Abstract
Purpose
Hepatocellular cancer (HCC) is highly resistant to chemotherapy and is associated with a poor prognosis. Chronic hepatitis C (HCV) infection is a major cause of HCC. However, the effect of viral proteins in mediating chemosensitivity in tumor cells is unknown. We postulated that HCV viral proteins could modulate therapeutic responses by altering host cell microRNA (miRNA) expression.
Experimental design
HepG2 malignant hepatocytes were stably transfected with full length HCV genome (Hep-394) or an empty vector (Hep-SWX). miRNA profiling was performed by using a custom microarray, and the expression of selected miRNAs was validated by real time PCR. Protein expression was assessed by western blotting, while caspase activation by a luminometric assay.
Results
The IC50 to sorafenib was lower in Hep-394 compared to Hep-SWX control cells. Alterations in miRNA expression occurred with 10 miRNAs > 2-fold down-regulated and 23 miRNAs > 2-fold up-regulated in Hep-394 cells compared to controls. Of these, miR-193b was over-expressed by 5-fold in Hep-394 cells. miR-193b was predicted to target Mcl-1, an anti-apoptotic protein that can modulate the response to sorafenib. The expression of Mcl-1 expression was decreased and basal caspase-3/7 activity and PARP cleavage were increased in Hep-394 cells compared to controls. Moreover, transfection with precursors to miR-193b decreased both Mcl-1 expression and the IC50 to sorafenib.
Conclusions
Cellular expression of full length HCV increases sensitivity to sorafenib by miRNA-dependent modulation of Mcl-1 and apoptosis. Modulation of miRNA responses may be a useful strategy to enhance response to chemotherapy in HCC.
Keywords: microRNA, HCC, sorafenib, HCV
INTRODUCTION
Chronic hepatitis C virus (HCV) infection is the major cause of hepatocellular carcinoma (HCC) in the United States (1). The incidence of HCV associated HCC is predicted to increase over the next several years as a result of the epidemic of chronic HCV (2). In patients with chronic HCV infection, HCC usually arises in the setting of cirrhosis or bridging fibrosis. However, the presence of HCV is an independent risk factor for HCC. The responses to chemotherapy in the treatment of HCC have been dismal (3,4). In part this reflects the lack of understanding of the mechanisms by which HCV contributes to tumor cell behavior. The multi-kinase inhibitor sorafenib has been recently approved for the treatment of advanced HCC (5). Subgroup analysis of the SHARP trial suggests that sorafenib may be more effective for HCV associated HCC, but this remains to be verified (6).
HCV can interact with host cells to modulate cell survival signaling, alter gene expression and induce cell transformation (7). Modulation of gene expression in the liver by HCV has been implicated in malignant transformation, and viral proteins are frequently detected in HCV associated cirrhotic liver and HCC cells (8,9). Moreover, HCV associated proteins have been shown to alter hepatocellular gene expression by transcriptional trans-regulation (10). Alteration of gene expression by HCV proteins as a mediator of resistance to interferon has also been reported (11). However, the contribution of HCV associated proteins to chemosensitivity in HCC cells remains unknown.
Small non-coding RNA termed miRNAs have been recently shown to be potent modulators of gene expression. The expression of host cell miRNAs can be modulated by HCV. Furthermore, deregulated miRNA expression has been shown to influence the replication potential of HCV in liver cells (12,13). Several recent studies have shown that the expression of miRNAs is altered in human HCC, implicating them in hepatocarcinogenesis (14-16). We have recently shown that tumor cell behavior can be modulated by altered miRNA expression (15,17). Thus altered hepatocyte miRNA expression may result in cellular phenotypic changes that can modulate the sensitivity to therapeutic agents. In the present study we sought to assess the effect of HCV proteins on miRNA expression and their contribution to therapeutic responses in HCC.
MATERIALS and METHODS
Cell lines and culture
HepG2 human hepatocellular cancer cells were stably transfected with expression vectors encoding tcDNA representing the entire open reading frame of HCV genotype 1b (Hep-394) or empty vector (Hep-SWX) and validated as expressing HCV proteins by immunoblotting as previously described (11,18). Huh-7 cells were infected with the replicon of HCV JFH-1 to achieve an HCV RNA level of 5 × 10 8 copies / mg of total RNA (19). The cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (GIBCO, Gaithersburg, MD) containing 600 μg/ml G418 (GIBCO). Normal human hepatocytes (HH) were obtained from Sciencell (San Diego, CA) and cultured following the manufacturer's instructions.
Cytotoxicity assay
Cells (10,000/well) were plated in 96-well plates (BD Biosciences, Rockville, MD). Sorafenib or diluent control were added at concentrations ranging from 10−9 to 10−3 M. Cell viability was assessed after 72 hours using the CellTiter 96 AQueous assay kit (Promega, Madison, WI) as previously described (20). The IC50 value, at which 50% of the cell growth was inhibited compared with control, was derived using the XLfit software (IDBS, Burlington, MA).
Apoptosis assays
Cells were plated in 35 mm dishes. At the specified time points cells were stained with 27 μg/ml DAPI and then visualized with a fluorescence microscope (Nikon, Melville NY). Apoptotic cells were defined using morphological criteria including nuclear and cytoplasmic shrinkage, nuclear fragmentation, chromatin condensation and apoptotic bodies. At least 300 cells in several different high power fields were counted, and the number of apoptotic cells reported as a percentage of total cells counted in the same fields. For caspase activation assay, cells (10,000/well) were seeded in 96-well plate and caspase 3/7 activity assessed after 72 hours using a luminometric assay (Caspase-Glo 3/7 assay, Promega, Madison, WI). Apoptotic cells were also detected by labeling DNA breaks through the Terminal Deoxynucleotide Transferase dUTP Nick End Labeling (TUNEL) assay (Invitrogen, Carlsbad, CA). Cells were processed as indicated by the manufacturer and analyzed by flow cytometry with a BD FACScalibur (Heidelberg, Germany).
MicroRNA isolation and expression profiling
RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA). Total RNA (5 μg) was reverse transcribed using biotin end-labeled random octamer oligonucleotide primers. Hybridization of biotin-labeled complementary DNA was performed using a custom miRNA microarray chip (OSU-CCC version 4.0), which includes 898 human miRNA genes and precursors, spotted in duplicates. The hybridized chips were washed and processed to detect biotin-containing transcripts by streptavidin–Alexa647 conjugate, scanned and quantitated using an Axon 4000B scanner (Axon Instruments, Downingtown, PA) and the GenePix 6.0 software (Axon Instruments, Downingtown, PA). Three replicates were tested for each sample.
