Abstract
Purpose
Due to variation of outcome among cases, we sought to examine whether overall survival in ovarian cancer was associated with common inherited variants in 227 candidate genes from ovarian cancer-related pathways including angiogenesis, inflammation, detoxification, glycosylation, one-carbon transfer, apoptosis, cell cycle regulation, and cellular senescence.
Experimental Design
Blood samples were obtained from 325 women with invasive epithelial ovarian cancer diagnosed at the Mayo Clinic from 1999 to 2006. During a median follow-up of 3.8 years (range, 0.1 – 8.6 years), 157 deaths were observed. Germline DNA was analyzed at 1,416 single nucleotide polymorphisms (SNPs). For all patients, and for 203 with serous subtype, we assessed the overall significance of each gene and pathway, and estimated risk of death via hazard ratios (HRs) and 95% confidence intervals (CIs), adjusting for known prognostic factors.
Results
Variation within angiogenesis was most strongly associated with survival time overall (p=0.03) and among patients with serous cancer (p=0.05), particularly for EIF2B5 rs4912474 (all patients HR 0.69, 95% CI 0.54-0.89, p=0.004), VEGFC rs17697305 (serous subtype HR 2.29, 95% CI 1.34-3.92, p=0.003), and four SNPs in VHL. Variation within the inflammation pathway was borderline significant (all patients, p=0.09), and SNPs in CCR3, IL1B, IL18, CCL2, and ALOX5 which correlated with survival time are worthy of follow-up.
Conclusion
An extensive multiple-pathway assessment found evidence that inherited differences may play a role in outcome of ovarian cancer patients, particularly in genes within the angiogenesis and inflammation pathways. Our work supports efforts to target such mediators for therapeutic gain.
Keywords: ovarian cancer, angiogenesis, survival, epidemiology, apoptosis
INTRODUCTION
Ovarian cancer is the fourth leading cause of cancer death among women in the United States (1). Paclitaxel (Taxol) and platinum-based chemotherapy (either cisplatin or carboplatin) represent standard therapy after surgical debulking (2). Unfortunately, even with modern chemotherapy, most patients with advanced disease relapse and die of ovarian cancer; ten-year overall survival remains around 30% (3-5). Clinical features such as age, stage, co-morbidities, or degree of debulking are known prognostic factors (6). However, additional variation in outcome among patients remains unexplained, some of which may be due to inherited factors (germline, as opposed to tumor or somatic changes) that vary among patients. Inherited variation may influence outcome through differential metabolism of chemotherapeutic agents (e.g., variants in ERCC1 and ABCB1 modifying chemo-response), variable immune response, or through a variety of other host-related factors. For example, inherited BRCA1 and BRCA2 mutations are known to correlate with ovarian cancer survival, and common variants in BRAF, KRAS, IL8, VEGF, CXCR2, and ADM may contribute to genetically-determined differences in ovarian cancer outcome (7, 8).
Here, we hypothesized that inherited variants in mechanistic ovarian cancer-related pathways may influence ovarian cancer survival time (Figure 1). In particular, variants related to tumor angiogenesis may influence outcome as a major contributor to pathogenesis; anti-angiogenic therapy with bevacizumab represents the most successful application of a biologically-targeted ovarian cancer therapy (9). Aberrant inflammatory processes can impact multiple tumorigenic pathways (10). Detoxifying molecules are key in response to oxidative stress (11) and the processing of chemotherapeutic agents. Alterations of cell cycle regulators cooperate in ovarian tumor development, and tumor molecular analyses show frequent mutation of these regulators (12). Host defense mechanisms that eliminate mutated cells through permanent growth arrest (cellular senescence) are considered crucial tumor suppressive mechanisms (13). Apoptotic pathways have been shown to be frequently altered in both tumor progression and drug resistance (14). Glycosylation (15-17), one carbon transfer (18), and other processes may also be susceptible to inherited changes which lead to differential ovarian cancer outcome. Controlling for known prognostic factors, the purpose of the current study was to interrogate informative inherited markers in large sets of candidate genes among these pathways in more than 300 prospectively-ascertained, consecutively-enrolled patients with invasive ovarian cancer seen at the Mayo Clinic in Rochester, MN. Identifying survival-associated inherited variants can help to elucidate the most important drivers of ovarian cancer, representing ideal targets for tailored therapeutics, and may also enhance our current prognostic capabilities.
Figure 1. Ovarian cancer pathways.
Role of angiogenesis, inflammation, detoxification, glycosylation, one-carbon transfer, apoptosis, cell cycle, and cellular senescence pathways in ovarian cancer development.
MATERIALS AND METHODS
Study Participants
Recruitment of patients from Mayo Clinic's gynecologic surgery and medical oncology departments, including administration of questionnaires and venipuncture, used established protocols (19). The protocol was approved by the Institutional Review Board, and all participants gave written informed consent. Eligible patients were women aged 20 years or older living in MN, IA, WI, IL, ND, or SD and ascertained within one year of a diagnosis of pathologically-confirmed primary invasive epithelial ovarian cancer. Three hundred and forty-four invasive patients recruited between December 14, 1999 and March 19, 2006 and diagnosed within the preceding year (median time from recruitment to diagnosis = 4 days) were studied, and 334 were successfully genotyped (see below). Prior to analysis, additional pathologic review excluded nine patients found to be of non-epithelial or non-ovarian origin, resulting in 325 analyzed patients. Data on clinical features of disease including histology, surgical outcomes, and chemotherapy were abstracted by an experienced research nurse with review by gynecologic and medical oncology clinicians. Epidemiologic risk factor data including family history, reproductive factors, and exposure to known and suspected ovarian cancer risk factors were obtained through administration of a 16-page written questionnaire. Data on vital status was obtained from several sources including the Mayo Clinic computerized medical record, the Mayo Clinic Tumor Registry, which follows patients annually for overall survival who were diagnosed or received initial treatment at Mayo Clinic, and the National Death Index. Follow-up data was obtained through July 31, 2008.
Gene Selection
With the goal of comprehensiveness, key genes within the pathways of angiogenesis (N=18 genes), apoptosis (N=18), cell cycle (N=39), cellular senescence (N=33), detoxification (N=15), glycosylation (N=26), inflammation (N=26), and one-carbon transfer (N=21) were identified in October 2005 using a number of sources including peer-reviewed published literature, the Cancer Genome Anatomy Project, Biokarta, and KEGG pathway databases. In addition, we included 25 genes previously shown to have a 50% or more reduction in expression in ovarian tumor versus normal tissue using the Affymetrix U133A GeneChip array (20). Finally, based on unpublished data and recent reports, a small number of genes related to NF-κB (NFKBIA, NFKBIB) and hormone metabolism (CYP19A1, SULT1E1) as well as the granulin-epithelin precursor (GRN) (21), and transcription elongation factor A (SII)-like 7 (TCEAL7) (22) were included. The complete list of 227 included genes is provided in Table 1, with additional detail in Table S1.
