Abstract
The nicotinic acetylcholine receptor (nAChR) is a member of the important Cys-loop ligand-gated ion channel superfamily that modulates neuronal excitability. After responding to their agonists, their actions are terminated either by removal of ligand or by fast and slow desensitization, processes that play an important role in modulating the duration of conducting states and hence of integrated neuronal behavior. We monitored structural changes occurring during fast and slow desensitization in the transmembrane domain of the Torpedo nAChR using time–resolved photolabeling with the hydrophobic probe 3-(trifluoromethyl)-3-(m-iodophenyl) diazirine (TID). After channel opening, TID photolabels a residue on the δ-subunit’s M2–M3 loop and a cluster of four residues on δM1 and δM2, defining an open state pocket [Arevalo, E. et al. (2005) J. Biol. Chem. 280, 13631–13640]. We now find that photolabeling of this pocket persists during the transition to the fast desensitized state, decreasing only with the transition to the slow desensitized state. In contrast, photoincorporation in the channel lumen at the conserved 9′ leucines on the second transmembrane helix (M2–9′) decreased successively during the resting to open and open to fast desensitized state transitions, implying that the local conformation is different in each state, a conclusion consistent with the hypothesis that there are separate gates for channel opening and desensitization. Thus, although during fast desensitization there is a conformation change in the channel lumen at the level of M2–9′, there is none in the regions of the δ-subunit’s M2–M3 loop and the interior of its M1–M4 helix bundle until slow desensitization occurs.
The Cys–loop ligand-gated ion channel superfamily, which includes nicotinic acetylcholine receptors (nAChRs), 5-hydroxytryptamine type 3 (5-HT3) receptors, γ-aminobutyric acid type A (GABAA) receptors, and glycine receptors, has been studied intensively. These channels modulate synaptic and extrasynaptic neuronal excitability in response to their agonists and their actions are terminated either by removal of ligand or by desensitization, a process that plays an important role in modulating the duration of ligand–gated conducting states and hence of integrated neuronal behavior (1–3). A primary challenge is not only to understand how agonist binding in the extracellular ligand–binding domain (LBD) triggers a conformation change that opens an ion pore at a gate some 50 Å distant in the transmembrane domain (TMD), but also how the channel then closes and the receptor desensitizes. Understanding of channel opening has been aided by crystallographic structures of the LBD from homologous molluscan acetylcholine binding proteins (reviewed in (4)) and a cryoelectron microscopy structure of the Torpedo acetylcholine receptor in the absence of agonist in the resting, closed state (5). Promisingly, two homologous prokaryotic channels have been crystallized recently (6–8), one of which may be in the open state, but these channels do not desensitize and there is no structure at comparable resolution of the desensitized state.
In contrast to the progress made in studies of channel opening (9, 10, 11, 12, 13), the structural basis of desensitization is poorly understood. Auerbach and Akk have proposed a two–gate model (14), in which there are structurally distinct activation and desensitization gates, an hypothesis that is supported by other lines of evidences (15, 16). Others have suggested that each subunit has only one desensitized structure and that the difference between fast and slow desensitization is merely in the number of subunits in the desensitized state (17). The LBD–TMD interface is implicated in desensitization because mutations therein affect open-channel lifetime and rate of desensitization in parallel (18).
An underlying problem with studying desensitized states is that they are nonconducting, and electrophysiological experiments infer their existence indirectly from the kinetics of the disappearance and reappearance of conducting states. The muscle type nAChR occurs in such abundance in Torpedo electroplaques that the application of complementary kinetic techniques that are not dependent on ion conduction have been possible. For example, fluorescence techniques have been employed to probe agonist–induced conformational changes using agonist site and noncompetitive inhibitor site probes (19, 20). This receptor is particularly convenient for probing conformational changes because the rates of opening, fast desensitization and slow desensitization assure that the peak occurrence of each successive state is well separated in time from the others (21–23; for example, see Fig. 4b in the latter reference).
