Summary
The structures of T7 RNA polymerase (T7 RNAP) captured in the initiation and elongation phases of transcription, as well as an intermediate stage provide insights into how this RNA polymerase protein can initiate RNA synthesis and synthesize 7 to 10 nucleotides of RNA while remaining bound to the DNA promoter site. Recently, the structures of T7 RNAP bound to it promoter DNA along with either a 7 nucleotide or 8 nucleotide transcript show an elongated product site resulting from a 40° or 45° rotation of the promoter and domain that binds it. The different functional properties of the initiation and elongation phases of transcription are illuminated from structures of the initiation and elongation complexes. Structural insights into the translocation of the product transcript of RNAP, its separation of the downstream duplex DNA and its removal of the transcript from the heteroduplex are provided by the structures of several states of nucleotide incorporation. A conformational change in the “fingers” domain that results from the binding or dissociation of incoming NTP or PPi appears to be associated with the state of translocation of T7 RNAP.
DNA dependent RNA polymerases are able to recognize their DNA promoter site, where the initiation of transcription occurs, and remain bound to it throughout the initiation phase of RNA synthesis and then transition to an elongation phase in which the initiation site is released. An important question is how these enzymes are able to accommodate the elongating heteroduplex product while remaining attached to the promoter site; furthermore, what is the nature of the structural transition from the initiation to the elongation phase and how is it achieved? A second question concerns the structural basis of promoter clearance upon transition to the elongation phase as well as the source of processivity of this phase. A third property of RNA polymerases to be addressed is how and when the incorporation of nucleotides into a growing primer chain results in translocation of the product duplex and a concomitant separation of the strands of the downstream duplex DNA.
The most complete description of the structural basis of the transition from the initiation phase of transcription to the elongation phase exists for T7 RNA polymerase1,2,3,4. The initiation phase of transcription by T7 RNAP exhibits the accumulation of the heteroduplex product in the active site that gradually displaces a portion of the polymerase that provides the specificity of T7 RNAP for the promoter site4.
T7 RNAP, like the cellular RNA polymerases, translocates along the DNA template as the duplex product is synthesized, and is able to open the downstream DNA duplex without requiring a separate helicase protein. Structural data on T7 RNA polymerase are consistent with a mechanism of translocation that is associated with the dissociation of PPi,5,6. There is an equilibrium among the conformational states associated with duplex translocation, and we can only establish that the most stable state is altered by the protein conformational changes associated with the binding or dissociation of the ligands PPi or NTP. Distinguishing between whether the product is being “pushed” by the conformational change or the change is capturing an equilibrium state of translocation generally requires knowing which of these two paths is the fastest, the rate of ligand induced conformational change or the one-dimensional diffusion process of the enzyme on the DNA, data which are generally unknown. Hence, the goal here is to describe what is currently known about the structural, biochemical and kinetic bases for understanding the mechanisms of translocation and strand separation.
The transition from the initiation to the elongation phase
T7 RNA polymerase initiates RNA synthesis after binding to a specific promoter DNA sequence and opening the DNA duplex; the enzyme remains bound to the promoter sequence throughout the initiation phase which involves the synthesis of an 8 to 12 nucleotide transcript. This phase, often referred to as the abortive synthesis phase, results in the synthesis and release of numerous short transcripts7. Only after the transition to the elongation phase does transcription proceed with complete processively until completion of the entire RNA transcript has been synthesized. Various models have been previously proposed to account for the enzyme s ability to remain bound to the promoter while synthesizing RNA. These included the inchworm model whereby the enzyme s active site catalyzing nucleotide incorporation progresses down the template while the promoter binding site remains fixed on the DNA, or the scrunching model where the product is accumulated in the polymerase active site.