Computational analysis
Average values of the replicate spots of each miRNA were background subtracted, normalized, and further analyzed. Normalization was performed using the global median method. We selected the miRNAs measured as present in at least as many samples as the smallest class in the data set (50%). Absent calls were thresholded to 4.5 (log2 scale) before statistical analysis, representing the average minimum intensity level detectable in the system. Differentially expressed miRNAs were identified using the Class Comparison Analysis of BRB tools version 3.6.0 (http://linus.nci.nih.gov/BRB-ArrayTools.html). This tool is designed to analyze data using the parametric tests t/F tests, and random variance t/F tests. The criterion for inclusion of a gene in the gene list is a p-value less than 0.05.
Real time PCR assay for mature miRNAs
Total RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA) and the expression of specific mature miRNAs was confirmed by real time PCR analysis using a TaqMan human MicroRNA Assay Kit (Applied Biosystems, Foster City, CA). miRNA expression was normalized to RNU6 expression.
Western Blotting
Immunoblot analysis was performed as previously described (15). The following primary antibodies were used: goat polyclonal anti-actin (1:400) (Santa Cruz Biotechnology, CA), mouse monoclonal anti-Mcl-1 (1:500) (Santa Cruz Biotechnologies, CA), rabbit polyclonal anti-PARP (1:500) (Santa Cruz Biotechnology, CA), mouse monoclonal anti-raf-1 (1:200) (Abcam, MA), rabbit polyclonal anti-p38 ( Santa Cruz Biotechnology, CA), rabbit polyclonal anti-phospho-p38 (1:250) (Abcam, MA), mouse monoclonal anti-c-Kit (1:200) (Santa Cruz Biotechnology, CA), rabbit polyclonal VEGF (1:500) (Upstate Millipore, MA).
Transfections
Transfections were performed by nuclear transfection using the Nucleofector system solution V program T28 (Amaxa Biosystems, Koln, Germany) and 100 nM of pre-miR-193b or control per-miR (Ambion, Austin, TX). Transfected cells were then re-suspended in culture media containing 10% FBS for 72 hours before study. For silencing experiments the anti-miR-193b and anti-miR control probes were obtained from Exiqon (Vedbaek, Denmark) and used at final concentration of 100 nM.
Luciferase assay
The intact recognition sequence between miR-193b and the 3'-UTR of Mcl-1 was cloned downstream of the firefly luciferase gene as follows. Total cDNA was obtained following reverse transcriptase using random primers. The following primers Mcl-1-3'UTR-F:5’-GGACTAGTTTTGGGAAGCTATGGAGGAG-3 ’ and Mcl-1-3’UTR-R:5’-TCCCCGCGGAGAGAGGAAAAGCTTCCCTTG-3’ were used for amplification of the 3'-UTR of Mcl-1. The product was then digested with SpeI and SacII (New England Biolabs Ipswich, MA) and cloned into a pGL3 Control Vector (Promega, Madison, WI) to generate the Mcl-1-WT reporter construct. A reporter construct with a deletion in the putative recognition sequence for miR-193b was also constructed (Mcl-1-MUT) by using the QuikChange site-directed mutagenesis kit (Stratagene), according to the manufacturer's instructions. Primers used for mutagenesis were as follows: Mcl-1-MUTF: 5 ’-CCTTGTTGAGAACAGGAAAGGCCAGGCAAGTC-3’Mcl-1 MUTR 5’-GACTTGCCTGGCCTTTCCTGTTCTCAACAAGG-3’. All constructs were verified by sequencing. Cells were co-transfected with 1 μg Mcl-1-WT or Mcl-1-MUT construct and 0.1 μg pRL-TK Renilla luciferase expression construct followed by a precursor miR-193b or control precursor at 100 nM final concentration using Lipofectamine (Invitrogen, Carlsabad, CA,). Correspondingly luciferase assays were performed 24 hours after transfection using the Dual Glo Assay system (Promega, Madison, WI) according to manufacturer's protocol in a multiwell plate luminometer (Veritas, Turner Biosystems, Sunnyvale CA).
miRNA locked nucleic acid (LNA) in situ hybridization
miR-193b detection was performed on a custom HCC tissue array by in situ hybridization (ISH) as previously described (21). The negative controls included omission of the probe and the use of a scrambled LNA probe. After in situ hybridization for miR-193b, the slides were analyzed for immunohistochemistry using the antibody against hepatitis C NS4 protein (dilution 1:25) (Chemicon Int. Temucula, CA). For immunohistochemistry, we used the Ultrasensitive Universal Fast Red system from Ventana Medical Systems (Tucson, AZ) after digestion in Protease 1 for four minutes. After co-localization, the data was analyzed with the Nuance software (Cambridge Research Institute, Woburn, MA) which converts the RGB-based signal to a fluorescent-based signal and, thus, allows concomitant analysis of hepatitis C and miR-193b in situ results.
Reagents
Sorafenib was obtained from LC Laboratories (Woburn, MA) and diluted in dimethyl sulfoxide (DMSO). All reagents obtained were of the highest purity available.
Statistical analysis
Results are expressed as mean ± standard error (SEM), unless indicated otherwise. Comparisons between groups were performed using the two-tailed Student's t test. Significance was accepted when p was less than 0.05.
RESULTS
HCV viral proteins alter chemosensitivity in malignant hepatocytes
To explore the effect of HCV proteins on chemosensitivity, we used HepG2 cells stably transfected to express the full length genome of HCV (Hep-394), or control empty vector (Hep-SWX). Sorafenib induced cytotoxicity in a concentration-dependent manner (Figure 1). The IC50 for sorafenib was 3.54 ± 0.89 μM in control Hep-SWX cells and was reduced by >50% to 1.07 ± 0.15 μM in Hep-394 cells (p=0.02). These findings indicated that the constitutive expression of viral proteins can modulate cytotoxicity in HepG2 cells.
Figure 1.
A: Hep-G2 cells, stably transfected with full length HCV (Hep-394) or empty vector (Hep-SWX), were plated in 96-well plates and incubated with varying concentrations of sorafenib. Cell viability was assessed by the CellTiter 96 AQueous assay kit after 72 hours of exposure to drug or diluent control. IC50 values were calculated after curve-fitting using the XLfit Software. Error bars indicate SEM (n=6). B: Cells were plated in 35-mm dishes and stained with DAPI at the indicated time points. Cells with morphological changes of apoptosis were counted using a fluorescence microscope and the number of apoptotic cells expressed as a percentage of the total cells. Error bars indicate SEM. * p = 0.03 (n=3). C: Hep-SWX and Hep-394 were incubated in serum free medium and caspase 3/7 activation was assessed after 72 hours using a luminometric assay. Error bars indicate SEM. * p = 0.003 (n=3). D: Cell lysates from Hep-SWX and Hep-394 cells were obtained and evaluated for the expression of PARP by western blotting. Representative blots are shown along with the average and SEM of the expression of cleaved PARP in Hep-394 cells relative to the expression in Hep-SWX from three separate experiments. p: 0.002. E: Hep-SWX and Hep-394 cells were incubated in serum-free medium with sorafenib (2μM) or DMSO for 72 hours and the apoptotic cells quantitated by TUNEL assay and flow cytometry. Bars represent mean and SEM of 4 independent experiments. * p< 0.05 relative to Hep-SWX for each condition. F: The expression of Mcl-1 was analyzed by immunoblot analysis. Representative blots are shown along with quantitative data representing the average and SEM from three separate experiments. The relative expression of Mcl-1 to actin is expressed relative to that in Hep-SWX cells. p < 0.001.