Time–resolved photolabeling has also been used to probe agonist–induced conformation changes (24–28). It has a resolution in the millisecond range, well suited to probing conformational changes as the receptor progresses from the open state through the desensitized states. We have used time–resolved photolabeling with the hydrophobic photolabel 3-(trifluoromethyl)-3-(m-iodophenyl) diazirine (TID) to systematically probe for structural changes on transient states of the nAChR. Within 1.5 ms of activation by rapid addition of agonist, a novel group of residues were robustly photolabeled (28). These residues, which were not detected in the equilibrium resting states, we termed activation–dependent to distinguish them from those channel lumen residues that are efficiently photolabeled in both the resting and open states. The former residues were located both at the extracellular end of the TMD of the δ-subunit between the transmembrane helices δM1 and δM2 and in the interface between the TMD and LBD on the extracellular δM2–M3 loop (Ile-288). The current study aimed to determine at what phase of desensitization this structural change is reversed and whether similar conformation changes could be detected on other nAChR subunits.
Experimental Procedures
Materials
nAChR-enriched membranes were isolated from Torpedo californica electric organs as described previously (29). The final membrane suspension was stored at −80°C in 38% sucrose, 0.02% NaN3 under argon. The specific activity of the binding sites was determined using an [3H]acetylcholine (PerkinElmer Life Sciences) binding assay and estimated at 0.5–2.1 nmol of Acetylcholine sites per milligram of protein as determined by micro-BCA assay (Pierce). 3-(Trifluoromethyl)3-m-([125I] iodophenyl)-diazirine ([125I]TID, 10 Ci/mmol) was obtained from GE Healthcare (Buckinghamshire, UK). Endoproteinase Lys-C (EndoLys-C), a lysine-specific protease, was obtained from Roche Applied Science, and Staphylococcus aureus endopeptidase Glu-C (V8 protease), a glutamate-specific protease, was from MP Biochemicals. TPCK-treated Trypsin, which is lysine and arginine specific, was from Worthington Biochemical Corporation (Freehold, NJ). All HPLC solvents were HPLC grade. Torpedo physiological saline (TPS) contains 250 mM NaCl, 5 mM KCl, 2 mM MgCl2, 5 mM sodium phosphate, pH 7.0, and 0.02% NaN3.
Time-resolved photolabeling of nAChR-enriched membranes
The method used was previously described (27, 28). Briefly, the loop of one of the two six-way sample valves was filled with 0.5 mL of nAChR-enriched membranes (4 mg protein/mL) equilibrated with 7 μM [125I]TID. The other six-way sample valve’s loop was filled with 0.5 mL of 20 mM carbamylcholine (carbachol). The contents of the loops were forced through the mixer by a pneumatic ram and after a designated incubation time, the mixed samples were expelled onto a rotating stainless steel disk (60 rpm) precooled in liquid nitrogen, where they were instantaneously (< 1 ms) frozen in a thin film. Incubation times were varied by changing the velocity of the ram and the lengths of the aging tube. The freeze-quenched samples were then irradiated (Black-Ray UV lamp model UVL-56) for 30 min at 366 nm at a distance of ~3 cm on the slowly rotating disk (3 rpm) in contact with liquid nitrogen. For the equilibrium condition (slow desensitized state), the nAChR-enriched membranes at 2 mg/mL were equilibrated with 3.5 μM [125I]TID and 10 mM of Carbachol and incubated for 1 h. Both loops of the two six-way valves were filled with 0.5 mL of this mixture and treated as previously described. To isolate nAChR subunits on a scale appropriate for the identification of photolabeled amino acids by Edman degradation (preparative photolabeling), six to eight samples per condition were obtained and pooled. An aliquot of each freeze-quenched sample was saved for protein concentration determination and for counting on a gamma counter.