The RNA polymerase from phage T7 (T7RNAP) is a 98,000 Dalton molecular weight, single subunit whose polymerase domain is homologous to that of the DNA polymerase I family of polymerases and is completely unrelated to the multisubunit RNA polymerases. It initiates transcription at a specific site by binding to a 17 base-pair promoter DNA that it recognizes through base specific interactions. These are made by side chains emanating from a beta-hairpin called the specificity loop that binds in a DNA major groove as well as a loop that interacts within the minor groove.1,8 The promoter binding site is formed in part by an N-terminal domain consisting of 300 amino acid residues whose structure is unique to this polymerase, as well as the β hairpin specificity loop, which is an insertion into the fingers domain of the polymerase. Comparison of the structure of the T7 RNAP bound to a 17 base-pair promoter DNA (nucleotides −1 to −17) with that of its complex to a promoter duplex containing a 5 nucleotide 5′ template overhang (nucleotides 1-5) and a 3 nucleotide transcript shows that the promoter recognition is identical, but the template strand has accumulated in the active site after the synthesis of 3 nucleotides in order to position the +4 template base appropriately for nucleotide insertion; thus, a small amount of “scrunching” of the template has occurred that allowed the polymerase catalytic site to move 3 nucleotides down the template strand.8 However, the cleft in which the 3 base-pair heteroduplex resides in the initiation complex is not sufficiently large to accommodate even a fourth base-pair in the post-translocation state. The possibility that the transcript peeled off the DNA after only three base-pairs was not consistent with biochemical data, such as the crosslinking of template and transcript strands, which strongly implied that the heteroduplex would be as long as 8 base-pairs.9 To understand the mechanism by which the T7 RNAP progresses from the initiation to the elongation phase it was necessary to obtain the structures of intermediate states4 in the transition from initiation to elongation, as well as that of the elongation phase.2,3
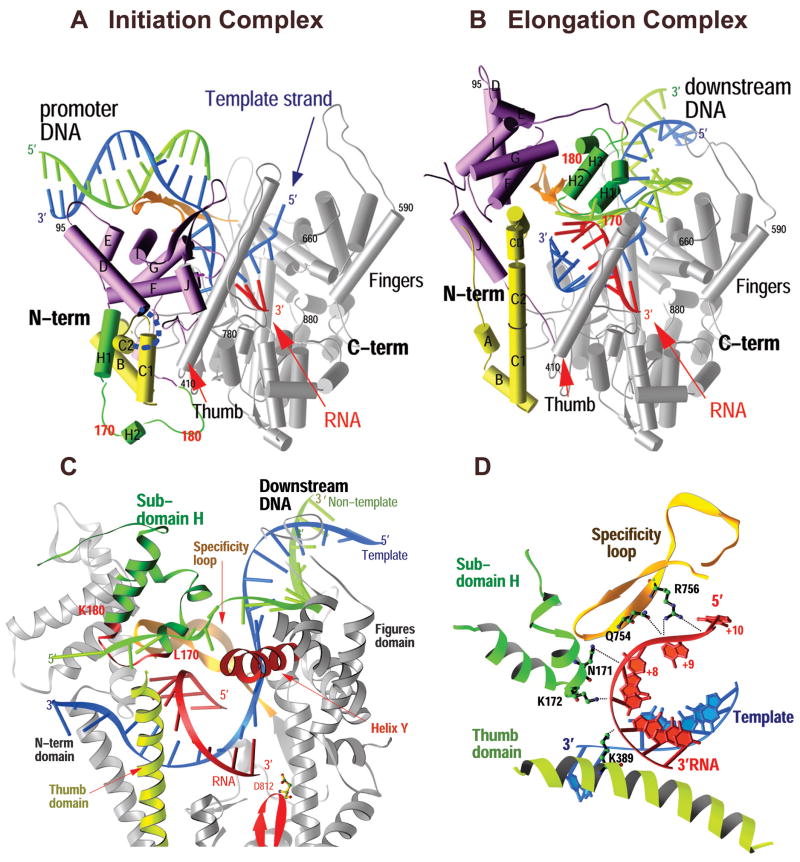
The structure of T7 RNAP captured in an elongation complex was able to accommodate all the biochemical data and show that the transition from initiation to elongation is accompanied by a massive conformational change, particularly in the N-terminal domain of the polymerase2,3. This conformational change results in the enlargement of the binding cleft for the heteroduplex product enabling it to accommodate up to 8 base-pairs of heteroduplex, the complete destruction of the promoter binding site and the formation of a tunnel through which the RNA transcript was seen to emerge after being separated from the template DNA.2 In addition to the 8 base-pair heteroduplex, four nucleotides of single stranded RNA transcript at its 5′ end are seen to be “peeled” off the template, consistent with biochemical data, 9,10 by an alpha-helix in the thumb domain as well as side chain interactions with a newly refolded H subdomain and the specificity loop (Figure, C & D). Furthermore, a 9 nucleotide bubble is formed in the substrate DNA as predicted from biochemical experiments of Huang and Sousa 10, stabilized on the one hand by the 8 base-pair heteroduplex and on the other by extensive interactions of the single stranded non-template strand with the protein (Figure 4.1D).2 Comparison of the initiation and elongation structures (Figure, A & B) showed that the N-terminal domain could be most conveniently thought of as consisting of 3 subdomains. In particular, an assembly of six α helices, moved as a rigid body through a rotation of 220° and a translation of 30 Å that both provided space for the elongating product and positioned the subdomain in the location formerly occupied by the promoter DNA. Perhaps the most dramatic structural arrangement observed in this transition is a loop of protein that moves from one side of the N-terminal domain to the complete opposite side of the 6-helix subdomain and refolds into a two helix H domain that forms part of the lid of the transcript tunnel through which the RNA product departs.2
Figure 4.