Expression of HCV proteins can modulate apoptosis
Induction of apoptosis is a mechanism of cytotoxicity induced by sorafenib. To examine the effect of HCV protein expression on apoptosis, we examined morphological and biochemical changes of apoptosis in HCC cells. The number of cells undergoing apoptosis as well as the expression of caspase 3/7 and PARP cleavage were increased under basal conditions in Hep-394 compared to Hep-SWX cells (Figure 1). Thus, expression of viral proteins can modulate the basal level of apoptosis and thereby potentially modulate responses to therapeutic agents. Indeed, an increased apoptotic rate was observed in Hep-394 cells compared to Hep-SWX in response to sorafenib (Figure 1). Cell cycle analysis by flow cytometry in cells treated with sorafenib or diluent control showed a greater reduction of cells in phase S induced by sorafenib in Hep-394 compared to Hep-SWX (supplementary Figure 1).
Sorafenib has been shown to induce apoptosis via a down-regulation of the anti-apoptotic protein Mcl-1 in both solid and hematological malignancies (22,23). The role of Mcl-1 as an important determinant of response to chemotherapy has been studied in many other cancers (24). Mcl-1 expression was significantly decreased in Hep-394 cells compared to Hep-SWX controls (Figure 1) indicating that viral proteins can modulate cellular expression of this protein. Sorafenib is a multi-kinase inhibitor that can target several pathways. Therefore, we assessed the effect of HCV on expression of several cellular targets in Hep-SWX and Hep-394 cells, as well as in Huh-7 cells stably infected with HCV. Mcl-1 was consistently reduced in both HCV expressing cell lines, while raf-1, c-kit and VEGF were not altered by HCV expression (supplementary Figure 2). These data suggest a role for alterations in Mcl-1 in mediating the altered sensitivity to sorafenib in HCV expressing cells. Indeed, Mcl-1 expression was reduced by sorafenib and was decreased in Hep-394 cells compared to Hep-SWX control cells after treatment with sorafenib (supplementary figure 2).
HCV viral proteins alter the expression of selected miRNAs
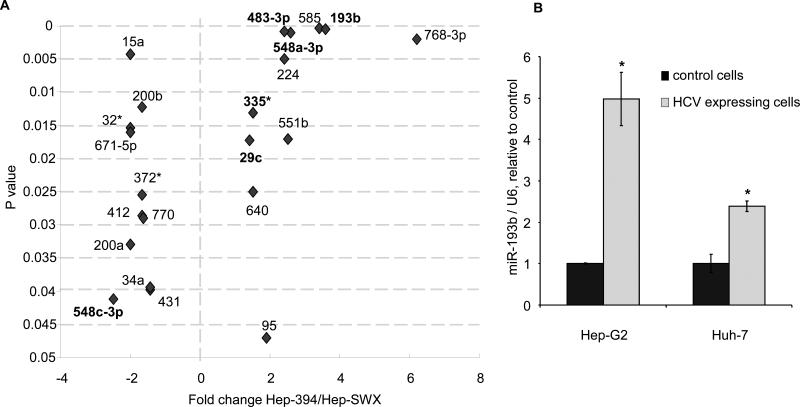
We postulated that the differential sensitivity to sorafenib in Hep-394 cells arose as a result of alterations in miRNA expression induced by HCV proteins. To test this hypothesis, we first profiled miRNA expression in Hep-394 and Hep-SWX cells using a custom microarray. Our analysis identified seventy-five miRNAs that were aberrantly expressed in Hep-394 cells compared to Hep-SWX cells, and thereby comprised a specific miRNA signature associated with HCV proteins. Of these, twenty-nine miRNAs were reduced in expression with a ratio ranging between 0.2 and 0.7 whereas forty-six miRNAs were increased in expression in Hep-394 with ratio ranging from 1.4 to 12.9. Of the latter, the expression of 23 miRNAs was more than 2-fold changed in Hep-394 cells compared to Hep-SWX controls (Table 1). In order to identify miRNA that may be associated with the cancer phenotype, we also profiled miRNA expression in non-malignant human hepatocytes and compared miRNA expression to those in Hep-SWX cells. Of the 75 miRNAs previously associated with HCV, we identified 22 miRNAs whose expression was not changed significantly in Hep-SWX HCC cells compared to normal human hepatocytes (Figure 2). This group thus includes miRNAs that are modulated by the expression of viral proteins, but are not altered in malignant cells. Amongst these, the expression of three miRNAs, miR-768-3p, miR-193b and miR-585 was altered by greater than three-fold by HCV viral protein expression.
Table 1. Differentially expressed microRNAs in Hep-394 vs Hep-SWX HCC cells.
miRNA that were altered in expression by >2-fold, with a p value < 0.05 are listed.
| microRNA |
Relative expression Hep-SWX / Hep-394 |
p-value |
microRNA |
Relative expression Hep-SWX / Hep-394 |
p-value |
|
|---|---|---|---|---|---|---|
| miR-130a | 12.98 | <0.0001 | miR-206 | 0.2 | 0.01 | |
| miR-99b | 7.88 | <0.0001 | miR-627 | 0.31 | 0.002 | |
| miR181b | 6.4 | 0.0006 | miR-196a | 0.37 | 0.001 | |
| miR-768-3p | 6.2 | 0.002 | miR-192 | 0.43 | 0.0006 | |
| miR-565 | 5.18 | <0.0001 | miR-570 | 0.47 | 0.01 | |
| miR-126* | 3.64 | <0.0001 | miR-657 | 0.47 | 0.001 | |
| miR-146a | 3.64 | 0.002 | miR-668 | 0.48 | 0.03 | |
| miR-193b | 3.62 | 0.0005 | miR-548c-3p | 0.48 | 0.04 | |
| miR-585 | 3.4 | 0.0004 | miR-425* | 0.49 | 0.04 | |
| miR-130b | 3.3 | 0.0005 | miR-345 | 0.5 | 0.008 | |
| miR-100 | 3.12 | 0.002 | ||||
| miR-146b-5p | 3.03 | 0.005 | ||||
| miR-181a | 2.9 | 0.002 | ||||
| miR-148b | 2.69 | 0.002 | ||||
| miR-548a-3p | 2.61 | 0.001 | ||||
| miR-551b | 2.55 | 0.01 | ||||
| miR-224 | 2.43 | 0.005 | ||||
| miR-483-3p | 2.4 | 0.0009 | ||||
| miR-106b | 2.31 | 0.005 | ||||
| miR-136 | 2.25 | 0.005 | ||||
| miR-660 | 2.19 | 0.0001 | ||||
| miR-135a | 2.17 | 0.0002 | ||||
| miR-663 | 2.14 | 0.004 |
Figure 2.
