Van Keuren ML, et al. Loss of Omi mitochondrial protease activity causes the neuromuscular disorder of mnd2 mutant mice. Nature. 2003;425:721–727. doi: 10.1038/nature02052. [DOI] [PubMed] [Google Scholar]
- 23.Martins LM, Morrison A, Klupsch K, Fedele V, Moisoi N, Teismann P, Abuin A, Grau E, Geppert M, Livi GP, et al. Neuroprotective role of the Reaper-related serine protease HtrA2/Omi revealed by targeted deletion in mice. Mol Cell Biol. 2004;24:9848–9862. doi: 10.1128/MCB.24.22.9848-9862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okada H, Suh WK, Jin J, Woo M, Du C, Elia A, Duncan GS, Wakeham A, Itie A, Lowe SW, et al. Generation and characterization of Smac/DIABLO-deficient mice. Mol Cell Biol. 2002;22:3509–3517. doi: 10.1128/MCB.22.10.3509-3517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottfried Y, Rotem A, Lotan R, Steller H, Larisch S. The mitochondrial ARTS protein promotes apoptosis through targeting XIAP. Embo J. 2004;23:1627–1635. doi: 10.1038/sj.emboj.7600155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chai J, Yan N, Huh JR, Wu JW, Li W, Hay BA, Shi Y. Molecular mechanism of Reaper-Grim-Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination. Nat Struct Biol. 2003;10:892–898. doi: 10.1038/nsb989. [DOI] [PubMed] [Google Scholar]
- 27.Meier P, Silke J, Leevers SJ, Evan GI. The Drosophila caspase DRONC is regulated by DIAP1. Embo J. 2000;19:598–611. doi: 10.1093/emboj/19.4.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shiozaki EN, Chai J, Rigotti DJ, Riedl SJ, Li P, Srinivasula SM, Alnemri ES, Fairman R, Shi Y. Mechanism of XIAP-mediated inhibition of caspase-9. Mol Cell. 2003;11:519–527. doi: 10.1016/s1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 29.Ryoo HD, Bergmann A, Gonen H, Ciechanover A, Steller H. Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat Cell Biol. 2002;4:432–438. doi: 10.1038/ncb795. [DOI] [PubMed] [Google Scholar]
- 30.Wilson R, Goyal L, Ditzel M, Zachariou A, Baker DA, Agapite J, Steller H, Meier P. The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat Cell Biol. 2002;4:445–450. doi: 10.1038/ncb799. [DOI] [PubMed] [Google Scholar]
- 31.Lee TV, Ding T, Chen Z, Rajendran V, Scherr H, Lackey M, Bolduc C, Bergmann A. The E1 ubiquitin-activating enzyme Uba1 in Drosophila controls apoptosis autonomously and tissue growth non-autonomously. Development. 2008;135:43–52. doi: 10.1242/dev.011288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wing JP, Schreader BA, Yokokura T, Wang Y, Andrews PS, Huseinovic N, Dong CK, Ogdahl JL, Schwartz LM, White K, et al. Drosophila Morgue is an F box/ubiquitin conjugase domain protein important for grim-reaper mediated apoptosis. Nat Cell Biol. 2002;4:451–456. doi: 10.1038/ncb800. [DOI] [PubMed] [Google Scholar]
- 33.Ribaya JP, Ranmuthu M, Copeland J, Boyarskiy S, Blair AP, Hay B, Laski FA. The deubiquitinase emperor’s thumb is a regulator of apoptosis in Drosophila. Dev Biol. 2009;329:25–35. doi: 10.1016/j.ydbio.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 34.Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P, Zobel K, Dynek JN, Elliott LO, Wallweber HJ, et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell. 2007;131:669–681. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 35.Vince JE, Wong WW, Khan N, Feltham R, Chau D, Ahmed AU, Benetatos CA, Chunduru SK, Condon SM, McKinlay M, et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell. 2007;131:682–693. doi: 10.1016/j.cell.2007.10.037. Charecterization of the mechanism by which IAP antagonist peptides and mimetics induce apoptosis by cIAP1/2 ubiquitin-mediated degardation. As IAP antagonist-based theraputics have entered clinical trials, these studies showed that the mode of apoptosis induction in treated cells is through the ubiquitination and degradation of cIAP1 and 2, and not through physcial competition between IAP-caspase and IAP-antagonist. [DOI] [PubMed] [Google Scholar]