SDS-PAGE
To evaluate the reproducibility of the subunit photolabeling in the multiple samples collected for each preparative photolabeling condition, an aliquot of each frozen sample was thawed in sample buffer and resolved by SDS-PAGE. The polypeptides from each sample were visualized by Coomassie blue stain and the labeled bands of interest were detected within the wet gel by phosphorimaging (2 h exposure at 25°C) using a Storm PhosphorImager (Amersham Biosciences). The samples from each labeling condition were then pooled, separated by SDS-PAGE and analyzed by phosphoimager as above. To accommodate the excessive volume resulting from pooling up to 8 freeze-quenching runs, each containing ~ 1 mL, special gels with deep wells (6–8 cm) and extended stacking gels (4 cm) were used. The resulting phosphoimages were used as a template to excise the subunits of interest. For the α-subunit, the excised bands were used for an “in gel” proteolytic digestion (see below). For the other subunits, the subunit bands were eluted passively for 3 days at room temperature in 12 mL of elution buffer (100 mM NH4HCO3, 0.1% SDS, 2.5 mM dithiothreitol, pH 8.4), filtered, concentrated in Vivaspin 15 mL concentrators (Vivascience, Inc., Edgewood, NY), and precipitated in 75% acetone (>12 h at −20°C). The precipitates were resuspended overnight in 200 μL of resuspension buffer (15 mM tris(hydroxymethyl)aminomethane, 0.5 mM EDTA, and 0.1% SDS, pH 8.1) at room temperature.
Proteolytic digestion
Labeled subunits were digested with specific endoproteases. For the α-subunits, the excised bands were digested “in gel” with the V8 protease as previously described (30). After the electrophoresis, the mapping gels were stained with GelCode Blue stain (Pierce) and proteolytic fragments of 20 kDa (αV8–20), beginning at αSer-173 and containing the M1, M2, and M3 transmembrane helices, were excised and recovered as described above. Both αV8–20 and an aliquot of the recovered δ-subunit were digested with EndoLys-C (0.75 U) in resuspension buffer for 2 weeks at room temperature. The αV8–20 digests were fractionated using reversed phase HPLC. The δ-subunit digests were fractionated by Tricine SDS-PAGE (28), and the 125I band of 10–14 kDa, which was identified by phosphorimaging, was excised and recovered as described above. Aliquots of the δ-subunit were also digested for three days with V8 protease (100% w/w), and the digests were fractionated by reversed phase HPLC. Aliquots of β-subunit, were digested with trypsin (w/w) by adding 4 volumes of digestion buffer (50 mM NH4HCO3, 0.5% Genapol C-100, pH 8.1) followed after 10 min by 0.1 volume of trypsin in 20 mM CaCl2.
Reversed-phase HPLC
The proteolytic fragments of the nAChR subunits were purified on an Agilent series 1100 HPLC with an inline degasser and column heater. The purifications were achieved at 40°C using a Brownlee Aquapore C-4 column (100 × 2.1 mm, 7 μm particle size) with a C-2 guard column. The aqueous phase (solvent A) was 0.08% trifluoroacetic acid, the organic phase (solvent B) was 60% acetonitrile, 40% 2-propanol, 0.05% trifluoroacetic acid, and the gradients are included in the HPLC plots as dashed lines. The elution of peptides was monitored by absorbance at 215 nm (Spectroflow 757, Kratos Analytical). The flow rates were 0.2 mL/min, and fractions of 0.5 mL were collected. All HPLC solvents were HPLC grade.