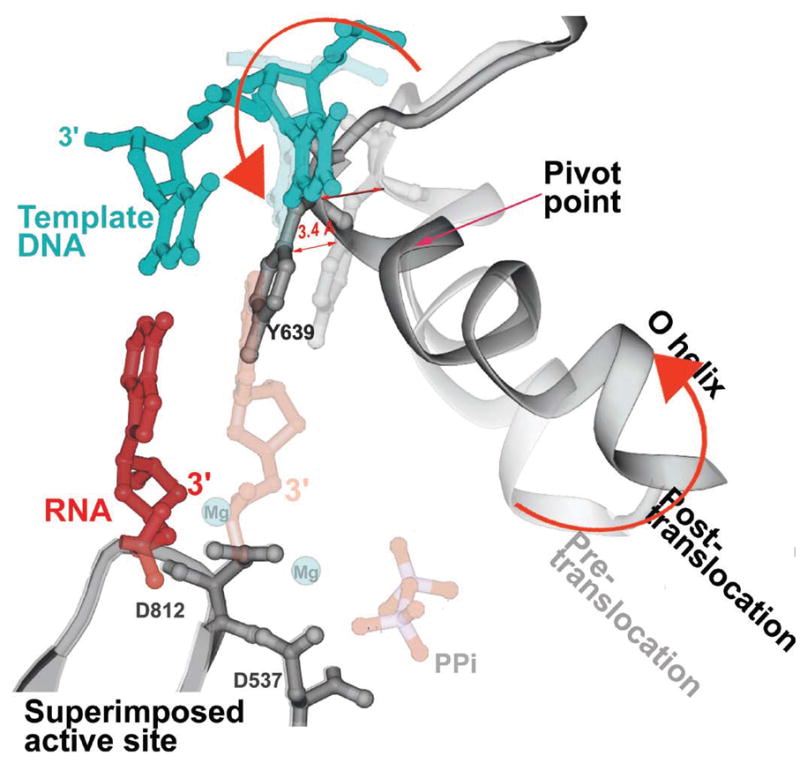
A superposition of the pre- and post-translocation structures at the active site showing the pivoted rotation undergone by the O helix that is associated with translocation (Yin & Steitz, 2004). In the pre-translocation complex (lighter colors), the O helix (light gray) is positioned in the closed conformation by PPi (light red), which is bound to the catalytic carboxyls through Mg. In this conformation, Y639 allows formation of the new base pair (light red and blue). After PPi release, the O helix rotates around Val634, which results in the positive end of the helix moving away from the active site while the other end of the helix moves Y639 by 3.4 Å into the position of the newly formed primer terminus resulting in translocation.
Figure 1.
Comparison of the structures of the T7 RNAP initiation and elongation complexes (A and B) and views of the transcription bubble (C and D).2 The initiation complex (A) and elongation complex (B) have been orientated equivalently by superimposing their palm domains. Helices are represented by cylinders and beta strands by arrows. The corresponding residues in the NH2-terminal domains of the two complexes that undergo major refolding are colored in yellow, green, and purple, and the COOH-terminal domain (residues 300 to 883) is colored in gray. The template DNA (blue), nontemplate DNA (green), and RNA (red) are represented with ribbon backbones. The proteolysis-susceptible region (residues 170 to 180) is a part of subdomain H (green) in the elongation complex and has moved more than 70 Å from its location in the initiation complex. The specificity loop (brown) recognizes the promoter during initiation and contacts the 5′ end of RNA during elongation, whereas the intercalating hairpin (purple) opens the upstream end of the bubble in the initiation phase and is not involved in elongation. The large conformational change in the NH2-terminal region of T7 RNAP facilitates promoter clearance. This figure was made with the program Ribbons. (C) Interactions of the transcription bubble and heteroduplex in the elongation complex with domain H (green and red) and specificity loop (brown). Proteolytic cuts within the red loop in subdomain H reduce elongation synthesis (21, 22). Thumb alpha-helix (yellow) and alpha-helix Y (orange) are analogously involved in strand separation. (D) Side chains from subdomain H (green), the specificity loop (brown), and the thumb that interact with the single-stranded 5′ end of the RNA transcript and facilitate its separation from the template. The DNA substrate in the initiation complex consisted of duplex promoter DNA from −1 to −17 and a 5′ nucleotide overhang of the template strand (nucleotides 1 to 5) at the 5′ end. The DNA used in the elongation complex contained 10 b.p. of downstream duplex DNA, a 10 nucleotide non-complementary DNA “bubble” and a 10 b.p upstream duplex DNA that is disordered in the structure.