- 36.Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, Ashkenazi A. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721–735. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 37.Lisi S, Mazzon I, White K. Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics. 2000;154:669–678. doi: 10.1093/genetics/154.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryoo HD, Gorenc T, Steller H. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev Cell. 2004;7:491–501. doi: 10.1016/j.devcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 39.Ditzel M, Broemer M, Tenev T, Bolduc C, Lee TV, Rigbolt KT, Elliott R, Zvelebil M, Blagoev B, Bergmann A, et al. Inactivation of effector caspases through nondegradative polyubiquitylation. Mol Cell. 2008;32:540–553. doi: 10.1016/j.molcel.2008.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schile AJ, Garcia-Fernandez M, Steller H. Regulation of apoptosis by XIAP ubiquitin-ligase activity. Genes Dev. 2008;22:2256–2266. doi: 10.1101/gad.1663108. The first study that showed that the RING domain of mammalian IAPs are important for regualtion of caspase activity. By using XIAP RING mutants, the authors demonstrated that it mediates the ubiquitination and in-activation of caspases and is a factor in tumor progression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eckelman BP, Salvesen GS, Scott FL. Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 2006;7:988–994. doi: 10.1038/sj.embor.7400795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shapiro PJ, Hsu HH, Jung H, Robbins ES, Ryoo HD. Regulation of the Drosophila apoptosome through feedback inhibition. Nat Cell Biol. 2008;10:1440–1446. doi: 10.1038/ncb1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olson MR, Holley CL, Yoo SJ, Huh JR, Hay BA, Kornbluth S. Reaper is regulated by IAP-mediated ubiquitination. J Biol Chem. 2003;278:4028–4034. doi: 10.1074/jbc.M209734200. [DOI] [PubMed] [Google Scholar]
- 44.Lotan R, Rotem A, Gonen H, Finberg JP, Kemeny S, Steller H, Ciechanover A, Larisch S. Regulation of the proapoptotic ARTS protein by ubiquitin-mediated degradation. J Biol Chem. 2005;280:25802–25810. doi: 10.1074/jbc.M501955200. [DOI] [PubMed] [Google Scholar]
- 45.Bertrand MJ, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J, Gillard JW, Jaquith JB, Morris SJ, Barker PA. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 46.Cheung HH, Plenchette S, Kern CJ, Mahoney DJ, Korneluk RG. The RING domain of cIAP1 mediates the degradation of RING-bearing inhibitor of apoptosis proteins by distinct pathways. Mol Biol Cell. 2008;19:2729–2740. doi: 10.1091/mbc.E08-01-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herman-Bachinsky Y, Ryoo HD, Ciechanover A, Gonen H. Regulation of the Drosophila ubiquitin ligase DIAP1 is mediated via several distinct ubiquitin system pathways. Cell Death Differ. 2007;14:861–871. doi: 10.1038/sj.cdd.4402079. [DOI] [PubMed] [Google Scholar]
- 48.Gyrd-Hansen M, Darding M, Miasari M, Santoro MM, Zender L, Xue W, Tenev T, da Fonseca PC, Zvelebil M, Bujnicki JM, et al. IAPs contain an evolutionarily conserved ubiquitin-binding domain that regulates NF-kappaB as well as cell survival and oncogenesis. Nat Cell Biol. 2008;10:1309–1317. doi: 10.1038/ncb1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuranaga E, Miura M. Nonapoptotic functions of caspases: caspases as regulatory molecules for immunity and cell-fate determination. Trends Cell Biol. 2007;17:135–144. doi: 10.1016/j.tcb.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 50.Feinstein-Rotkopf Y, Arama E. Can’t live without them, can live with them: roles of caspases during vital cellular processes. Apoptosis. 2009 doi: 10.1007/s10495-009-0346-6. A comprehensive review on the many roles of caspases outside the realm of cell death. [DOI] [PubMed] [Google Scholar]
- 51.Tokuyasu KT, Peacock WJ, Hardy RW. Dynamics of spermiogenesis in Drosophila melanogaster. I. Individualization process. Z Zellforsch Mikrosk Anat. 1972;124:479–506. doi: 10.1007/BF00335253. [DOI] [PubMed] [Google Scholar]
- 52.Fuller MT. In: The Development of Drosophila melanogaster. Arias MBaAM., editor. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- 53.Arama E, Agapite J, Steller H. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev Cell. 2003;4:687–697. doi: 10.1016/s1534-5807(03)00120-5. [DOI] [PubMed] [Google Scholar]
- 54.Arama E, Bader M, Srivastava M, Bergmann A, Steller H. The two Drosophila cytochrome C proteins can function in both respiration and caspase activation. Embo J. 2006;25:232–243. doi: 10.1038/sj.emboj.7600920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huh JR, Vernooy SY, Yu H, Yan N, Shi Y, Guo M, Hay BA. Multiple apoptotic caspase cascades are required in nonapoptotic roles for Drosophila spermatid individualization. PLoS Biol. 2004;2:E15. doi: 10.1371/journal.pbio.0020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muro I, Berry DL, Huh JR, Chen CH, Huang H, Yoo SJ, Guo M, Baehrecke EH, Hay BA. The Drosophila caspase Ice is important for many apoptotic cell deaths and for spermatid individualization, a nonapoptotic process. Development. 2006;133:3305–3315. doi: 10.1242/dev.02495. [DOI] [PubMed] [Google Scholar]