Sequence analysis
Most HPLC fractions of interest were pooled and drop-loaded onto Biobrene-treated glass fiber filters at 45°C. Fractions containing either αM4 or δM1 were absorbed onto a PVDF filter using the ProSorb absorption system (Applied Biosystems, Foster City, CA) following the manufacturer’s procedure. N-Terminal sequence analysis of isolated nAChR subunit fragments was performed using an Applied Biosystems Procise 492 protein sequencer modified such that 5/6 of each cycle were collected for gamma counting and the other 1/6 was used for amino-acid analysis. The pmol of phenylthiohydantoin (PTH)-derivatized amino acids in each cycle was determined by chromatographic peak heights. The initial amount (I0) and repetitive yield (R) for detected peptides were determined by a nonlinear least squares fit (Sigma Plot, Jandel Scientific) of the equation, f(x) = I0·Rx, where f(x) is the pmol of the amino acid in cycle x. Due to known problems with quantifying their PTH-derivatives, serines, histidines, tryptophans, and cysteines, although they were plotted, were omitted from the fit. The efficiency of the [125I]TID photoincorporation into a specific residue in cycle x was determined by the equation (cpmx − cpmx−1)/5· I0· Rx. For some samples, sequencing was interrupted and the material on the filter was treated with o-phthalaldehyde (OPA) as described (31, 32). OPA reacts with primary amines preferentially over secondary amines (i.e. proline), and may be used at any sequencing cycle to block Edman degradation of peptides not containing an N-terminal proline. The cpm detected in each cycle of Edman degradation was back-corrected for 125I decay to the date of labeling.
Although the output from the Edman analysis in cpm/pmol is quantitative, there are many variables that may introduce errors during the time–resolved photolabeling, digestion and purification steps. An analysis of errors in this and our previous time resolved work, excluding data with < 4 cpm/pmol, shows that the average standard deviation is 28 ± 16 (St. Dvn.) % of the mean cpm/pmol.
Results
Experimental strategy
We focused on conformation changes in three domains: the center of the pore around M2–9′; the region contralateral to the pore at the extracellular end of M2 (18′ & 22′), and the M2–M3 loop. We chose photolabeling times to coincide with peaks in the populations of the resting, open, fast desensitized and slow desensitized states based on evidence from rapid agonist–induced cation flux and fluorescent agonist binding in Torpedo acetylcholine receptor rich vesicles (22, 23, 33). Upon binding of agonist the resting state receptors convert to the open state in tens of microseconds (34). Passage to the fast desensitized state takes place with a time constant of ~150–300 ms and is complete by 1 s, while that to the slow desensitized state is complete in tens of seconds (23). Our experiments focused on changes occurring between pairs of states because at none of the four times points examined will all the receptors be in a single state. For example, in the absence of agonist the resting state is in equilibrium with some 15% of receptors that are desensitized (19). This desensitized state is poorly photolabeled by TID and its population does not change significantly in the first second after addition of agonist, so it may be ignored when considering changes in photolabeling with time. Furthermore, studies with a fluorescent agonist suggest that TID does not perturb the kinetics of desensitization significantly (35), and this is confirmed in electrophysiological studies (28).
We determined the relative change in photoincorporation of [125I]TID into nAChRs for three agonist–induced conformation changes: resting to open; open to fast desensitized, and fast desensitized to slow desensitized. The two conformations associated with each state transition were photolabeled on the same day under the same conditions, except for the time of incubation with agonist, and were subsequently processed in parallel. For the δ-subunit, we were interested in changes during all these transitions, whereas for the other subunits we concentrated on the first two because at the subunit level there is no change between the fast and slow desensitized states (28). Although conformations may coexist (see above), we use the terms open, fast desensitized and slow desensitized states to refer to receptors photolabeled after exposure to 10 mM carbachol for 10–15 ms, 1 s and 1 hour respectively. In all cases the membranes were pre-equilibrated with TID.
Photolabeling in the δM2 helix and δM2–δM3 loop
For each photolabeling condition, we isolated for sequence analysis the δ subunit fragment that begins at δMet-257, the N-terminus of δM2 (referred to as δM2–1′) and extends through δM3. The fragment was isolated from an EndoLys–C subunit digest by Tricine SDS-PAGE and reverse phase HPLC (28). The HPLC fractionations for the three state transitions are shown in Fig. S1A–C, where S denotes supplementary material throughout this manuscript, and the sequencing results are given in the three sections below. For each photolabeling experiment, the efficiency of [125I]TID photoincorporation at an individual position was quantified (in cpm/pmol) in order to compare labeling at other positions within the fragment from the same sample and from the second labeling condition performed in parallel.