The structure of the elongation complex immediately explains both promoter clearance and the processivity of the elongation phase. Promoter clearance is achieved because the promoter binding site is completely destroyed (Figure 2, A & B). Not only is the 6-helix domain in the position formerly occupied by the promoter, but the anti-parallel beta strand specificity loop that recognizes the promoter in the initiation complex has now moved to form part of the lid of the transcript exit tunnel and is seen to interact with the departing transcript,2 consistent with biochemical crosslinking data.9 High processivity is explained by the formation of this transcript exit tunnel that surrounds the transcript and thus functions analogously to the sliding clamp in DNA replication. Once the transcript is threaded through the tunnel, it can no longer dissociate from the enzyme until transcription is terminated.
Figure 2.
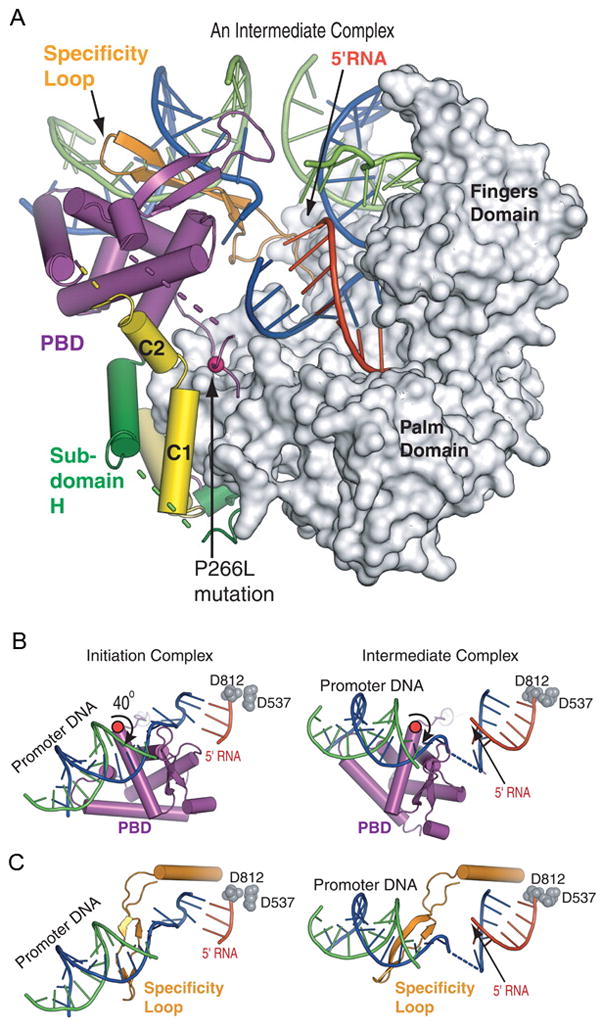
(A) The T7 RNAP in the 7-nt RNA intermediate complex is bound to both promoter and downstream DNA.4 The PBD has rotated by 40° away from the C-terminal domain, avoiding a steric clash with the transcript and allowing for 7 bp of heteroduplex to form in the active site.
(B, C) Promoter and PBD movements during the transition. (B) A view looking down onto the promoter bound to the PBD. A 40° rotation of the PBD away from the active site occurs around an axis that passes through a flexible loop of residues 198 to 204. The catalytic aspartic acid residues (D812 and D537) represent the active site. (C) The same view as in (B) without the PBD but showing the specificity loop, which also rotates.