Table 1. Distribution of demographic and clinical characteristics of ovarian cancer patients (N=325) and relation to overall survival.
| N (%) or Median (Range) | Hazard Ratio (95% CI) | P-value | |
|---|---|---|---|
| Vital status at last follow-up | |||
| Alive | 168 (52%) | ||
| Deceased | 157 (48%) | n.a. | n.a. |
| Length of follow-up, months | 45 (1 - 103) | n.a. | n.a. |
| Diagnosis Year | |||
| 1999-2000 | 48 (15%) | ||
| 2001 | 29 (9%) | ||
| 2002 | 63 (19%) | per-year HR | |
| 2003 | 51 (16%) | 0.96 (0.88, 1.07) | 0.50 |
| 2004 | 64 (20%) | ||
| 2005-2006 | 70 (22%) | ||
| Age at diagnosis (median 61, range 28 - 91) | 64 (20%) | Per category trend HR | |
| <50 | 84 (26%) | <0.001 | |
| 50-59 | 82 (25%) | 1.20 (1.05, 1.36) | |
| 60-69 | 66 (20%) | ||
| 70-79 | 29 (9%) | ||
| 80+ | |||
| Race | |||
| White | 320 (98%) | REF | 0.06 |
| Non-white | 5 (2%) | 0.39 (0.14, 1.05) | |
| State of residence | |||
| MN | 162 (50%) | Non-MN | |
| IA | 53 (16%) | v MN | |
| IL | 15 (5%) | 0.07 | |
| WI | 47 (14%) | 1.33 (0.97, 1.83) | |
| SD/ND | 48 (15%) | ||
| Body mass index, (kg/m2) | 27.7 (17.4, 60.1) | 0.97 (0.95, 1.00) | 0.06 |
| First degree family history of breast or ovarian cancer | |||
| No | 245 (77%) | REF | 0.76 |
| Yes | 75 (23%) | 0.95 (0.66, 1.36) | |
| Unknown | 5 | ||
| Prior cancer | |||
| No | 278 (86%) | REF | |
| Yes | 47 (14%) | 0.74 (0.46, 0.19) | 0.22 |
| Pre-surgery CA125, units | 591 (2.3, 45k) | 1.50 (1.20, 1.87) | <0.001 |
| Site of primary debulking surgery | |||
| Mayo Clinic | 294 (90%) | REF | 0.76 |
| Elsewhere | 31 (10%) | 0.92 (0.54, 1.56) | |
| Stage | |||
| I | 53 (17%) | Stage III/IV | |
| II | 21 (7%) | v. Stage I/II | <0.001 |
| III | 187 (59%) | ||
| IV | 58 (18%) | 7.67 (3.76, 15.6) | |
| Unknown | 6 | ||
| Histology | |||
| Serous | 203 (64%) | REF | |
| Endometrioid | 58 (18%) | 0.50 (0.30, 0.82) | <0.001 |
| Clear cell | 21 (7%) | 1.21 (0.63, 2.31) | |
| Mucinous | 9 (3%) | 0.18 (0.26, 1.31) | |
| Mixed epithelial | 28 (9%) | 0.56 (0.28, 1.10) | |
| Other/unknown | 6 | ||
| Grade | |||
| 1 | 13 (4%) | 3+4 v. 1+2 | |
| 2 | 40 (13%) | 4.57 (2.33, 8.98) | <0.001 |
| 3 | 151 (48%) | ||
| 4 | 113 (36%) | ||
| Debulking | |||
| Optimal (≤ 1 cm remaining) | 251 (81%) | REF | <0.001 |
| Sub-optimal (> 1 cm remaining) | 57 (19%) | 2.67 (1.89, 3.64) | |
| Unknown | 17 | ||
| Peritoneal cytology | |||
| Positive | 211 (73%) | 3.24 (1.95, 5.41) | <0.001 |
| Negative | 77 (27%) | REF | |
| Unknown | 37 | ||
| Ascites | |||
| Yes | 153 (64%) | REF | <0.001 |
| No | 85 (36%) | 0.33 (0.21, 0.53) | |
| Unknown | 87 | ||
| Chemotherapy | |||
| Yes | 297 (91%) | REF | 0.66 |
| No/unknown | 28 (9%) | 0.87 (0.47-1.61) | |
| Chemotherapeutic agent | |||
| Platinum and taxane | 271 (83%) | Both v. neither | |
| Platinum only | 8 (2%) | 0.55 | |
| Taxane only | 3 (1%) | 0.87 (0.54, 1.39) | |
| Neither platinum nor taxane | 43 (13%) |
Unadjusted univariate hazard ratios from Cox proportional hazards regression analysis, CI=confidence interval; bold indicates p<0.05; vital status as of July 30, 2008; length of follow-up calculated among 168 living patients; non-white participants include two African-American participants, two Asian participants, and two participants of other ethnicities; body mass index HR represents risk per-unit increase; participants with prior non-ovarian cancer included 21 breast cancers, 13 non-melanoma skin cancers, six melanomas, and seven other cancers; HRs for pre-surgery CA125 based on log10(pre-surgery CA125); platinum agents include cisplatin and carboplatin; taxane agents include paclitaxel and docetaxel.
SNP Selection
SNP selection strategies varied slightly across pathways due to logistical issues related to panel development. Generally, for all pathways except glycosylation and inflammation, tagSNPs were identified via analysis of unrelated Caucasian genotypes from the Hapmap Consortium's release 21a (genome build 34) (23), Perlegen Sciences (genome build 34) (24), SeattleSNPs (http://pga.mbt.washington.edu, genome build 35) and Panel 2 of the National Institute for Environmental Health Science SNPs (www.egp.gs.washington.edu, genome build 35). SNPs for each gene were chosen from each source from the region within 5 kb of the most 5′ and 3′ exons of each gene. tagSNPs were identified by applying the ldSelect algorithm (25) to group (i.e., bin) SNPs with minor allele frequency (MAF) ≥ 0.05 and pair-wise linkage disequilibrium (LD) threshold of r2 ≥ 0.8. Following binning, we selected tagSNPs for analysis from the source with the greatest number of SNPs with MAF ≥ 0.05 and the greatest number of LD bins that also met criteria for predicted likelihood of successful genotyping using the Illumina Golden Gate Assay™ quality score metrics. In addition to the tagSNPs, we included all additional putative-functional SNPs (within 1 kb upstream, 5′ UTR, 3′ UTR or non-synonymous) with MAF ≥ 0.05 identified in Ensembl version 34. For CDC25B, CUL1, E2F3, and SMARCA2, SNPs tagging at least one other SNP were selected using pairwise binning in Tagger for SNPs in HapMap with Caucaisan MAF ≥ 0.05, r2 ≥ 0.8, within 10 kb of genes (26). For glycosylation genes, a tagSNP approach was not used; rather all non-intronic SNPs within 10 kb of each gene with a MAF > 0.05 were attempted. For inflammation genes, SNPs within 10 kb of each gene with MAF > 0.05 among the CEU samples of Hapmap Consortium's release 21a (genome build 34) (23) were binned if r2 ≥ 0.8, and all non-synonymous SNPs were included. For CYP1A1, variants were selected using resequencing data among 60 Caucasian-American subjects (27) via both the tagSNP method (MAF ≥ 0.05, r2 ≥ 0.8) (25) and a haplotype-tagging method (MAF ≥ 0.02, haplotype frequency ≥ 0.01, r2 ≥ 0.9). Finally, each SNP was evaluated for compatibility with Illumina Golden Gate Assay™ ensuring all designability scores ≥ 0.6 and no SNP pairs within 60 bp. A complete annotated list of 1,536 attempted SNPs is provided in Table S2.
Genotyping
Genotyping for these and 2,041 other samples (population controls, borderline patients, invasive patients recruited elsewhere, and laboratory standards) was performed at the Mayo Clinic using the Illumina GoldenGate™ BeadArray assay. DNA was extracted from 10 to 15 mL fresh peripheral blood using the Gentra AutoPure LS Purgene salting out methodology (Gentra, Minneapolis, MN) and stored at 4°C. Samples were bar-coded to ensure accurate and reliable processing and plated with duplicates and lab standards. Following DNA activation, incubation, amplification, and automated genotype calling, quality control exclusions were performed using all data. Of 2,368 samples, we excluded 87 samples with poor clustering, two samples with low call rates (<95%) and two duplicated samples with an unusually high number of discrepancies; no excluded samples were from cases eligible for the current analysis. Of the 1,536 attempted SNPs, we excluded 44 SNPs which failed completely (or nearly completely), six SNPs due to low call rates (<95%), and seven monomorphic SNPs among all samples. Of the remaining 1,479 SNPs, the mean per-SNP call rate was 99.72%, the mean per-sample call rate was 98.64%, and reproducibility of duplicates was > 99.99%. Prior to the current analysis, additional review excluded 45 tagSNPs found to be incorrect due to a chromosome 7 misalignment (ABCB1, CDK6, PPP1R3A, NOS3), eight SNPs found to be > 10 kb from the gene of interest in current genome build 36.3 (GSTM1 rs366631; TIMP1 rs2283736; MGAT5 rs1257189, rs1257187; AKT1 rs4983559; TFDP2 rs733996, rs7644383, rs11720300), and ten SNPs with observed MAF < 0.01 in the current sample subset (ADH4 rs13112924, RBL1 rs1780703, TCEAL7 rs17340307, ERCC3 rs1803541, CUL1 rs6958001, IL4R rs1801275, SHMT1 rs2168781, BCL2 rs4987792, FUT6 rs364637 and TIMP3 rs1962223). Thus, 1,416 SNPs are included here.