The resting to open state transition
To confirm our previous data for this state transition (28) and provide a point of comparison, the peptide isolated by HPLC as shown in Fig. S1A and beginning at δMet-257 was subjected to 25 cycles of Edman degradation. The major photoincorporation in the pore region was at δM2–9′ Leu-265 (Fig. 1A) with minor labeling at δM2–13′ Val-269 and δM2–16′ Leu-272. In agreement with our previous work, the efficiency of [125I]TID incorporation at Leu-265 decreased approximately 2–fold between the resting and open states, from 115 to 68 cpm/pmol.
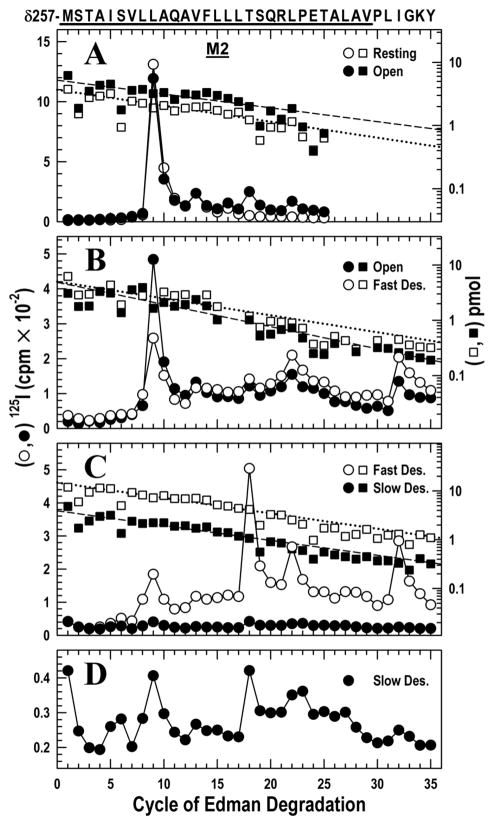
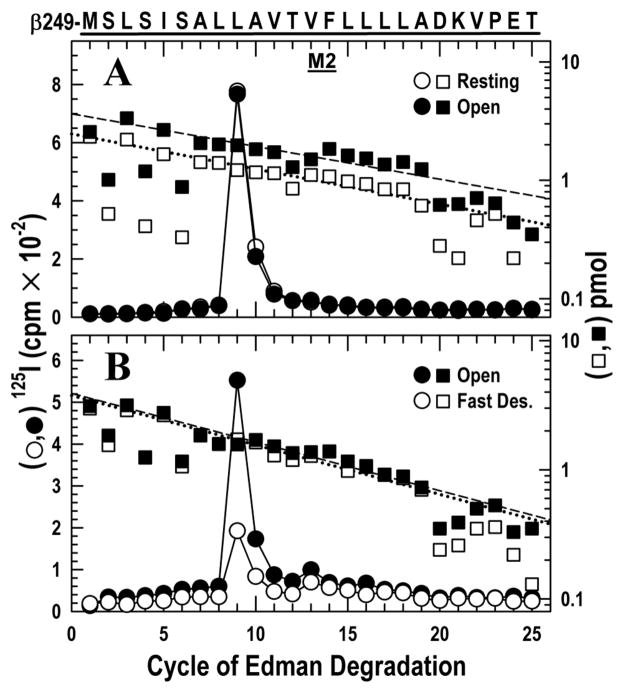
Figure 1. Photoincorporation of [125I]TID in the δM2 helix and δM2–M3 loop in the resting, open, fast desensitized and slow desensitized states of Torpedo nAChR.
nAChR-enriched membranes (4 mg/mL) were equilibrated with 7 mM [125I]TID and then rapidly mixed with an equal volume of buffer (resting), or agonist (Carbachol, 20 mM; 10 mM final concentration) for 15 ms (open state), 1 s (fast desensitized state) or ≥1 hour (slow desensitized state), then frozen in less than 1 ms, and finally photolabeled for 30 min (see Experimental Procedures). In each case, the fragment beginning at δMet-257 was isolated from EndoLys-C digests of δ subunits by SDS-PAGE and reversed phase HPLC as described in the Experimental Procedures (Fig. S1). The panels show 125I (○,●; left axis) and PTH-amino acids (□, ■; right axis) released during the sequencing of δ-subunit peptides. The sequence beginning at the N-terminus of δM2 at Met-257 is shown on the top axis with the M2 transmembrane helix highlighted with a dark bar.