The mechanism by which this remarkable conformational change is achieved is now emerging. Biochemical experiments designed to identify at which stages of the initiation phase particular structural rearrangements occur have been done by crosslinking domains whose positions change in the initiation to elongation transition. Formation of a disulfide crosslink between the six helix domain and the fingers domain that would restrict movement of these domains from the initiation state structure results in synthesis of transcripts of length five or six nucleotides, only one or two more than the initiation state would appear able to accommodate.11 Another disulfide crosslink between residue 128 in the helical domain and residue 393 in the fingers domain allows production of transcripts of lengths 7 and 8.11 These and other crosslinking experiments as well as Fe-BABE DNA cleavage studies led Sousa and coworkers12 to conclude that most of the major conformational changes in the transition from initiation to elongation do not occur during the elongation of the transcript from 3 to 8 nucleotides, but rather occur after the transcript reaches 8-9 nucleotides. Extension to this length also coincides with the release of the promoter.
Structural insights into the first phase of the transition from initiation to elongation were obtained from the structures of the T7 RNA polymerase complexed with a 33 base-pair primer-template DNA containing a 17 base-pair promoter, a transcription bubble of six noncomplementary bases followed by a 10 base-pair downstream complementary region and either a 7 or 8 nucleotide transcript were determined at 3.0 Å and 5 Å resolution. They establish the nature of the conformational changes that occur upon elongation of the transcript from 3 to 8 nucleotides.4 Attempts to capture this structure by using wild-type T7 RNAP resulted in structures that were identical to the initiation complex structure and lacked the added RNA transcript, presumably due to the instability of the intermediate complex relative to an aborted complex that reverts to the initiation state structure. To address this problem, the formation of a stable initiation to elongation intermediate complex was achieved using a mutant polymerase (P266L) that makes fewer abortive transcripts during the initiation phase of transcription.13
The structure of the intermediate complex containing a 7 nucleotide transcript reveals a left-handed 40° rotation of the six helix promoter binding domain (PBD), the specificity loop, and the bound promoter about an axis passing through residues 198 to 204, consistent with biochemical data indicating that T7 RNAP maintains promoter contacts during abortive synthesis (Fig. 2A and B). Due to the conformational change consisting largely of a rigid body rotation, the contacts between the PBD, specificity loop and promoter DNA in the structure of the intermediate complex remain the same as those in the initiation complex. The rotation, however, enlarges the product containing cleft in the active site to enable the accommodation of the 7 nucleotide elongated RNA. Furthermore, the PBD rotates by an additional 5° to 45° during the transcription of 8 nucleotides of RNA. The C2 alpha-helix axis undergoes a rotation relative to the C1 helix that places it about midway between its positions in the initiation and elongation complexes. In contrast to the significant changes observed in the position of the PBD and C2 helix, there is little movement of subdomain H during the first part of the transition from initiation to elongation.
It appears likely, though not experimentally proven, that as the RNA transcript elongates from 3nt to 7nt, the PBD may undergo a stepwide rotation to reach the position observed in the structure of the 7 nucleotide intermediate complex. What conformational changes occur in the second stage of the transition after the RNA is extended beyond 8 nucleotides is not established. It appears to involve the loss of promoter contacts and larger structural changes in the specificity loop, the PBD and subdomain H.