A: The ratio of expression of miRNAs in Hep-394 cells relative to Hep-SWX cells is shown, plotted against their p-values. Only those miRNAs that were not differentially expressed in non-malignant human hepatocytes compared to malignant Hep-SWX HCC cells are shown. miR-193b, miR-768-3p and miR-585 were the most significant up-regulated miRNAs in HCV-expressing cells compared to controls. miRNAs that are predicted to target Mcl-1 by at least one target prediction database are shown in bold. B: microRNA expression was evaluated by real time PCR. The expression of miR-193b was normalized to that of U6 RNA. Error bars indicate SEM. * p < 0.001 (n=3) relative to controls.
miRNA modulation of Mcl-1 in HepG2 cells
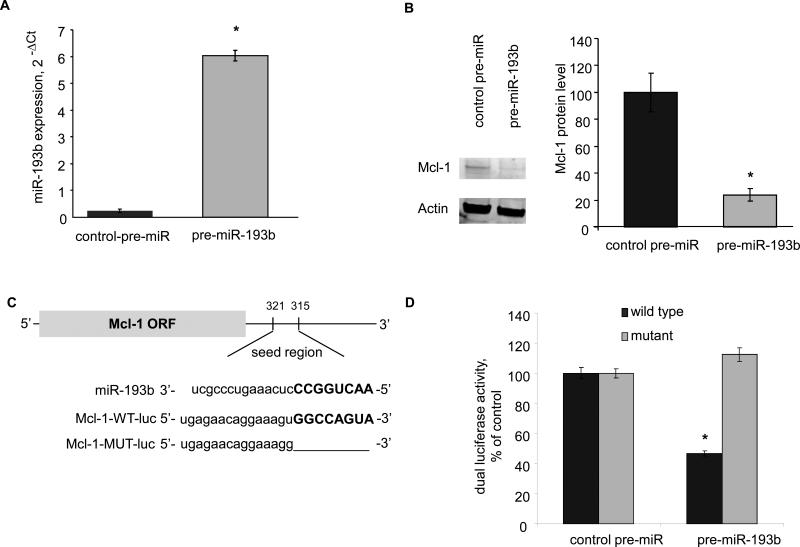
We next investigated the involvement of miRNAs in modulation of Mcl-1 expression by HCV proteins. The HCV associated miRNAs (Figure 2) were queried against several miRNA target prediction databases (Miranda, PicTar, Targetscan, miRbase) (25-28) to identify those with the potential to target Mcl-1. Mir-193b and miR-483-3p were predicted by more than one database. We focused on miR-193b given the high fold change and significance (Figure 2). First, we verified the expression of miR-193b by quantitative real-time PCR. The expression of miR-193b was increased by 5.0 ± 0.6 fold, in Hep-394 cells compared to control Hep-SWX cells and by 2.7 fold in HCV-infected Huh-7 cells compared to non infected controls (Figure 2). To further investigate the role of this miRNA, we transfected Hep-SWX cells with 100 nM of precursor-miR-193b or control-precursor-miRNA constructs. The efficacy of these constructs to modulate their target miRNA expression was verified using real-time PCR after 72 hours (Figure 3). We then assessed Mcl-1 expression in transfected cells by semi-quantitative immunoblot analysis. Mcl-1 protein was reduced in cells transfected with pre-miR-193b (Figure 3). To study whether the effect on Mcl-1 protein expression was the result of a direct interaction between miR-193b and the 3’UTR of Mcl-1, we used a reporter construct based assay in which the 3’UTR of Mcl-1 was cloned downstream of the luciferase gene. The miR-193b recognition site in the 3’UTR of Mcl-1 is illustrated in Figure 3. Hep-SWX cells were transfected with the luciferase reporter construct (Mcl-1-WT-luc), the renilla as loading control and the precursor to miR-193b or control pre-miRNA. Luciferase activity was reduced by 53.0 ± 1.8 % in cells transfected with pre-miR-193b compared to controls. As confirmation of these data, no changes were observed in the luciferase activity when we used constructs with deletions in the putative recognition site (Mcl-1-MUT-luc) that prevented miR-193b from interacting with the 3'UTR of Mcl-1. These data indicate that HCV proteins can increase the expression of miR-193b in HepG2 cells, and that an increase in miR-193b can alter Mcl-1 expression in HepG2 cells.
Figure 3.
A: Hep-SWX cells were transfected with control-pre-miR and pre-miR-193b at 100 nM. After 72 hours, the expression of miR-193b was quantitated by real time PCR. Error bars indicate SEM.* p < 0.001 (n=3). B: Immunoblot analysis was performed in transfected Hep-SWX cells. Representative blots are shown along with quantitative data. Error bars indicate SEM (n=3). The relative expression of Mcl-1 to actin is expressed as a percentage of that in Hep-SWX cells. * p < 0.001. C: The location of target sites of miR-193b in the Mcl-1 3'-UTR are shown. The sequence of the mutated Mcl-1 3’-UTR construct with mutations to delete the putative miR-193b binding site is also displayed. D: Hep-SWX cells were transfected with the Renilla luciferase expression construct pRL-tk, the luciferase construct Mcl-1-WT-luc or Mcl-1-MUT-luc (in which the Mcl-1 target site was deleted) and miR-193b or control precursor. The data represent mean and SEM from 3 determinations from four independent transfections. * p < 0.001 compared to the respective controls.