A, nAChRs were photolabeled in the resting (○, □) and open (●, ■) states. For sequencing the resting and open state samples, 9,230 and 16,140 cpm of 125I were loaded onto filters respectively. The primary sequence began at δMet-257 (δM2–1′) (□, Io = 3.7 pmol, R = 94%; ■ Io = 5.2 pmol, R = 95%), with a secondary sequence beginning at δAsn-437 in both samples at ~0.3 pmol. Peaks of 125I release (○,●) were detected in the resting state at cycles 9, 13, 16 and in the open state at cycles 9, 13, 16, 18 and 22, corresponding respectively to residues δLeu-265, δVal-269, δLeu-272, δThr-274 and δLeu-278.
B, nAChRs were photolabeled in the open (●, ■) and the fast desensitized (○, □) states. For sequencing the open and fast desensitized state samples, 24,021 and 28,221 cpm of 125I were loaded onto filters respectively. The fragment beginning at δMet-257 was present (■: Io = 4.9 pmol, R = 91%; □: Io = 5.0 pmol, R = 93%) along with the fragment beginning at δAsn-437 at ~ 8 pmol in both samples, which contains the M4 helix beginning at cycle 20. Peaks of 125I release were observed in both samples at cycles 9, 13, 18, 22 and 32. Residues in M4 are labeled inefficiently by TID (δSer-457 at δMet-467 at < 1 cpm/pmol) because they are at the lipid interface (37) and they do not contribute to the peaks of 125I release observed here.
C, nAChRs were photolabeled in the fast (○,□) and slow desensitized (●, ■) states. For sequencing the fast desensitized and slow desensitized state samples, 15,450 and 2,370 cpm of 125I were loaded onto filters respectively. The primary sequence began at δMet-257 (□, Io = 15 pmol, R = 93%; ■, Io = 3.9 pmol, R = 93%), with the fragment beginning at δAsn-437 also present at ~ 2 pmol in each sample. Peaks of 125I release occurred in the same cycles as in B.
D, The data from C for the slow desensitized state are replotted on an expanded scale.
Upon transition to the open state, photoincorporation was observed into two additional residues located contralateral to the channel lumen (cycles 18 and 22). These residues, δM2–18′ Thr-274 and δM2–22′ Leu-278, were labeled with similar efficiency of 14 and 9 cpm/pmol, respectively, in the open state and not labeled in the resting state. This confirms our previous study (28).
The open to fast desensitized state transition
As above, the peptide beginning with δMet-257 was isolated (Fig. S1B) and then subjected to 35 cycles of Edman degradation (Fig. 1B). This state transition caused photoincorporation in the channel lumen to decrease 3–fold at δM2–9′ Leu-265, from 39 to 12 cpm/pmol.
In contrast, in the two residues contralateral to the channel lumen, the transition from the open to the fast desensitized state resulted in little change in photoincorporation: δM2–18′ Thr-274, 7 vs. 5, and δM2–22′ Leu-278, 11 vs. 11 cpm/pmol. The δM2–M3 loop δIle-288 (cycle 32) behaved similarly (65 vs. 48 cpm/pmol). The higher levels of photoincorporation in the δM2–M3 loop vs. δM2 was observed previously in the open state (28).