Translocation by T7 RNA polymerase
The structural basis of heteroduplex product translocation and separation of downstream duplex DNA by the monomeric T7 RNA polymerase has been illuminated by high resolution crystal structures of the four states of nucleotide incorporation by T7 RNAP in the elongation phase of DNA synthesis.5 These structures include those of the enzyme in a post-translocated product complex2, a complex with the post-translocated product with the incoming nucleotide bound in a pre-insertion site14, the structure of a ternary complex with the incoming NTP positioned for incorporation and finally the product complex in a pre-translocation state including bound pyrophosphate.5 Two of these states, the binary complex with primer-template DNA and the ternary complex including the incoming deoxy-NTP, have been determined for several DNA polymerase I enzymes as well15–19, and the structures of these states are largely identical to the corresponding complexes with the T7 RNA polymerase. Furthermore, the initiation complex of T7 RNAP with a 3 nucleotide transcript, but without PPi, is in the post-translocated state. The conclusion from these studies is that a conformational change in the fingers domain of the pol I family polymerases from an “open” to a “closed” state is stabilized by the binding of the NTP (or dNTP) to the O helix followed by its positioning in the insertion site, and the change from a “closed” to an “open” state is associated with release of the product pyrophosphate. Further, it is the conformational change resulting from the release of the pyrophosphate product that is associated both with translocation of the heteroduplex product and with strand separation of the downstream DNA.5
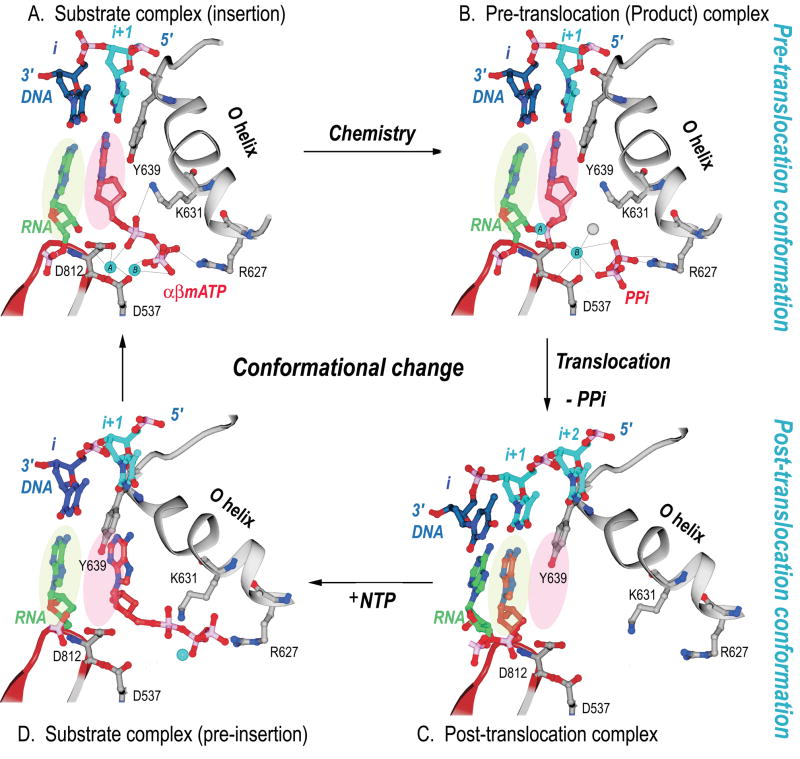
The structures of the four stages of nucleotide incorporation by T7 RNA polymerase have been determined for the enzyme during its elongation phase of RNA synthesis (Figure 3). The structure of the complex after nucleotide incorporation and pyrophosphate dissociation is in the post-translocation state and the base that is to template the next incoming nucleotide is located in a binding pocket outside the active site.17,19 The substrate NTP binds initially to a distinct “open” conformation of the enzyme at a site located on the O helix. It is there apparently base-paired to the next template base, but still not in a position for insertion.14 A conformational change that can be described as the rotation of a 5 helix assembly about a pivot point then occurs that results in the formation of a closed complex in which the incoming nucleotide is properly positioned for insertion and the templating base moves from a preinsertion pocket to pair with this incoming base.5 This conformational change is stablized by the ionic links that are able to form in the closed conformation between the three phosphates of the NTP and the two magnesium ions bound to the enzyme on one side and an arginine residue of the O helix on the other. The structure of T7 RNAP in the pre-translocation product complex that forms in the presence of pyrophosphate (Fig. 3B) is identical to the structure of the enzyme in the ternary complex (Fig. 3A). The enzyme continues to be held in the closed conformation after nucleotide incorporation, once again, by the pyrophosphate product forming an ionic crosslink between a magnesium ion bound to the active site carboxylates and an arginine on the O helix. Dissociation of pyrophosphate completes this cycle and results in the formation of an open complex accompanied by translocation of the product heteroduplex.
Figure 3.
Structural changes at the active site of T7 RNAP during a single nucleotide addition cycle.5 This figure shows the O helix with its phosphate binding K631 and R627, a β turn-β motif bearing the metal binding catalytic D812 and D537, template nucleotides in blue, the RNA primer terminus in green, as well as the P and N sites in green and pink ovals. (A) The NTP (red) is bound to the N site in position to be inserted with its metal bound triphosphate moiety crosslinking the O helix to the active site aspartic acid residues. Template nucleotide i+1 (light blue) forms a base pair with the correct incoming nucleotide. (B) The product of the phosphoryl transfer reaction shows a Mg ion (blue) bound to PPi (red), which crosslinks D537 to R627, thereby maintaining RNAP in an identical conformation as in the substrate complex. The 3′ end of RNA remains in the N site in a pre-translocation state. (C) Release of Mg-PPi results in the loss of the link between the O helix and D537, which promotes the rotation of the O helix and translocation of the 3′ end of the RNA to the P site. The RNAP conformational change also places Y639 into the N site and positions the i+2 template nucleotide into the flipped-out, pre-insertion position. (D) A modeled NTP preinsertion complex with NTP bound to the post-translocated RNAP. Although the base binding site is blocked by the side chain of Y639, the triphosphate binding site on the O helix is accessible.