miR-193b alters sensitivity to chemotherapy and induces apoptosis
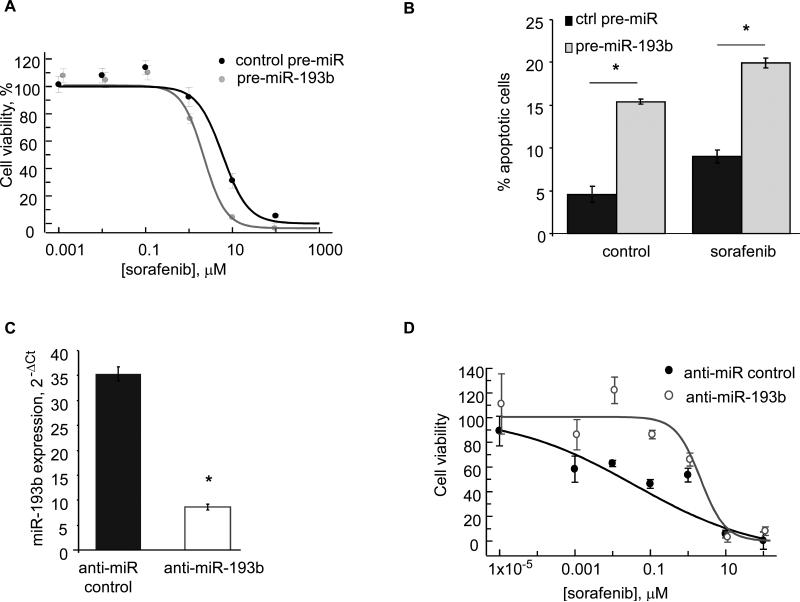
Alterations in Mcl-1 can modulate sensitivity to sorafenib. To evaluate the impact of alterations in miR-193b on sensitivity to sorafenib, Hep-SWX cells and Huh-7 cells were transfected with pre-miR-193b or control-pre-miRNA and the IC50 to sorafenib was determined after 72 hours. In Hep-SWX cells transfected with precursors to miR-193b, the IC50 to sorafenib was decreased compared to cells transfected with control precursors (2.1 ± 0.1 vs 5.6 ± 1.0 μM) (Figure 4). Notably, IC50 to sorafenib was reduced by 28.0 ± 1.7 % (p: 0.0001) in Huh-7 cells transfected with pre-miR-193b (Supplementary Figure 3). Thus, these observations were consistent with our previously noted effects of this miRNA on Mcl-1 expression. To verify whether pre-miR-193b could sensitize HepG2 cells to sorafenib through the enhancement of apoptosis we measured apoptosis in Hep-SWX cells transfected with either control precursor-miR or pre-miR-193b. Forty-eight hours after the transfection, cells were treated with 2 μM sorafenib for 72 hours. Transfection with premiR-193b sensitized cells to sorafenib-induced apoptosis (Figure 4). Next, we studied whether the inhibition of miR-193b in Hep-394 cells could alter the sensitivity of these cells to sorafenib. A reduction of 4-fold in miR-193b expression was observed in Hep-394 cells transfected with anti-miR-193b compared to controls. Consistently with our previous data the IC50 to sorafenib was increased from 0.4 to 1.5 μM (p: 0.01) (Figure 4). Together, these data suggest that pre-miR-193b might modulate sensitivity of HepG2 cells to sorafenib by altering cellular apoptosis.
Figure 4.
A: Hep-SWX cells were transfected with control-pre-miR and pre-miR-193b at 100 nM and plated in 96-well plates. After 72 hrs, sorafenib was added at the indicated concentrations. Error bars indicate SEM. B: Hep-SWX cells, transfected with 100 nM control-precursor-miR or pre-miR-193b were incubated with 2 μM sorafenib or DMSO and the number of cells undergoing apoptosis was counted after 72 hours using fluorescence microscopy after staining with DAPI. Error bars indicate SEM. * p < 0.01 (n=3). C: Hep-394 cells were transfected with anti-miR control or anti-miR-193b at 100 nM. After 72 hours RNA was collected and real time PCR was performed to assess the expression of miR-193b. Bars represent mean and SEM of 4 replicates. * p = < 0.001. B: Hep-394 cells were transfected with anti-miR control or anti-miR-193b and cell viability assessed after 72 hours. Error bars indicate SEM. Transfection with anti-miR-193b reduced the sensitivity of Hep-394 cells to sorafenib.
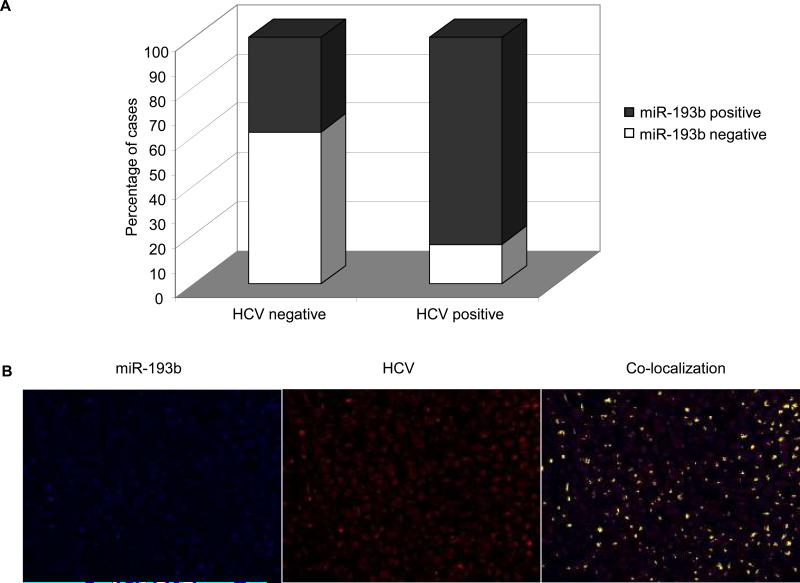
miR-193b expression is increased in HCV positive HCC tissues
We next analyzed the expression of miR-193b by in situ hybridization in a series of 57 HCC tissues. Mir-193b was expressed in 36 samples. Next, we assessed the expression of HCV by immunohistochemistry. Thirty one samples were positive for HCV. miR-193b and HCV were both positive in 46% of HCC, while they were both negative in 28% of cases. More interestingly, HCV often co-localized with miR-193b in the same cells as shown in figure 5. These data are in line with previous findings showing an increase of miR-193 in hepatitis infected cirrhotic liver compared to non infected-non cirrhotic liver (29). Furthermore, analysis of the raw data by Jiang et al (29) showed an increased mean miR-193 expression in HCV positive HCC samples compared to HCC of other etiologies (4.2 ± 0.4 vs 2.8 ± 0.4).
Figure 5.
Paraffin-embedded, formalin-fixed HCC tissues were incubated with LNA-anti-miR-193b and antibody against HCV. A: The proportion of cases of HCC positive for miR-193b and HCV expression is depicted in the columns. B: Picture of a representative case was taken with the Nuance system that converts the RGB-based signal to a fluorescent-based signal. Blue indicates miR-193b, red indicates HCV and yellow indicate overlapping (co-localization) of blue and red.