Statistical Analysis
We used Cox proportional hazards regression (28) to estimate hazard ratios (HRs) and 95% confidence intervals (CIs) for the associations with overall survival, adjusted for known prognostic factors. In order to adjust for known factors, a clinical risk score was created and used as a covariate in genetic models. The risk score was created from the linear combination of the following variables multiplied by their Cox regression coefficients (i.e., xβ): race (white, other), age (<50, 50-59, 60-69, 70-79, 80+), location of debulking surgery (Mayo, elsewhere), stage (III/IV, I/II, missing), debulking (optimal, suboptimal, missing), ascites (yes, no, missing), grade (1 or 2, 3 or 4, missing), histology (serous, endometrioid, clear cell, mucinous, mixed, other or unknown), pre-surgery CA125 (log10), and chemotherapy (taxane, platinum). Using a single variable for the risk score allowed for reduced degrees of freedom over inclusion of all individual prognostic factors in Cox models (29-31). The clinical risk score was highly predictive of survival time (HR 2.7, 95% CI 2.2 - 3.3). Genotype specific survival curves from a Cox model were created using the median clinical risk score as a baseline covariate.
Association testing proceeded for SNPs, for genes, and for pathways. First, for each SNP, a primary association using an ordinal coding (i.e., 0, 1, 2 copies of minor allele) was tested (analogous to the Armitage test for trend for binary endpoints), and HRs with 95% CIs were estimated per-allele as well as for heterozygotes and minor allele homozygotes compared to major allele homozygotes separately. Gene level tests were completed using principal component analysis (PCA) in a manner similar to that described previously (32). Briefly, PCs were created using linear combinations of ordinally-scaled SNPs (based on number of minor alleles), and the smallest set of resulting PCs that explained at least 90% of the SNP variance within the gene was then included in Cox models. The rationale for this approach over inclusion of the entire collection of SNPs in a multivariate model is the resulting reduction in degrees of freedom, avoidance of model fitting issues to due multi-collinearity from LD, and potential improvement in statistical power. Finally, to assess overall significance of a pathway, we computed the tail strength (33) of the set of gene level tests from each pathway and performed permutation testing based on 1,000 simulations of our entire association-testing process. Tail strength is closely related to the false discovery rate and assesses the relative strength of the collection of an observed set of empirical p values. A positive tail strength indicates more associations than expected by chance, and a negative tail strength indicates fewer associations than expected by chance. Analyses were repeated on the subset of serous patients only (N=203), due to possible genetic heterogeneity of associations by histological subtype.
As a conservative approach, overall interpretation of associations began with examination at the pathway level, followed next by assessment of gene- and SNP-level results. This tiered testing approach was carried out in the spirit of Fisher's protected least significance difference test; individual associations on a lower hierarchy level or tier were not considered statistically significant in the absence of significance at higher tier(s). At the pathway and gene levels, we focused on results significant at p=0.10, and, at the SNP level, we focused on results significant at p=0.05. Because of this tiered interpretation of results and the exploratory nature of this analysis, results were not corrected for multiple testing. Plots of LD displaying r2 for each gene based on 847 Caucasian participants (patients studied here, borderline patients, and population controls) were created using Haploview v 4.1 (34).
RESULTS
Characteristics of 325 invasive epithelial ovarian cancer patients and their relation to survival are shown in Table 1. The majority of patients had tumors of serous histology (63%) and stage III/IV disease (77%) consistent with the typical distributions of invasive disease (35). Most patients underwent primary debulking surgery at Mayo Clinic (90%) with ≤ 1 cm tumor remaining (81%) and had ascites (64%) and positive cytology (73%) at initial surgery. One hundred and fifty-seven patients (48%) died during the observation time; among 168 surviving patients, median follow-up time was 3.8 years (range, 0.1-8.6 years).
Angiogenesis Pathway
Covariate-adjusted pathway analysis revealed that overall variation among genes in the angiogenesis pathway (18 genes, 130 SNPs tested) was the strongest inherited predictor of overall survival among all patients (empirical p=0.03) and for the 203 patients with serous subtype (empirical p=0.05; Table 3, Table S3). These p-values are based on 1,000 permutations of the analytical process used. Four angiogenesis genes, VHL (encoding the von Hippel-Lindau tumor suppressor), EIF2B5 (encoding the eukaryotic translation initiation factor 2B, subunit 5ε), VEGFC (encoding the vascular endothelial growth factor [VEGF] C), and MMP3 (encoding matrix metallopeptidase 3), were correlated with survival (p<0.10) among all patients or among patients with serous subtype (Table 3). Notably, the summed variation at four SNPs in VHL was significantly associated with survival among all patients (p=0.001) and those with serous subtype (p=0.003). Three VHL SNPs showed similar associations with improved survival among minor allele carriers (e.g., rs265318 per-allele HR 0.66, 95% CI, 0.44-0.99; Figure 2; Table 4) and were moderately correlated (r2>0.2, Figure S1). Consistent with improved survival, at VHL SNP rs17610448, no deaths were observed among the 14 heterozygotes although seven deaths were expected (Figure 2).
Table 3. Ovarian cancer survival pathway-level and gene-level results, covariate adjusted p-values.
| Pathway (N Genes)/Gene | All Patients (N=325) | Serous Subtype (N=203) |
|---|---|---|
| Angiogenesis (N=18) | 0.03 | 0.05 |
| VHL | 0.0001 | 0.0003 |
| EIF2B5 | 0.03 | 0.16 |
| VEGFC | 0.05 | 0.03 |
| MMP3 | 0.08 | 0.08 |
| Inflammation (N=28) | 0.09 | 0.12 |
| CCR3 | 0.01 | 0.02 |
| IL1B | 0.02 | 0.16 |
| IL18 | 0.02 | 0.01 |
| CRP | 0.08 | 0.07 |
| IL1A | 0.08 | 0.26 |
| CCL2 | 0.25 | 0.04 |
| ALOX5 | 0.55 | 0.01 |
| Detoxification (N=15) | 0.19 | 0.27 |
| CYP1A1 | 0.02 | 0.34 |
| Glycosylation (N=26) | 0.22 | 0.22 |
| ST6GALNAC5 | 0.01 | 0.01 |
| GALNT2 | 0.05 | 0.03 |
| FUT9 | 0.05 | 0.04 |
| FUT8 | 0.06 | 0.04 |
| GALNT6 | 0.14 | 0.01 |
| ST8SIA5 | 0.17 | 0.01 |
| One-Carbon Metabolism (N=21) | 0.63 | 0.71 |
| MBD4 | 0.04 | 0.06 |
| MTHFR | 0.08 | 0.16 |
| MTHFD1L | 0.23 | 0.05 |
| Apoptosis (N=18) | 0.77 | 0.82 |
| CASP6 | 0.06 | 0.23 |
| CASP7 | 0.12 | 0.04 |
| Cell Cycle (N=39) | 0.81 | 0.86 |
| E2F6 | 0.007 | 0.003 |
| RBL2 | 0.005 | 0.01 |
| CCNE2 | 0.03 | 0.03 |
| CCND3 | 0.12 | 0.05 |
| Cellular Senescence (N=33) | 0.88 | 0.59 |
| IGF2 | 0.10 | 0.25 |
| SHC1 | 0.10 | 0.10 |
| SMARCA4 | 0.13 | 0.02 |
| BCL2 | 0.27 | 0.08 |
| Under-Expressed Genes (N=25) | 0.95 | 0.60 |
| P2RX5 | 0.08 | 0.03 |
| DKK3 | 0.09 | 0.38 |
| FGF5 | 0.23 | 0.002 |
| TPM1 | 0.76 | 0.09 |
Results adjusted for clinical risk score composed of race (white, other), age (<50, 50-59, 60-69, 70-79, 80+), location of debulking surgery (Mayo, elsewhere), stage (III/IV, I/II, missing), debulking (optimal, suboptimal, missing), ascites (yes, no, missing), grade (1 or 2, 3 or 4, missing), histology (serous, endometrioid, clear cell, mucinous, mixed, other or unknown), pre-surgery CA125 (log10), and chemotherapy (taxane, platinum); bold indicates p<0.05; sorted in order of pathway-level significance and gene-level significance among all patients; all pathways are shown, but only genes with p<0.10 are listed; single-SNP significance is provided forCCR3 because only one SNP was examined.