The conformational change in the enzyme associated with the return to the open state, consists largely of a rotation of a 5-helix bundle that includes the O helix about a pivot axis. Rotation of the bundle to the “open” conformation results in a 3.5 Å movement of a tyrosine residue (Y639) that becomes stacked on the primer-template bases. This change indeed stabilizes the translocated position of the product duplex (Figure 4). The position of this tyrosine in this open state of the enzyme not only sterically precludes the return of the heteroduplex to its pre-translocation position, but also blocks the insertion site for the next incoming NTP. Only after the NTP bound to the pre-insertion site on the O helix in the open state promotes the formation of the closed state does the tyrosine side chain move out of the way thereby allowing the incoming NTP base and the template base to move in to the insertion site and replace it. Since this tyrosine is conserved in all Pol I family polymerases, a similar mechanism of translocation is suggested. The translocated state may be associated with the open conformation being more stable than the closed conformation after dissociation of the pyrophosphate and loss of the cognate electrostatic crosslinks. In the open conformation the rotated 5 helix bundle buries hydrophobic surface area which presumably provides the “spring” energy to stabilize formation of the open conformation and the associated translocation upon PPi dissociation.
Interestingly, the single molecule studies of Thomen et al.20 estimate the equilbirum constant between the pre- and post-translocation states in the binary complex of the enzyme with DNA (i.e. bound to neither incoming nucleotide nor pyrophosphate) corresponds to a 3 to1 preference for the post-translocated state at this stage of the reaction, a value compatible with the structural data. Also, chemical cleavage studies show a distribution of transcript states that favor pre-translocation in the presence of PPi and post-translocation in the presence of NTP.12
While the equilibrium that exists between the pre-translocated, “closed” product state in the absence of PPi and the translocated, “open” state forms the translocated product state in the absence of PPi in three separate structures of T7 RNAP,1,2,3 the structures of three binary substrate complexes with A family DNA polymerases16,19,17 and the B family φ 29 DNA polymerase.6 Also, the structures of all A and B family apo-DNA polymerases, as well as three structures of T7 RNAP without substrates are in the open conformation that is associated with the translocated state. Although the DNA substrate can diffuse between the pre- and post-translocation states in the closed conformation of the enzymes, it cannot exist in the pre-translocation state in the open form due to the position of Y639. Since the templating base is likewise blocked from the nucleotide insertion site by Y639, the open to closed state conformational change must occur prior to catalysis, though it is not necessarily the rate limiting step. While the equilibrium constant between the pre- and post-translocated states cannot be determined by X-ray crystallography, the exclusive observation of the translocated state in all A-family polymerase product complexes in the absence of PPi suggests that it is significantly energetically favored. These structures appear to describe a plausible pathway for translocation and delivery of the NTP to the incorporation site. The binding of the incoming nucleotide to the translocated open state and its further conversion into a closed translocation state will stabilize the complex and prevent the primer terminus from entering the pre-translocation position, unless the NTP dissociates. Thus, the binding of NTP can be described as stabilizing the translocated state.
Acknowledgments
Funding for research on polymerases in the T.A.S. laboratory was provided by NIH grant GM57510.