The fast to slow desensitized state transition
Following the procedures above, the peptide beginning with δMet-257 was isolated (Fig. S1C) and sequenced (Fig. 1C). In the two residues contralateral to the channel lumen and the one in the δM2–M3 loop, the transition from the fast to the slow desensitized state was accompanied by a marked decrease in photoincorporation: δM2–18′ Thr-274, 19 vs. 3; δM2–22′ Leu-278, 8 vs. 1, and δM2–M3 loop δIle-288, 24 vs. 2 cpm/pmol. Although the decrease in photoincorporation is nearly an order of magnitude, the pattern of photoincorporation in the slow desensitized state is seen to be similar to that in fast desensitized state when displayed with an expanded scale (Fig. 1D). Thus, the rapid structural changes that took place in these regions of the nAChR during channel opening and that remained present in the fast desensitized state are finally reversed when the slow desensitized state is attained tens of seconds later, but the pattern is distinct from that in the resting state.
Within the lumen of the ion channel (δM2–9′) photoincorporation was so low (2 vs. 1 cpm/pmol for the fast and slow desensitized state respectively) that we hesitate to draw a conclusion. The low release at cycle 9 in the fast desensitized state is likely anomalous (compare to Fig. 1B). Furthermore, release later in the same sequencing run at cycle 32 (δIle-288) was normal and is independently corroborated in Fig. 2C.
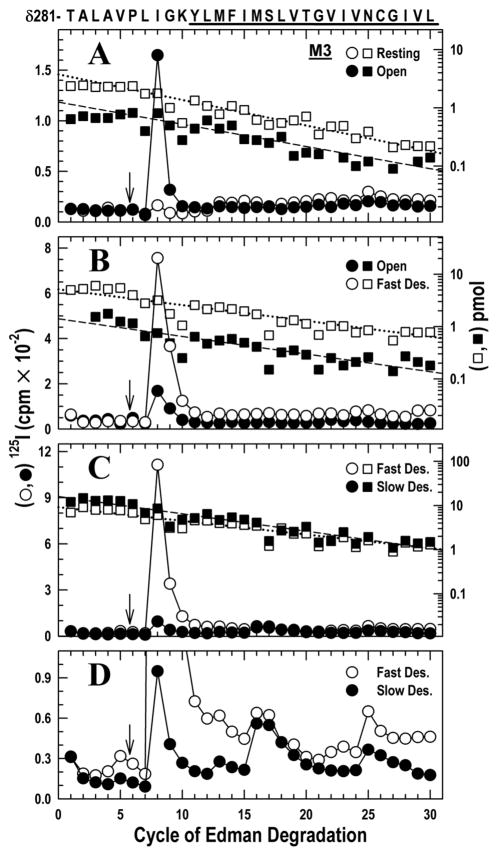
Figure 2. Photoincorporation of [125I]TID in the δM2–M3 loop and δM3 helix in the resting, open, fast and slow desensitized states of Torpedo nAChR.
Photolabeled nAChRs were from the experiments described in Fig. 1. The panels show 125I (○, ●; left axis) and PTH-amino acids (□, ■; right axis) released during the sequencing. The sequence of the δ-subunit fragment beginning at δThr-281 (δM2–25′), near the C-terminus of δM2 and continuing through the δM2–δM3 loop into δM3 (highlighted with a dark bar), is shown on the top axis. Aliquots of δ subunits were digested in solution with V8 protease. The digests were fractionated by reversed phase HPLC from which fractions containing the desired peptide were pooled (Fig. S2A–C) for sequencing. To restrict the sequencing to the fragment beginning at δThr-281, each sequencing filter was treated with OPA after 5 cycles of Edman degradation (i.e. when δPro-286 was the N-terminal residue) (↓) to block other peptides that do not contain a proline at that cycle.
A, nAChRs were photolabeled in the resting (○, □) and open (●, ■) states. For sequencing the resting and open state samples, 4,650 and 2,750 cpm of 125I were loaded onto filters respectively. The fragment beginning at δThr-281 was the only sequence remaining after the OPA treatment (□, Io = 3.8 pmol, R = 90 %; ■, Io = 1.3 pmol, R = 92 %). A prominent release of 125I was detected in cycle 8 (δIle-288) only in the open state sample. There were minor peaks of 125I release in cycles 13 and 25 (δMet-293 and δAsn-305).