Figure 2. Covariate-adjusted survival curves by genotype.
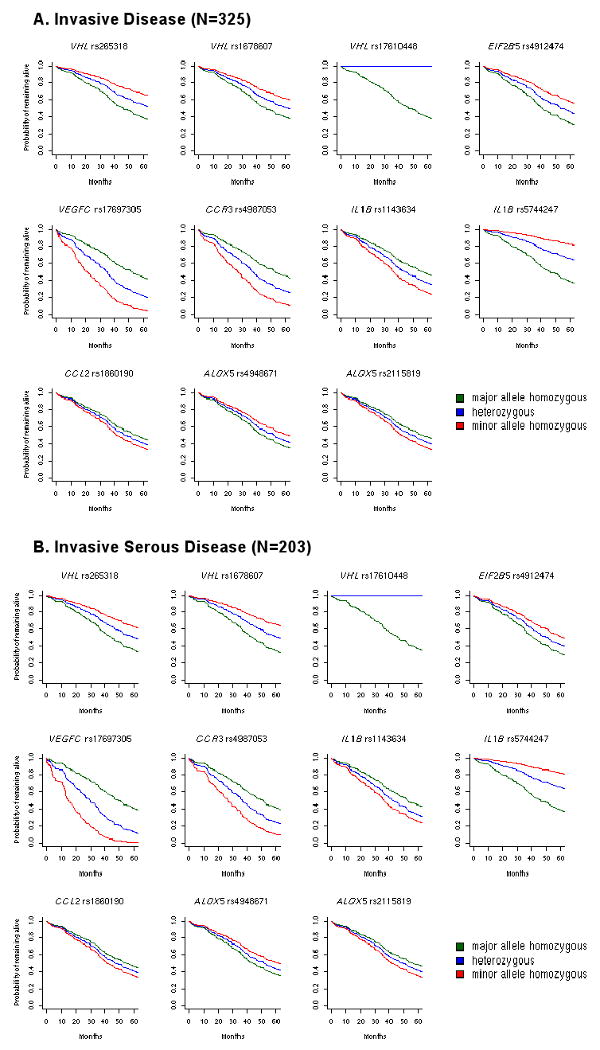
Genotype-specific survival curves from Cox models using the median clinical risk score as a baseline covariate.
Table 4. Ovarian cancer survival SNP-level results, hazard radios and confidence intervals.
| All Patients | Serous Subtype | ||||
|---|---|---|---|---|---|
| Gene | SNP | Per allele HR (95% CI) | p-value | Per allele HR (95% CI) | p-value |
| Angiogenesis | |||||
| VHL | rs265318 | 0.66 (0.44-0.99) | 0.04 | 0.67 (0.43-1.04) | 0.07 |
| rs1678607 | 0.72 (0.50-1.03) | 0.08 | 0.63 (0.42-0.97) | 0.03 | |
| rs1642742 | 0.87 (0.68-1.10) | 0.25 | 0.87 (0.66-1.15) | 0.33 | |
| rs17610448 | n.e. | n.e. | n.e. | n.e. | |
| EIF2B5 | rs2292611 | 1.06 (0.79-1.43) | 0.69 | 0.98 (0.69-1.38) | 0.90 |
| rs865808 | 1.19 (0.97-1.47) | 0.10 | 1.08 (0.84-1.38) | 0.55 | |
| rs843360 | 1.22 (0.99-1.51) | 0.06 | 1.17 (0.92-1.49) | 0.20 | |
| rs4912474 | 0.69 (0.54-0.89) | 0.004 | 0.76 (0.57-1.00) | 0.05 | |
| VEGFC | rs1385736 | 0.91 (0.70-1.18) | 0.47 | 0.96 (0.72-1.30) | 0.81 |
| rs12510099 | 0.82 (0.56-1.19) | 0.29 | 0.89 (0.57-1.41) | 0.63 | |
| rs17697305 | 1.87 (1.14-3.07) | 0.01 | 2.29 (1.34-3.92) | 0.003 | |
| rs17697515 | 0.76 (0.49-1.18) | 0.22 | 0.88 (0.51-1.51) | 0.64 | |
| rs1485766 | 0.74 (0.59-0.93) | 0.01 | 0.74 (0.57-0.96) | 0.02 | |
| rs11947611 | 1.02 (0.83-1.27) | 0.83 | 1.01 (0.79-1.29) | 0.94 | |
| rs7693545 | 1.03 (0.82-1.30) | 0.78 | 1.02 (0.78-1.33) | 0.88 | |
| rs3775194 | 0.98 (0.77-1.24) | 0.84 | 0.95 (0.72-1.24) | 0.70 | |
| rs1564922 | 0.83 (0.59-1.18) | 0.30 | 0.77 (0.50-1.19) | 0.24 | |
| rs4557213 | 0.80 (0.55-1.17) | 0.25 | 0.72 (0.44-1.16) | 0.18 | |
| Inflammation | |||||
| CCR3 | rs4987053 | 1.64 (1.13-2.38) | 0.01 | 1.61 (1.07-2.41) | 0.02 |
| IL1B | rs7596461 | 1.23 (0.87-1.73) | 0.25 | 1.12 (0.73-1.71) | 0.61 |
| rs7596684 | 1.28 (0.98-1.66) | 0.07 | 1.28 (0.93-1.75) | 0.12 | |
| rs1143643 | 0.84 (0.67-1.06) | 0.14 | 0.90 (0.70-1.17) | 0.45 | |
| rs1143634 | 1.36 (1.04-1.78) | 0.02 | 1.36 (0.98-1.88) | 0.06 | |
| rs16944 | 0.83 (0.65-1.06) | 0.13 | 0.85 (0.63-1.14) | 0.27 | |
| rs4848306 | 0.90 (0.72-1.13) | 0.36 | 0.90 (0.69-1.17) | 0.45 | |
| rs13032029 | 0.91 (0.73-1.15) | 0.44 | 0.92 (0.71-1.20) | 0.55 | |
| rs10169916 | 0.82 (0.64-1.05) | 0.11 | 0.85 (0.63-1.14) | 0.27 | |
| IL18 | rs243908 | 1.22 (0.99-1.51) | 0.06 | 1.32 (1.03-1.69) | 0.03 |
| rs5744280 | 1.01 (0.80-1.27) | 0.95 | 0.94 (0.72-1.22) | 0.63 | |
| rs549908 | 1.23 (0.99-1.52) | 0.06 | 1.31 (1.02-1.68) | 0.03 | |
| rs5744258 | 1.02 (0.80-1.32) | 0.85 | 1.09 (0.81-1.46) | 0.59 | |
| rs5744256 | 1.02 (0.80-1.32) | 0.85 | 1.09 (0.81-1.46) | 0.59 | |
| rs5744247 | 0.45 (0.27-0.75) | 0.003 | 0.39 (0.21-0.73) | 0.003 | |
| rs4937113 | 0.97 (0.78-1.21) | 0.81 | 1.00 (0.77-1.30) | 1.00 | |
| rs2043055 | 1.09 (0.87-1.37) | 0.45 | 1.02 (0.79-1.32) | 0.86 | |
| rs1946519 | 0.91 (0.73-1.14) | 0.41 | 0.90 (0.68-1.18) | 0.45 | |
| rs1293344 | 1.13 (0.91-1.41) | 0.26 | 1.20 (0.92-1.56) | 0.18 | |
| rs360712 | 1.11 (0.88-1.40) | 0.37 | 1.13 (0.85-1.49) | 0.41 | |
| rs11214108 | 0.54 (0.34-0.85) | 0.01 | 0.46 (0.27-0.79) | 0.01 | |
| CCL2 | rs17652343 | 1.06 (0.82-1.37) | 0.63 | 0.96 (0.72-1.30) | 0.80 |
| rs11652585 | 1.32 (0.91-1.91) | 0.14 | 1.59 (1.09-2.33) | 0.02 | |
| rs8068314 | 1.32 (0.91-1.91) | 0.14 | 1.59 (1.09-2.33) | 0.02 | |
| rs2857653 | 0.88 (0.67-1.17) | 0.38 | 0.80 (0.57-1.11) | 0.18 | |
| rs1860190 | 1.18 (0.93-1.48) | 0.17 | 1.39 (1.07-1.82) | 0.01 | |
| rs1024611 | 1.09 (0.86-1.39) | 0.48 | 1.19 (0.90-1.58) | 0.21 | |
| rs3760396 | 1.05 (0.81-1.37) | 0.69 | 0.95 (0.70-1.29) | 0.76 | |
| rs4586 | 1.18 (0.94-1.49) | 0.15 | 1.40 (1.08-1.82) | 0.01 | |
| ALOX5 | rs1864414 | 1.10 (0.80-1.51) | 0.55 | 1.24 (0.87-1.77) | 0.23 |
| rs3824612 | 1.11 (0.88-1.41) | 0.37 | 1.30 (0.99-1.70) | 0.06 | |
| rs745986 | 1.16 (0.88-1.53) | 0.29 | 1.17 (0.86-1.61) | 0.31 | |
| rs4948671 | 0.82 (0.65-1.03) | 0.09 | 0.67 (0.51-0.88) | 0.003 | |
| rs7099684 | 1.02 (0.79-1.31) | 0.87 | 0.90 (0.67-1.20) | 0.46 | |
| rs2115819 | 1.20 (0.96-1.50) | 0.12 | 1.54 (1.18-2.02) | 0.002 | |
| rs10900213 | 0.88 (0.71-1.10) | 0.26 | 0.74 (0.58-0.95) | 0.02 | |