DISCUSSION
Pharmacological treatment options for HCC continue to evolve and therapeutic agents which have some benefit have recently become available or are being developed. Selection of treatment for HCC, however, has not been based on etiological considerations but rather on tumoral factors such as size, number and extension, as well as patient factors such as performance status and underlying liver disease (30). However, HCC is a heterogeneous cancer and can arise as a result of causes as diverse as environmental toxins, chronic infections, or metabolic derangements within the liver (4). Genomic expression profiling studies in HCC have identified varying gene expression profiles for HCC associated with HBV or HCV infection (31-33) thereby suggesting that hepatocarcinogenesis may reflect distinct molecular mechanisms based on the underlying etiological agent. Such changes may be expected to affect a broad range of cellular effects within host cells.
The relationship between HCV and miRNA expression is of considerable interest given that miRNAs can target many host cell genes. A distinct group of miRNAs that are associated with HCV was identified in our studies. Of these, miR-193b was one of the most significantly altered in expression. Randall et al. studied HCV-dependent modulation of miRNA expression and showed an up-regulation for miR-322, miR-197, miR-532-5p and miR-374 in HCV-expressing Huh-7 cells. miR-21, miR-130 and miR-122 were unaffected by HCV (13). miR-122 is highly prevalent in the liver, and is down-regulated in HCC. Consistently with Randall et al, we also did not notice any effect of viral protein expression on miR-122 expression. The differences in miRNA expression between their studies and our observations may be due to differences in cell lines or in miRNA assays. By a cloning strategy, Tuschl and colleagues identified an increased frequency of several miRNAs such as miR130a, 181b and 26a associated with the expression of HCV in Huh-7 cells. Other miRNAs, such as miR-16, miR-192 and miR-196b were reduced similar to our findings (34). Studies of miRNA expression in liver tissues of HCV-infected patients by Varnholdt et al. showed increased expression of miR-122, miR-100, miR-10a, miR198 and miR-145 in HCC tissues when compared to normal adjacent tissues, suggesting that the underlying HCV infection can modulate the expression of miRNAs in cancer (35). However, it is not possible to speculate on any direct HCV-induced effects in these studies as comparisons with HCV negative HCCs were not reported. The mechanism by which HCV proteins can modulate the expression of miRNAs is also of interest. HCV core protein can suppress the activity of Dicer, the enzyme involved in the procession of miRNAs and thus alter the production of mature miRNAs (36). The non structural NS5B protein, as a viral RNA-dependent RNA polymerase, could use miRNAs as primers to synthesize dsRNA and then amplify the process of RNA interference (37).
Anti-cancer drugs exert their effect, at least in part, by triggering apoptosis (38). In vitro studies showed that HCV proteins can cause massive apoptosis in HCC cells by activation of the mitochondrial pathways of apoptosis and sensitization to TRAIL or Fas ligand and can inhibit DNA replication in normal hepatocytes (32,39). Our studies show a propensity for HCV viral protein expressing cells to undergo apoptosis and alter sensitivity to therapeutic agents such as sorafenib. Thus, targeting the modulation of apoptosis by HCV is a reasonable approach to enhance therapeutic drug sensitivity in HCC. Whether the same apoptotic activation occurs in non transformed hepatocytes is less clear. Moreover, immune mechanisms are key determinants of viral clearance in HCV infection as variation in genes involved in the immune response can contribute to the ability to clear the virus (40).
The relationship of miRNAs to chemotherapy responses is likely to be complex. For example, we have shown that gemcitabine can modulate the expression of miRNAs (17). Indeed miRNA expression has been shown to correlate with chemo-sensitivity (41). Interferon can exert anti-HCV effects by inducing miRNAs that have sequence predicted targets within the HCV genomic RNA (42). Thus, further study of the miRNA responses to chemotherapeutic agents is necessary. Identifying miRNAs that are modulated by chemotherapeutic agents and that may have additional benefits on viral replication is likely to be helpful in the treatment of HCV associated HCC.
Several lines of evidence indicate that Mcl-1 is an important mediator of chemo-sensitivity in HCC. Mcl-1 is over-expressed in HCC cell lines and in 50% of human HCC tissues compared to normal adjacent tissues (43,44). Over-expression of Mcl-1 is associated with chemoresistance (43,45). Modulation of Mcl-1 has been implicated in HCC cell death by sorafenib but the mechanisms by which sorafenib modulates Mcl-1 has not been elucidated (22,23). Recent studies have shown that Mcl-1 expression can be modulated by miRNAs (46,47). Therapeutic approaches that manipulate miRNA expression are being developed, and use of miR-193b mimetics that alter the expression of miR-193b and target Mcl-1 will provide a novel and exciting approach to modulate chemo-sensitivity in HCC. Based on our studies the expression of miR-193b warrants further evaluation as a potentially useful biomarker of response to sorafenib in HCV-associated HCC.
The potential contribution of the presence of HCV to chemotherapy responses is likely to become of importance as new agents become available. The recent success with sorafenib has prompted interest in identifying new agents for HCC. Previously, Leung et al. noted a correlation between positive HCV serology and response to polychemotherapy regimens (48). In large multi-center phase II and phase-III trials that proved the activity of sorafenib in HCC, more than a half of the patients were infected with HCV (5,49). In a randomized controlled trial of sorafenib for hepatocellular cancer, patients were not stratified based on HCV status. However, a subgroup analysis in HCV-positive patients showed a median overall survival of 14 months in patients treated with sorafenib compared to 7.9 months in the placebo group, with similar trends in the time to progression (6). Interestingly the median overall survival in the placebo group was similar between the overall population and the HCV positive sub-population, suggesting that sorafenib might achieve better results specifically in the HCV sub-group. Based on our observations, future trials of chemotherapeutic agents in HCC should be stratified according to their hepatitis C status.
STATEMENT OF TRANSLATIONAL RELEVANCE.
In these studies we show that HCV proteins can modulate the expression of miRNA and alter the sensitivity of hepatocellular carcinoma (HCC) cells to sorafenib. HCC is highly resistant to chemotherapy. Although sorafenib has been shown to have some efficacy, responses remain poor. The findings of this study indicate that HCV expressing cells might respond better to sorafenib due to modulation of apoptosis by miRNA that are induced by viral proteins. These observations provide the rationale for stratification of HCC patients according to their HCV positivity in therapeutic trials. Moreover, modulation of miR-193b may be a reasonable strategy to improve therapeutic responses of HCC that warrants further evaluation.