Abbreviations
- NTP
nucleotide triphosphate
- PBD
promoter binding domain
- PDB
Protein Data Bank
- T7RNAP
bacteriophage T7 DNA dependent RNA polymerase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cheetham GMT, Steitz TA. Structure of a transcribing T7 RNA polymerase initiation complex. Science. 1999;286:2305. doi: 10.1126/science.286.5448.2305. [DOI] [PubMed] [Google Scholar]
- 2.Yin YW, Steitz TA. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002;298:1387. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- 3.Tahirov TH, Temiakov D, Anikin M, Patlan V, McAllister WT, Vassylyev DG, Yokoyoma S. Structure of a T7 RNA polymerase elongation complex at 2.9 Å resolution. Nature. 2002;420:43. doi: 10.1038/nature01129. [DOI] [PubMed] [Google Scholar]
- 4**.Durniak K, Bailey S, Steitz TA. The structure of a transcribing T7 RNA polymerase in transition from initiation to elongation. Science. 2008;322:553–557. doi: 10.1126/science.1163433. The crystal structures of T7 RNAP bound to substrate DNA and either a 7 nt or 8nt transcript are described. They show a 40° and 45° rotation of the promoter binding domains resulting in an enlargement of the heteroduplex product binding cleft. They represent intermediate states in the transition from the initiation to elongation states. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin YW, Steitz TA. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004;116:393. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 6*.Berman AJ, Kamtekar S, Goodman S, Lázaro JM, de Vega M, Blanco L, Salas M, Steitz TA. Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases. EMBO J. 2007;26:3493–3505. doi: 10.1038/sj.emboj.7601780. These structures of ternary complex of phi29 DNA polymerase show conformational changes in the fingers domain of this B-family polymerase that are analogous to those of the A-family polymerases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brieba LG, Sousa R. T7 promoter release mediated by DNA scrunching. EMBO J. 2001;20:6826. doi: 10.1093/emboj/20.23.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheetham GMT, Jeruzalmi D, Steitz TA. Structural basis for initiation of transcription from an RNA polymerase-promoter complex. Nature. 1999;399:80–83. doi: 10.1038/19999. [DOI] [PubMed] [Google Scholar]
- 9.Temiakov D, Mentesana PE, Ma K, Mustaev A, Borukhov S, McAllister WT. The specificity loop of T7 RNA polymerase interacts first with the promoter and then with the elongating transcript, suggesting a mechanism for promoter clearance. Proc Natl Acad Sci USA. 2000;97:14109. doi: 10.1073/pnas.250473197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang J, Sousa R. T7 RNA polymerase elongation complex structure and movement. J Mol Biol. 2000;303:347. doi: 10.1006/jmbi.2000.4150. [DOI] [PubMed] [Google Scholar]
- 11.Ma K, Temiakov D, Anikin M, McAllister WT. Probing conformational changes in T7 RNA polymerase during initiation and termination by using engineered disulfide crosslinks. Proc Natl Acad Sci USA. 2005;102:17612. doi: 10.1073/pnas.0508865102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo Q, Sousa R. Translocation by T7 RNA polymerase: A sensitively poised Brownian ratchet. J Mol Biol. 2006;358:241. doi: 10.1016/j.jmb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Guillerez J, Lopez PJ, Proux F, Launay H, Dreyfus M. A mutation in T7 RNA polymerase that facilitates promoter clearance. Proc Natl Acad Sci USA. 2005;102:5958. doi: 10.1073/pnas.0407141102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Temiakov D, Patlan V, Anikin M, McAllister MT, Yokoyama S, Vassylyev DG. Structural basis for substrate selection by T7 RNA polymerases. Cell. 2004;116:381. doi: 10.1016/s0092-8674(04)00059-5. [DOI] [PubMed] [Google Scholar]
- 15.Doublié SD, Ellenberger T. The mechanism of action of T7 DNA polymerase. Curr Opin Struct Biol. 1998;8:704. doi: 10.1016/s0959-440x(98)80089-4. [DOI] [PubMed] [Google Scholar]
- 16.Doublié S, Tabor S, Long AJ, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature. 1998;391:251. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998;17:7514. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson SJ, Taylor JS, Beese LS. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc Natl Acad Sci USA. 2003;100:3895. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiefer JR, Mao C, Braman JC, Beese LS. Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature. 1998;391:304. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]
- 20.Thomen P, Lopez PJ, Heslot F. Unraveling the mechanism of RNA-polymerase forward motion by using mechanical force. Phys Rev Letters. 2005;94:128102. doi: 10.1103/PhysRevLett.94.128102. [DOI] [PubMed] [Google Scholar]
- 21.Ideda RA, Richardson CC. Interactions of a proteolytically nicked RNA polymerase of bacteriophage T7 with its promoter. J Biol Chem. 1987;262:3800. [PubMed] [Google Scholar]
- 22.Muller DK, Martin CT, Coleman JE. Processivity of proteolytically modified forms of T7 RNA polymerase. Biochem. 1988;27:5763. doi: 10.1021/bi00415a055. [DOI] [PubMed] [Google Scholar]