| rs10900215 | 1.08 (0.80-1.47) | 0.62 | 1.05 (0.73-1.50) | 0.79 | |
| rs4948672 | 1.11 (0.89-1.38) | 0.37 | 1.04 (0.82-1.33) | 0.74 | |
| rs12264801 | 1.11 (0.90-1.38) | 0.33 | 1.31 (1.02-1.69) | 0.04 | |
| rs892691 | 1.07 (0.82-1.39) | 0.63 | 0.97 (0.72-1.30) | 0.83 | |
| rs1565096 | 1.00 (0.79-1.28) | 0.98 | 0.96 (0.74-1.25) | 0.77 | |
| rs1487562 | 1.13 (0.87-1.49) | 0.36 | 1.22 (0.90-1.65) | 0.21 | |
| rs702366 | 0.81 (0.66-1.01) | 0.06 | 0.82 (0.65-1.05) | 0.12 | |
| rs2291427 | 1.12 (0.89-1.39) | 0.33 | 1.07 (0.84-1.37) | 0.58 | |
| rs4948674 | 0.81 (0.66-1.01) | 0.06 | 0.76 (0.59-0.97) | 0.03 | |
Adjusted hazard ratios (HR), confidence intervals (CI), and per-allele (trend) p-values are shown which incorporate a clinical risk score composed of race (white, other), age (<50, 50-59, 60-69, 70-79, 80+), location of debulking surgery (Mayo, elsewhere), stage (III/IV, I/II, missing), debulking (optimal, suboptimal, missing), ascites (yes, no, missing), grade (1 or 2, 3 or 4, missing), histology (serous, endometrioid, clear cell, mucinous, mixed, other or unknown), pre-surgery CA125 (log10), and chemotherapy (taxane, platinum); bold indicates p<0.05 (per minor allele results); sorted in order of pathway-level significance (Table 3) and gene-level significance (Table 4) among all patients, and SNP chromosomal order (Table S2); only pathways with p<0.10 are listed, and only SNPs in genes with gene-level p<0.05 are listed.
At EIF2B5, patients who carried minor alleles at the 3′ downstream SNP rs4912474 (tagging rs843358 V587I and 3′ downstream rs844540 and rs4575925) had better survival than non-carriers after adjustment for known prognostic factors (HR 95% CI, 0.69 0.54-0.89, p=0.004). This was apparent for all patients and remained borderline significant among patients with serous subtype (p=0.05, Table 4). In VEGFC among patients with serous subtype, carriers of the minor allele at rs17697305 had poorer survival (HR 2.29, 95% CI 1.34-3.92, p=0.003, Figure 2), whereas carriers of the rs1485766 minor allele had improved survival (HR 0.74, 95% CI 0.57-0.96, p=0.02, Table 4). These appear to be two independent signals (r2=0.02; Figure S1).
Inflammation Pathway
Next in statistical significance, variation within the inflammation pathway (26 genes, 154 SNPs tested) showed borderline statistical association with survival in all patients (empirical p=0.09) and serous ovarian cancer patients, adjusting for known prognostic factors (empirical p=0.12, Table 3). Among inflammation genes, seven individual genes were risk factors for survival at p<0.10 in either all patients or patients with serous cancer (Table 4). These include CCR3 encoding chemokine receptor 3, IL1B encoding interleukin 1 β, IL18 (encoding interleukin 18), CRP (encoding C-reactive protein), IL1A (encoding interleukin 1 α), CCL2 (encoding chemokine ligand 2), and ALOX5 (encoding arachidonate 5-lipoxygenase) (Table S1). The strongest gene-level result for patients with serous tumors was IL18 (p=0.006). Of these genes, CCR3 and IL18 were significant at p<0.05 in both patient groups.
At the SNP level, only one CCR3 SNP was analyzed (synonymous rs4987053), and carriers of the minor allele demonstrated poorer survival (all patients, HR 1.64, 95% CI 1.13-2.38, p=0.01; serous only, HR 1.61, 95% CI 1.07-2.41, p=0.02; Figure 2). At IL18, 12 SNPs were analyzed and two correlated SNPs (intronic rs5744247and 5′ upstream rs11214108, r2=0.54; Figure S1) showed significantly decreased survival (p<0.01) among all patients and among patients with serous cancer (serous subtype, HR 0.39, 95% CI 0.21-0.73). In addition, minor alleles in two other correlated IL18 SNPs (3′ downstream rs243908 and synonymous rs549908, r2=0.98; Figure S1) were associated with poorer survival among patients with serous cancer (0.03), and borderline significant covariate-adjusted increased risk among all patients (p=0.06). Interestingly, these two SNPs were independent of the former two correlated IL18 SNPs (Figure S1). Among patients with serous subtype, several intronic SNPs within ALOX5 (Figure S1) were associated with survival (rs2115819, HR 1.54, 95% CI 1.18-2.02, Figure 2, Table 4).