Supplementary Material
Supplementary figure legends
Supplementary Figure 1. Hep-SWX and Hep-394 cells were incubated in serum-free medium with sorafenib (2uM) or DMSO for 72 hours and the DNA content analyzed by Tunel assay and flow cytometry after staining with propidium iodide. Bars represent mean and SEM of 4 independent experiments. * p< 0.05. In Hep-394 sorafenib decreased the number of cells in phase S. The serum starvation might be responsible for the alteration of phase G0/1 and G2.
Supplementary Figure 2. A: Hep-SWX and Hep-394 cells were incubated in serum-free medium with sorafenib (2uM) or DMSO for 72 hours and cell lysates obtained and evaluated for the expression of the indicated proteins. Representative blots are shown along with quantitative data showing the average and SEM of protein expression relative to the expression in Hep-SWX incubated with DMSO. * p < 0.05 (n=3). C-kit expression was not detectable in Hep-SWX and Hep-394 cells in line with previous findings showing lack of c-kit mRNA in HepG2 cells (50,51) B: Cell lysates from HCV infected and control Huh-7 cells were evaluated for the expression of the indicated proteins by western blotting analysis. Representative blots are shown along with quantitative data representing average and SEM of protein expression relative to controls. * p < 0.05 (n=3).
Supplementary Figure 3. Huh-7 cells were transfected with control-pre-miR and pre-miR-193b at 100 nM for 72 hours and plated in 96-well plates. A: miR-193b expression was quantitated by real time PCR. Error bars indicate SEM.* p<0.05 (n=3). B: Sorafenib was added at scalar concentrations and cell viability assessed by the CellTiter 96 AQueous assay kit after 72 hours. IC50 values were calculated after curve-fitting using the XLfit Software. Error bars indicate SEM. .* p<0.05 (n=7).
Acknowledgments
Financial support: American Italian Cancer Foundation (NV)
REFERENCES
- 1.Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45:529–38. doi: 10.1016/j.jhep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB, Marrero JA, Rudolph L, Reddy KR. Diagnosis and treatment of hepatocellular carcinoma. Gastroenterology. 2008;134:1752–63. doi: 10.1053/j.gastro.2008.02.090. [DOI] [PubMed] [Google Scholar]
- 3.Trinchet JC, Ganne-Carrie N, Nahon P, N'Kontchou G, Beaugrand M. Hepatocellular carcinoma in patients with hepatitis C virus-related chronic liver disease. World J Gastroenterol. 2007;13:2455–60. doi: 10.3748/wjg.v13.i17.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 5.Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–90. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 6.Bolondi L, Caspary W, Bennouna J, et al. Clinical benefit of sorafenib in hepatitis C patients with hepatocellular carcinoma (HCC): Subgroup analysis of the SHARP trial. Proc Am Soc Clin Oncol GastroIntestinal Cancer Symposium. 2008 (abstr # 129) [Google Scholar]
- 7.Shao RX, Hoshida Y, Otsuka M, et al. Hepatic gene expression profiles associated with fibrosis progression and hepatocarcinogenesis in hepatitis C patients. World J Gastroenterol. 2005;11:1995–9. doi: 10.3748/wjg.v11.i13.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sansonno D, Cornacchiulo V, Racanelli V, Dammacco F. In situ simultaneous detection of hepatitis C virus RNA and hepatitis C virus-related antigens in hepatocellular carcinoma. Cancer. 1997;80:22–33. [PubMed] [Google Scholar]
- 9.Uchida T, Shikata T, Tanaka E, Kiyosawa K. Immunoperoxidase staining of hepatitis C virus in formalin-fixed, paraffin-embedded needle liver biopsies. Virchows Arch. 1994;424:465–9. doi: 10.1007/BF00191430. [DOI] [PubMed] [Google Scholar]
- 10.Feitelson MA, Sun B, Satiroglu Tufan NL, et al. Genetic mechanisms of hepatocarcinogenesis. Oncogene. 2002;21:2593–604. doi: 10.1038/sj.onc.1205434. [DOI] [PubMed] [Google Scholar]
- 11.Aizaki H, Saito S, Ogino T, et al. Suppression of interferon-induced antiviral activity in cells expressing hepatitis C virus proteins. J Interferon Cytokine Res. 2000;20:1111–20. doi: 10.1089/107999000750053780. [DOI] [PubMed] [Google Scholar]
- 12.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–81. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 13.Randall G, Panis M, Cooper JD, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci U S A. 2007;104:12884–9. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murakami Y, Yasuda T, Saigo K, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–45. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 15.Meng F, Henson R, Wehbe-Janek H, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gramantieri L, Ferracin M, Fornari F, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–9. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 17.Meng F, Henson R, Lang M, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–29. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 18.Aizaki H, Harada T, Otsuka M, et al. Expression profiling of liver cell lines expressing entire or parts of hepatitis C virus open reading frame. Hepatology. 2002;36:1431–8. doi: 10.1053/jhep.2002.36937. [DOI] [PubMed] [Google Scholar]
- 19.Masaki T, Suzuki R, Murakami K, et al. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J Virol. 2008;82:7964–76. doi: 10.1128/JVI.00826-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park J, Tadlock L, Gores GJ, Patel T. Inhibition of interleukin 6-mediated mitogen-activated protein kinase activation attenuates growth of a cholangiocarcinoma cell line. Hepatology. 1999;30:1128–33. doi: 10.1002/hep.510300522. [DOI] [PubMed] [Google Scholar]
- 21.Nuovo GJ, Elton TS, Nana-Sinkam P, et al. A methodology for the combined in situ analyses of the precursor and mature forms of microRNAs and correlation with their putative targets. Nat Protoc. 2009;4:107–15. doi: 10.1038/nprot.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu C, Bruzek LM, Meng XW, et al. The role of Mcl-1 downregulation in the proapoptotic activity of the multikinase inhibitor BAY 43-9006. Oncogene. 2005;24:6861–9. doi: 10.1038/sj.onc.1208841. [DOI] [PubMed] [Google Scholar]
- 23.Rahmani M, Davis EM, Bauer C, Dent P, Grant S. Apoptosis induced by the kinase inhibitor BAY 43-9006 in human leukemia cells involves down-regulation of Mcl-1 through inhibition of translation. J Biol Chem. 2005;280:35217–27. doi: 10.1074/jbc.M506551200. [DOI] [PubMed] [Google Scholar]
- 24.Warr MR, Shore GC. Unique biology of Mcl-1: therapeutic opportunities in cancer. Curr Mol Med. 2008;8:138–47. doi: 10.2174/156652408783769580. [DOI] [PubMed] [Google Scholar]