- 57.Arama E, Bader M, Rieckhof GE, Steller H. A ubiquitin ligase complex regulates caspase activation during sperm differentiation in Drosophila. PLoS Biol. 2007;5:e251. doi: 10.1371/journal.pbio.0050251. Describes other E3 ligases (than IAPs) that regulate caspase activity. This study charecterzed a Cullin3-based complex that controls caspase activation in differentiating spermatids. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kissel H, Georgescu MM, Larisch S, Manova K, Hunnicutt GR, Steller H. The Sept4 septin locus is required for sperm terminal differentiation in mice. Dev Cell. 2005;8:353–364. doi: 10.1016/j.devcel.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 59.Yatsenko AN, Roy A, Chen R, Ma L, Murthy LJ, Yan W, Lamb DJ, Matzuk MM. Non-invasive genetic diagnosis of male infertility using spermatozoal RNA: KLHL10 mutations in oligozoospermic patients impair homodimerization. Hum Mol Genet. 2006;15:3411–3419. doi: 10.1093/hmg/ddl417. [DOI] [PubMed] [Google Scholar]
- 60.Williams DW, Kondo S, Krzyzanowska A, Hiromi Y, Truman JW. Local caspase activity directs engulfment of dendrites during pruning. Nat Neurosci. 2006;9:1234–1236. doi: 10.1038/nn1774. [DOI] [PubMed] [Google Scholar]
- 61.Kuo CT, Zhu S, Younger S, Jan LY, Jan YN. Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron. 2006;51:283–290. doi: 10.1016/j.neuron.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 62.Nikolaev A, McLaughlin T, O’Leary DD, Tessier-Lavigne M. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature. 2009;457:981–989. doi: 10.1038/nature07767. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Bingol B, Schuman EM. Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature. 2006;441:1144–1148. doi: 10.1038/nature04769. Demonstarted the ability of the proteaosme to move to their point of action thus mediating locolized degradation. By showing that proteasomes are sequstered to the site of excitation in neurons, this study highlighted the possibility that proteasome locolization could be another level of regulation by the UPS. [DOI] [PubMed] [Google Scholar]
- 64.Zhong L, Belote JM. The testis-specific proteasome subunit Pros{alpha}6T of D. melanogaster is required for individualization and nuclear maturation during spermatogenesis. Development. 2007 doi: 10.1242/dev.004770. This study demonstarted that proteasomes themselves are subject to regulation by subunit rearrengement, which is vital to a subset of cellular processes including caspase activation. [DOI] [PubMed] [Google Scholar]
- 65.Gesellchen V, Kuttenkeuler D, Steckel M, Pelte N, Boutros M. An RNA interference screen identifies Inhibitor of Apoptosis Protein 2 as a regulator of innate immune signalling in Drosophila. EMBO Rep. 2005;6:979–984. doi: 10.1038/sj.embor.7400530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leulier F, Lhocine N, Lemaitre B, Meier P. The Drosophila inhibitor of apoptosis protein DIAP2 functions in innate immunity and is essential to resist gram-negative bacterial infection. Mol Cell Biol. 2006;26:7821–7831. doi: 10.1128/MCB.00548-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ribeiro PS, Kuranaga E, Tenev T, Leulier F, Miura M, Meier P. DIAP2 functions as a mechanism-based regulator of drICE that contributes to the caspase activity threshold in living cells. J Cell Biol. 2007;179:1467–1480. doi: 10.1083/jcb.200706027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hauser HP, Bardroff M, Pyrowolakis G, Jentsch S. A giant ubiquitin-conjugating enzyme related to IAP apoptosis inhibitors. J Cell Biol. 1998;141:1415–1422. doi: 10.1083/jcb.141.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lotz K, Pyrowolakis G, Jentsch S. BRUCE, a giant E2/E3 ubiquitin ligase and inhibitor of apoptosis protein of the trans-Golgi network, is required for normal placenta development and mouse survival. Mol Cell Biol. 2004;24:9339–9350. doi: 10.1128/MCB.24.21.9339-9350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hao Y, Sekine K, Kawabata A, Nakamura H, Ishioka T, Ohata H, Katayama R, Hashimoto C, Zhang X, Noda T, et al. Apollon ubiquitinates SMAC and caspase-9, and has an essential cytoprotection function. Nat Cell Biol. 2004;6:849–860. doi: 10.1038/ncb1159. [DOI] [PubMed] [Google Scholar]
- 71.Vernooy SY, Chow V, Su J, Verbrugghe K, Yang J, Cole S, Olson MR, Hay BA. Drosophila Bruce can potently suppress Rpr- and Grim-dependent but not Hid-dependent cell death. Curr Biol. 2002;12:1164–1168. doi: 10.1016/s0960-9822(02)00935-1. [DOI] [PubMed] [Google Scholar]
- 72.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]