Other Pathways
Analysis of the detoxification pathway resulted in a positive tail strength, although not statistically significant in either case group (empirical p>0.10, Table S3). Only one detoxification gene showed significant correlation with survival (CYP1A1, encoding cytochrome P450, family 1, subfamily A, polypeptide 1, p=0.02 among all patients). Carrying the minor allele at the 5′ CYP1A1 rs2470893 was associated with improved survival (HR 0.69, 95%CI 0.53-0.89, p=0.005) (Table S2). Finally, we note that considering genes across all pathways, 14 genes (6.2%) were associated with survival among all patients (p<0.05), and 19 genes (8.4%) were associated with survival among those with serous subtype (p<0.05; Table 4). These results suggest that there was modest evidence for a greater number of associated genes than expected by chance at p=0.05 and that subset analysis of patients with serous ovarian cancer may have strengthened the homogeneity of patients.
DISCUSSION
In this study of consecutively-recruited, richly-annotated epithelial ovarian cancer patients from a single institution, we found that inherited inter-individual differences in angiogenesis, and, to a lesser extent, inflammation, may influence survival time after adjustment for other prognostic factors. A unique aspect of our approach was the clinical, demographic, and treatment homogeneity of the patients studied which, when combined with high data completeness, allowed us to assess the role of inherited variants above and beyond known clinical factors. To our knowledge, this is the broadest investigation of the influence of host genetic pathways on ovarian cancer outcome to date.
Vascularization and expression of proangiogenic factors has been shown to correlate with prognosis of ovarian tumors (36). Bevacizumab, an antibody targeting VEGF, has recently shown significant promise in the treatment of ovarian cancer (9). We found that variation in VHL was associated with survival among all patients (p=0.0001) and among patients with serous ovarian cancer (p=0.0003); certain VHL SNPs were associated with improved survival (e.g., rs1678607 and serous subtype, HR 0.63, 95% CI 0.42-0.97, p=0.03). This gene encodes a component of an E3 ubiquitin ligase that targets the hypoxia-inducible factor (HIF) transcription factor for destruction in the presence of oxygen (Figure S2) (37). SNPs in HIF1A (encoding HIF-1α) were also assessed here, and minor alleles at rs966824 were associated with poorer survival (all patients HR 0.51, 95% CI 0.24-1.09, p=0.08; serous subtype HR 0.38, 95% CI 0.16-0.95, p=0.04); analysis of SNP-SNP or gene-gene interactions may be warranted. VHL mutations lead to stabilization of these HIFs and transcription of angiogenesis genes; however loss of functionality of VHL in endothelium may also be a contributing factor in angiogenesis and survival time through a HIF-VEGF-independent mechanism (38).
We found that VEGFC rs17697305 genotypes were associated with poorer survival among all patients (HR 1.87, 95 CI% 1.14-3.07 p=0.01) and among patients with serous subtype (HR 2.29, 95% CI 1.34-3.92, p=0.003). VEFG-C plays a key role in lymphangiogenesis and the promotion of tumor invasion (39), and expression and/or serum levels of VEFG-C have been reported to be prognostic in other cancers (40). Interestingly, although prior studies of VEGFA SNPs reported association with progression-free survival among 53 ovarian cancer patients with recurrent disease (8) and suggested interactions between SNPs in a collection of 563 ovarian cancer patients, we observed no correlation with survival time and any of the five VEGFA SNPs assessed here. In the angiogenesis gene EIF2B5, rare alleles at rs49121474 were associated here with improved survival (p=0.004). EIF2B5 encodes a guanine nucleotide-exchange factor that regulates the translation initiation phase of protein synthesis, promoting angiogenesis and tumor growth (41). Finally, we hypothesize that associations seen in MMP3 may be due to a variable ability to create and breakdown the extracellular matrix which is critical to angiogenic processes.
Our finding, although of borderline statistical significance, that SNPs in the inflammation pathway (notably CCR3, IL1B, and IL18) may be prognostic factors is consistent with work showing that immune factors play a role in ovarian carcinogenesis (42, 43). Here, rs4987053 in the chemokine receptor CCR3 was associated with poorer survival among all patients (HR 1.64 95% CI 1.13-2.38, p=0.01) and among women with serous ovarian cancer (HR 1.61 95% CI 1.07-2.41, p=0.02). CCR3 responds and binds to a variety of chemokines, including eotaxin (CCL11), eotaxin-3 (CCL26), MCP-3 (CCL7), MCP-4 (CCL13), and RANTES (CCL5) and is a component of an immune complex that could be a prognostic factor for relapse-free survival in ovarian cancer (44). IL1B encodes a member of the interleukin 1 cytokine family that is an important mediator of the inflammatory response and is involved in a variety of cellular activities, including cell proliferation, differentiation, and apoptosis. Associations with recurrence in other cancers (45) have been reported. IL-18 is a pro-inflammatory cytokine that belongs to the IL-1 family of ligands. Elevated serum levels of circulating Il-18 and rare alleles at rs187238 have been linked to metastatic disease in ovarian cancer patients (46, 47).
In summary, this study reports on the largest number of polymorphisms, genes, and pathways analyzed for associations with inherited variation and ovarian cancer survival to date. Strengths of this work include the breadth and depth of the pathways assessed and the use of recommended targeting of tagging SNPs in addition to putative functional SNPs within genes (48). Use of a clinic-based population provided the ability to account for detailed clinical data, which are not routinely available in population-based studies, while restriction to patients living within a defined surrounding geographic region minimized the potential for the referral bias sometimes seen in clinic-based studies. Centralized pathology review, high-quality genotyping, covariate adjustment, and conservative interpretation of results are also strengths of this study. A limitation of this work is that time to recurrence was not assessed; thus recurring patients who did not pass away were censored. This, in part, reflects the lack of a single accepted definition of disease recurrence in ovarian cancer (i.e., CA-125 vs palpable or radiographic evidence of disease). An additional limitation of this analysis is its restriction to 325 patients; thus, replication of the most-promising associations found here will be necessary in other patient series. In conclusion, we provide evidence that inherited variation in angiogenesis and inflammation pathways may impact outcome in ovarian cancer. Additional research is needed to exploit these observational findings into avenues for personalized cancer treatment.
Supplementary Material
Table 2. List of investigated genes (N=227) and number of SNPs analyzed by pathway.