- 25.Miranda KC, Huynh T, Tay Y, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–17. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 26.Krek A, Grun D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 27.Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006;342:129–38. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 28.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 29.Jiang J, Gusev Y, Aderca I, et al. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419–27. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Llovet JM, Di Bisceglie AM, Bruix J, et al. Design and endpoints of clinical trials in hepatocellular carcinoma. J Natl Cancer Inst. 2008;100:698–711. doi: 10.1093/jnci/djn134. [DOI] [PubMed] [Google Scholar]
- 31.Llovet JM, Chen Y, Wurmbach E, et al. A molecular signature to discriminate dysplastic nodules from early hepatocellular carcinoma in HCV cirrhosis. Gastroenterology. 2006;131:1758–67. doi: 10.1053/j.gastro.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 32.Tang W, Lazaro CA, Campbell JS, et al. Responses of nontransformed human hepatocytes to conditional expression of full-length hepatitis C virus open reading frame. Am J Pathol. 2007;171:1831–46. doi: 10.2353/ajpath.2007.070413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoon SY, Kim JM, Oh JH, et al. Gene expression profiling of human HBV- and/or HCV-associated hepatocellular carcinoma cells using expressed sequence tags. Int J Oncol. 2006;29:315–27. [PubMed] [Google Scholar]
- 34.Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varnholt H, Drebber U, Schulze F, et al. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223–32. doi: 10.1002/hep.22158. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Kato N, Jazag A, et al. Hepatitis C virus core protein is a potent inhibitor of RNA silencing-based antiviral response. Gastroenterology. 2006;130:883–92. doi: 10.1053/j.gastro.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 37.Lipardi C, Wei Q, Paterson BM. RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell. 2001;107:297–307. doi: 10.1016/s0092-8674(01)00537-2. [DOI] [PubMed] [Google Scholar]
- 38.Lei XY, Zhong M, Feng LF, et al. siRNA-mediated Bcl-2 and Bcl-xl gene silencing sensitizes human hepatoblastoma cells to chemotherapeutic drugs. Clin Exp Pharmacol Physiol. 2007;34:450–6. doi: 10.1111/j.1440-1681.2007.04593.x. [DOI] [PubMed] [Google Scholar]
- 39.Zhu H, Dong H, Eksioglu E, et al. Hepatitis C virus triggers apoptosis of a newly developed hepatoma cell line through antiviral defense system. Gastroenterology. 2007;133:1649–59. doi: 10.1053/j.gastro.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 40.Thomas DL, Thio CL, Martin MP, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blower PE, Chung JH, Verducci JS, et al. MicroRNAs modulate the chemosensitivity of tumor cells. Mol Cancer Ther. 2008;7:1–9. doi: 10.1158/1535-7163.MCT-07-0573. [DOI] [PubMed] [Google Scholar]
- 42.Pedersen IM, Cheng G, Wieland S, et al. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–22. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fleischer B, Schulze-Bergkamen H, Schuchmann M, et al. Mcl-1 is an anti-apoptotic factor for human hepatocellular carcinoma. Int J Oncol. 2006;28:25–32. [PubMed] [Google Scholar]
- 44.Sieghart W, Losert D, Strommer S, et al. Mcl-1 overexpression in hepatocellular carcinoma: a potential target for antisense therapy. J Hepatol. 2006;44:151–7. doi: 10.1016/j.jhep.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 45.Schulze-Bergkamen H, Fleischer B, Schuchmann M, et al. Suppression of Mcl-1 via RNA interference sensitizes human hepatocellular carcinoma cells towards apoptosis induction. BMC Cancer. 2006;6:232. doi: 10.1186/1471-2407-6-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–40. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Su H, Yang JR, Xu T, et al. MicroRNA-101, Down-regulated in Hepatocellular Carcinoma, Promotes Apoptosis and Suppresses Tumorigenicity. Cancer Res. 2009 doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- 48.Leung TW, Tang AM, Zee B, et al. Factors predicting response and survival in 149 patients with unresectable hepatocellular carcinoma treated by combination cisplatin, interferon-alpha, doxorubicin and 5-fluorouracil chemotherapy. Cancer. 2002;94:421–7. doi: 10.1002/cncr.10236. [DOI] [PubMed] [Google Scholar]
- 49.abou-Alfa GK, Schwartz L, Ricci S, et al. Phase II study of sorafenib in patients with advanced hepatocellular carcinoma. J Clin Oncol. 2006;24:4293–300. doi: 10.1200/JCO.2005.01.3441. [DOI] [PubMed] [Google Scholar]
- 50.Jabari S, Meissnitzer M, Quint K, et al. Cellular plasticity of trans- and dedifferentiation markers in human hepatoma cells in vitro and in vivo. Int J Oncol. 2009;35:69–80. doi: 10.3892/ijo_00000314. [DOI] [PubMed] [Google Scholar]
- 51.Mansuroglu T, Baumhoer D, Dudas J, et al. Expression of stem cell factor receptor c-kit in human nontumoral and tumoral hepatic cells. Eur J Gastroenterol Hepatol. 2009;21:1206–11. doi: 10.1097/MEG.0b013e328317f4ef. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure legends
Supplementary Figure 1. Hep-SWX and Hep-394 cells were incubated in serum-free medium with sorafenib (2uM) or DMSO for 72 hours and the DNA content analyzed by Tunel assay and flow cytometry after staining with propidium iodide. Bars represent mean and SEM of 4 independent experiments. * p< 0.05. In Hep-394 sorafenib decreased the number of cells in phase S. The serum starvation might be responsible for the alteration of phase G0/1 and G2.
Supplementary Figure 2. A: Hep-SWX and Hep-394 cells were incubated in serum-free medium with sorafenib (2uM) or DMSO for 72 hours and cell lysates obtained and evaluated for the expression of the indicated proteins. Representative blots are shown along with quantitative data showing the average and SEM of protein expression relative to the expression in Hep-SWX incubated with DMSO. * p < 0.05 (n=3). C-kit expression was not detectable in Hep-SWX and Hep-394 cells in line with previous findings showing lack of c-kit mRNA in HepG2 cells (50,51) B: Cell lysates from HCV infected and control Huh-7 cells were evaluated for the expression of the indicated proteins by western blotting analysis. Representative blots are shown along with quantitative data representing average and SEM of protein expression relative to controls. * p < 0.05 (n=3).
Supplementary Figure 3. Huh-7 cells were transfected with control-pre-miR and pre-miR-193b at 100 nM for 72 hours and plated in 96-well plates. A: miR-193b expression was quantitated by real time PCR. Error bars indicate SEM.* p<0.05 (n=3). B: Sorafenib was added at scalar concentrations and cell viability assessed by the CellTiter 96 AQueous assay kit after 72 hours. IC50 values were calculated after curve-fitting using the XLfit Software. Error bars indicate SEM. .* p<0.05 (n=7).