| Pathway | Genes | N SNPs Analyzed (N Attempted) |
|---|---|---|
| Angiogenesis | EIF2B5, ELAVL1, HIF1A, KDR, MMP1, MMP2, MMP26, MMP3, MMP9, NOS3, PDGFD, PDGFRA, TIMP1, TIMP2, TIMP3, VEGFA, VEGFC, VHL | 130 (147) |
| Apoptosis | APAF1, ATM, BAD, BAX, BCL2L1, BID, CASP10, CASP3, CASP6, CASP7, CASP8, CASP9, CYC1, EIF2S1, PARP1, PTK2, STAT1, TLN1 | 118 (122) |
| Cell Cycle | ABL1, CCNA1, CCNA2, CCNB1, CCNB2, CCND1, CCND2, CCND3, CCNE1, CCNE2, CCNG1, CCNG2, CDC2, CDC25A, CDC25B, CDK2, CDK4, CDK6, CDK7, CDKN1A, CDKN1B, CDKN2A, CDKN2B, CDKN2C, CDKN2D, CUL1, E2F1, E2F2, E2F3, E2F4, E2F5, E2F6, PLK1, RB1, RBL1, RBL2, SKP2, TFDP1, TFDP2 | 222 (244) |
| Cellular Senescence | ACTB, AKT1, ARID1A, BCL2, EGFR, ERCC3, FHIT, HRAS, IGF1, IGF1R, IGF2, KRAS, MYC, NF1, NR3C1, PIK3CB, POLR2A, PRKCA, SHC1, SMARCA2, SMARCA4, SMARCB1, SMARCC1, SMARCC2, SMARCE1, SOS1, SRC, TBP, TEP1, TERT, TNKS, TP53, XRCC5 | 211 (223) |
| Detoxification | ADH1A, ADH1B, ADH1C, ADH4, ADH5, ADH6, CYP1A1, CYP1A2, EPHX1, GSTM1, GSTP1, NAT1, NAT2, NQO1, NQO2 | 108 (114) |
| Glycosylation | FUT10, FUT3, FUT5, FUT6, FUT7, FUT8, FUT9, GALNT1, GALNT14, GALNT2, GALNT3, GALNT5, GALNT6, GALNT7, MGAT2, MGAT3, MGAT5, POFUT1, ST3GAL1, ST3GAL3, ST3GAL4, ST3GAL5, ST6GAL1, ST6GALNAC5, ST8SIA2, ST8SIA5 | 95 (101) |
| Inflammation | ALOX12, ALOX15, ALOX5, CCL11, CCL2, CCL3, CCR3, CRP, CXCL16, IL10, IL15RA, IL18, IL1A, IL1B, IL1RN, IL4R, IL6, IL6R, IL7R, IL8RA, IL8RB, IL9, PTGS1, PTGS2, TLR2, TNF | 154 (163) |
| One-carbon transfer | AHCYL1, ALDH1L1, DHFR, DNMT1, DNMT3A, DNMT3B, DPYD, FOLR1, MAT2B, MBD4, MGMT, MTHFD1, MTHFD1L, MTHFD2, MTHFR, MTHFS, MTR, MTRR, SHMT1, SLC19A1, TYMS | 179 (187) |
| Under-expressed | ABCB1, ACTG1, ARPC4, COL8A1, DKK3, DUSP1, EHD2, ELOVL4, F3, FGF2, FGF5, FOXD1, GALK1, HNT, KCNG1, LAMA4, MAPRE2, NRP2, P2RX5, PAPPA, PPP1R3A, PPP3CC, TBX3, TPM1, ULBP2 | 120 (154) |
| Miscellaneous | CYP19A1, GRN, NFKBIA, NFKBIB, SULT1E1, TCEAL7 | 79 (81) |
| 1,416 (1,536) |
Under-expressed genes from Dressman et al., J Clin Oncol, 25: 517-525, 2007 and Lancaster et al., Int J Gynecol Cancer, 16: 1733-1745, 2006; N analyzed SNPs excludes 50 (3%) which failed genotyping and 70 (5%) with incorrect genomic coordinates or low MAF.
Acknowledgments
Financial Support This work was supported by the National Cancer Institute (R01-CA86888, R01-CA122443, P50-CA136393-01A1), the Fred C. and Katherine B. Andersen Foundation, and the Mayo Foundation.
We thank Elaine A. Elliott for project management; Michele M. Schmidt, Zachary S. Fredericksen, and Stephanie S. Anderson for data management; Karin M. Goodman for recruitment and abstraction; Katelyn E. Goodman for assistance with manuscript preparation; and Vijayalakshmi Shridhar for advice on gene selection.
Footnotes
Presented in part on April 22, 2009 at the 100th Annual Meeting of the American Association for Cancer Research in Denver, CO, USA.
STATEMENT OF TRANSLATIONAL RELEVANCE: Ovarian cancer is the most lethal of gynecologic malignancies, yet much remains unknown about reasons for the observed differences in outcome across patients. Angiogenesis represents a critical mechanism for therapeutic intervention. Here, we report for the first time that inherited variation in angiogenesis genes correlates with survival time among 325 women seen at the Mayo Clinic. The potential for development of targeted therapeutics may lead to improve outcome in this deadly disease.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Cannistra SA. Cancer of the ovary. N Engl J Med. 2004;351:2519–29. doi: 10.1056/NEJMra041842. [DOI] [PubMed] [Google Scholar]
- 3.Hoskins WJ, Bundy BN, Thigpen JT, Omura GA. The influence of cytoreductive surgery on recurrence-free interval and survival in small-volume stage III epithelial ovarian cancer: a Gynecologic Oncology Group study. Gynecol Oncol. 1992;47:159–66. doi: 10.1016/0090-8258(92)90100-w. [DOI] [PubMed] [Google Scholar]
- 4.McGuire V, Jesser CA, Whittemore AS. Survival among U.S. women with invasive epithelial ovarian cancer. Gynecol Oncol. 2002;84:399–403. doi: 10.1006/gyno.2001.6536. [DOI] [PubMed] [Google Scholar]
- 5.Barnholtz-Sloan JS, Schwartz AG, Qureshi F, Jacques S, Malone J, Munkarah AR. Ovarian cancer: changes in patterns at diagnosis and relative survival over the last three decades. Am J Obstet Gynecol. 2003;189:1120–7. doi: 10.1067/s0002-9378(03)00579-9. [DOI] [PubMed] [Google Scholar]
- 6.Aletti GD, Gallenberg MM, Cliby WA, Jatoi A, Hartmann LC. Current management strategies for ovarian cancer. Mayo Clin Proc. 2007;82:751–70. doi: 10.4065/82.6.751. [DOI] [PubMed] [Google Scholar]
- 7.Quaye L, Gayther SA, Ramus SJ, et al. The effects of common genetic variants in oncogenes on ovarian cancer survival. Clin Cancer Res. 2008;14:5833–9. doi: 10.1158/1078-0432.CCR-08-0819. [DOI] [PubMed] [Google Scholar]
- 8.Schultheis AM, Lurje G, Rhodes KE, et al. Polymorphisms and clinical outcome in recurrent ovarian cancer treated with cyclophosphamide and bevacizumab. Clin Cancer Res. 2008;14:7554–63. doi: 10.1158/1078-0432.CCR-08-0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nimeiri HS, Oza AM, Morgan RJ, et al. Efficacy and safety of bevacizumab plus erlotinib for patients with recurrent ovarian, primary peritoneal, and fallopian tube cancer: a trial of the Chicago, PMH, and California Phase II Consortia. Gynecol Oncol. 2008;110:49–55. doi: 10.1016/j.ygyno.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inoue J, Gohda J, Akiyama T, Semba K. NF-kappaB activation in development and progression of cancer. Cancer Sci. 2007;98:268–74. doi: 10.1111/j.1349-7006.2007.00389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Storz P. Reactive oxygen species in tumor progression. Front Biosci. 2005;10:1881–96. doi: 10.2741/1667. [DOI] [PubMed] [Google Scholar]
- 12.D'Andrilli G, Giordano A, Bovicelli A. Epithelial ovarian cancer: the role of cell cycle genes in the different histotypes. Open clinical cancer journal. 2008;2:7–12. doi: 10.2174/1874189400802010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiantore MV, Vannucchi S, Mangino G, et al. Senescence and cell death pathways and their role in cancer therapeutic outcome. Curr Med Chem. 2009;16:287–300. doi: 10.2174/092986709787002691. [DOI] [PubMed] [Google Scholar]
- 14.Hajra KM, Tan L, Liu JR. Defective apoptosis underlies chemoresistance in ovarian cancer. Adv Exp Med Biol. 2008;622:197–208. doi: 10.1007/978-0-387-68969-2_16. [DOI] [PubMed] [Google Scholar]
- 15.Saldova R, Wormald MR, Dwek RA, Rudd PM. Glycosylation changes on serum glycoproteins in ovarian cancer may contribute to disease pathogenesis. Dis Markers. 2008;25:219–32. doi: 10.1155/2008/601583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baldus SE, Engelmann K, Hanisch FG. MUC1 and the MUCs: a family of human mucins with impact in cancer biology. Critical reviews in clinical laboratory sciences. 2004;41:189–231. doi: 10.1080/10408360490452040. [DOI] [PubMed] [Google Scholar]
- 17.Ho SB, Niehans GA, Lyftogt C, et al. Heterogeneity of mucin gene expression in normal and neoplastic tissues. Cancer Res. 1993;53:641–51. [PubMed] [Google Scholar]
- 18.Yap TA, Carden CP, Kaye SB. Beyond chemotherapy: targeted therapies in ovarian cancer. Nat Rev Cancer. 2009;9:167–81. doi: 10.1038/nrc2583. [DOI] [PubMed] [Google Scholar]
- 19.Sellers TA, Schildkraut JM, Pankratz VS, et al. Estrogen bioactivation, genetic polymorphisms, and ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:2536–43. doi: 10.1158/1055-9965.EPI-05-0142. [DOI] [PubMed] [Google Scholar]
- 20.Lancaster JM, Dressman HK, Clarke JP, et al. Identification of genes associated with ovarian cancer metastasis using microarray expression analysis. Int J Gynecol Cancer. 2006;16:1733–45. doi: 10.1111/j.1525-1438.2006.00660.x. [DOI] [PubMed] [Google Scholar]
- 21.Jones MB, Michener CM, Blanchette JO, et al. The granulin-epithelin precursor/PC-cell-derived growth factor is a growth factor for epithelial ovarian cancer. Clin Cancer Res. 2003;9:44–51. [PubMed] [Google Scholar]
- 22.Chien J, Staub J, Avula R, et al. Epigenetic silencing of TCEAL7 (Bex4) in ovarian cancer. Oncogene. 2005;24:5089–100. doi: 10.1038/sj.onc.1208700. [DOI] [PubMed] [Google Scholar]
- 23.Frazer KA, Ballinger DG, Cox DR, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hinds DA, Stuve LL, Nilsen GB, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–9. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 25.Carlson CS, Eberle MA, Kruglyak L, Nickerson DA. Mapping complex disease loci in whole-genome association studies. Nature. 2004;429:446–52. doi: 10.1038/nature02623. [DOI] [PubMed] [Google Scholar]
- 26.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 27.Olson JE, Ingle JN, Ma CX, et al. A comprehensive examination of CYP19 variation and risk of breast cancer using two haplotype-tagging approaches. Breast Cancer Res Treat. 2007;102:237–47. doi: 10.1007/s10549-006-9324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cox DR. Regression models and life tables (with discussion) Journal of Royal Statistical Society, Series B. 1972;34:187–220. [Google Scholar]
- 29.Habermann TM, Wang SS, Maurer MJ, et al. Host immune gene polymorphisms in combination with clinical and demographic factors predict late survival in diffuse large B-cell lymphoma patients in the pre-rituximab era. Blood. 2008;112:2694–702. doi: 10.1182/blood-2007-09-111658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang SS, Maurer MJ, Morton LM, et al. Polymorphisms in DNA repair and one-carbon metabolism genes and overall survival in diffuse large B-cell lymphoma and follicular lymphoma. Leukemia. 2008 doi: 10.1038/leu.2008.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cerhan JR, Wang S, Maurer MJ, et al. Prognostic significance of host immune gene polymorphisms in follicular lymphoma survival. Blood. 2007;109:5439–46. doi: 10.1182/blood-2006-11-058040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gauderman WJ, Murcray C, Gilliland F, Conti DV. Testing association between disease and multiple SNPs in a candidate gene. Genet Epidemiol. 2007;31:383–95. doi: 10.1002/gepi.20219. [DOI] [PubMed] [Google Scholar]
- 33.Taylor J, Tibshirani R. A tail strength measure for assessing the overall univariate significance in a dataset. Biostatistics. 2006;7:167–81. doi: 10.1093/biostatistics/kxj009. [DOI] [PubMed] [Google Scholar]
- 34.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 35.Fleming GF, Ronnett BM, Seidman JE, et al., editors. Epithelial Ovarian Cancer. 5. Lippencott; 2009. [Google Scholar]
- 36.Rasila KK, Burger RA, Smith H, Lee FC, Verschraegen C. Angiogenesis in gynecological oncology-mechanism of tumor progression and therapeutic targets. Int J Gynecol Cancer. 2005;15:710–26. doi: 10.1111/j.1525-1438.2005.00132.x. [DOI] [PubMed] [Google Scholar]
- 37.Krieg M, Haas R, Brauch H, Acker T, Flamme I, Plate KH. Up-regulation of hypoxia-inducible factors HIF-1alpha and HIF-2alpha under normoxic conditions in renal carcinoma cells by von Hippel-Lindau tumor suppressor gene loss of function. Oncogene. 2000;19:5435–43. doi: 10.1038/sj.onc.1203938. [DOI] [PubMed] [Google Scholar]
- 38.Champion KJ, Guinea M, Dammai V, Hsu T. Endothelial function of von Hippel-Lindau tumor suppressor gene: control of fibroblast growth factor receptor signaling. Cancer Res. 2008;68:4649–57. doi: 10.1158/0008-5472.CAN-07-6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Issa A, Le TX, Shoushtari AN, Shields JD, Swartz MA. Vascular endothelial growth factor-C and C-C chemokine receptor 7 in tumor cell-lymphatic cross-talk promote invasive phenotype. Cancer Res. 2009;69:349–57. doi: 10.1158/0008-5472.CAN-08-1875. [DOI] [PubMed] [Google Scholar]
- 40.Alabi AA, Suppiah A, Madden LA, Monson JR, Greenman J. Preoperative serum levels of serum VEGF-C is associated with distant metastasis in colorectal cancer patients. Int J Colorectal Dis. 2009;24:269–74. doi: 10.1007/s00384-008-0622-x. [DOI] [PubMed] [Google Scholar]
- 41.Fogli A, Boespflug-Tanguy O. The large spectrum of eIF2B-related diseases. Biochem Soc Trans. 2006;34:22–9. doi: 10.1042/BST20060022. [DOI] [PubMed] [Google Scholar]
- 42.Knutson KL, Krco C, Goodman K, et al. T cell immunity to the folate receptor alpha is prevalent in women with breast or ovarian cancer. J Clin Onc. 2006;24:4254–61. doi: 10.1200/JCO.2006.05.9311. [DOI] [PubMed] [Google Scholar]
- 43.Zhang L, Conejo-Garcia JR, Katsaros D, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348:203–13. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- 44.Levina V, Nolen BM, Marrangoni AM, et al. Role of eotaxin-1 signaling in ovarian cancer. Clin Cancer Res. 2009;15:2647–56. doi: 10.1158/1078-0432.CCR-08-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lurje G, Hendifar AE, Schultheis AM, et al. Polymorphisms in interleukin 1 beta and interleukin 1 receptor antagonist associated with tumor recurrence in stage II colon cancer. Pharmacogenet Genomics. 2009;19:95–102. doi: 10.1097/FPC.0b013e32831a9ad1. [DOI] [PubMed] [Google Scholar]
- 46.Lissoni P, Brivio F, Rovelli F, et al. Serum concentrations of interleukin-18 in early and advanced cancer patients: enhanced secretion in metastatic disease. J Biol Regul Homeost Agents. 2000;14:275–7. [PubMed] [Google Scholar]
- 47.Bushley AW, Ferrell R, McDuffie K, et al. Polymorphisms of interleukin (IL)-1alpha, IL-1beta, IL-6, IL-10, and IL-18 and the risk of ovarian cancer. Gynecol Oncol. 2004;95:672–9. doi: 10.1016/j.ygyno.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 48.Maitland ML, Ratain MJ, Cox NJ. Interpreting P values in pharmacogenetic studies: a call for process and perspective. J Clin Oncol. 2007;25:4513–5. doi: 10.1200/JCO.2007.12.7803. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