B, nAChRs were photolabeled in the open (●, ■) and the fast desensitized (○, □) states. For sequencing the open and fast desensitized state samples, 6,880 and 24,263 cpm of 125I were loaded onto filters respectively. The fragment beginning at δThr-281 was the only sequence remaining after the OPA treatment (■, Io = 1.4, R = 93 %; □, Io = 5.2 pmol, R = 92 %). A prominent release of 125I was detected in cycle 8 (δIleu-288) in the resting and fast desensitized state, with minor release occurring at cycles 13, 22 and 25 (δMet-293, δVal-302 and δAsn-305).
C, nAChRs were photolabeled in the fast (○,□) and slow desensitized (●, ■) states. For sequencing the fast desensitized and slow desensitized state samples, 23,900 and 6,850 cpm of 125I were loaded onto filters respectively. The fragment beginning at δThr-281 was the only sequence remaining after the OPA treatment (□, Io = 9.1 pmol, R = 93%; ■, Io = 16 pmol, R = 91%). The prominent 125I releases in cycle 8 (δIle-288) in the fast desensitized state fell dramatically during slow desensitization.
D, Because the amount of peptide is higher in C than in A & B, the low photoincorporation in δM3 (cycles 13, 22 and 25; δMet-293, δVal-302 and δAsn-305) is clear when the data is plotted on an expanded axis.
Photolabeling in the δM2–δM3 loop and δM3 helix
To provide a more complete characterization of the state dependence of photolabeling in the δM2–δM3 loop and to characterize labeling within δM3, the δ subunit was digested with V8 protease, which cleaves at δGlu-280, and the digest was fractionated by reversed phase HPLC (36). Although the hydrophobic HPLC fractions with photoincorporated 125I (Fig. S2A) include other hydrophobic fragments containing δM1, δM2, and δM4, these can be N-terminal blocked by treatment with OPA, as described above, after the 5th cycle, leaving only the δThr-281 fragment, now starting at δPro-286, to be sequenced (31, 36). The advantage of this strategy is that δIle-288 is assayed as the third residue after the OPA treatment rather than as the 32nd residue of the sequencing run as in the samples digested with EndoLys-C (Fig. 1). The results are given in the three sections below.
The resting to open state transition
In the open state sample there was a single sharp 125I release in the 8th cycle (Fig. 2A), consistent with [125I]TID incorporation into δIle-288 on the δM2–M3 loop. The large magnitude of the photoincorporation contrasts with the very weak labeling in the resting state (49 vs. 1 cpm/pmol), consistent with previous observations (28).
The open to fast desensitized state transition
Sequence analysis (Fig. 2B) revealed once again a single peak of 125I release in the 8th cycle, corresponding to δIle-288, but this time it was evident in both the open and fast desensitized states. After correction for the difference in the quantity of peptide being sequenced, the efficiency of photolabeling was similar in both states, (38 vs. 53 cpm/pmol respectively). This suggests that the structural change in the δM2–M3 loop initiated by opening remains present when the channel is closed by fast desensitization, consistent with the findings in Fig. 1B.
The fast to slow desensitized state transition
Photoincorporation into δIle-288 decreased dramatically upon slow desensitization from 43 to 2 cpm/pmol (Fig. 2C), a low level similar to that in the resting state. Because a larger amount of peptide was sequenced in this run, the relatively inefficient labeling in cycles 13, 16, and 25 is evident when the data are replotted with an expanded scale (Fig. 2D). Photoincorporation into these residues in the fast and slow desensitized states respectively were: δMet-293 (0.1 vs. 0.4 cpm/pmol); δMet-296 (1 vs. 2 cpm/pmol), and δAsn-305 (4 vs. 2 cpm/pmol). The latter residue is also apparent in Fig 2A at comparable levels (6 vs. 5 cpm/pmol) for the resting and open states respectively. The lack of clear state–dependence is not unexpected because some of these residues were reported previously to be photolabeled by [125I]TID from the lipid interface (37).
Comparison of photolabeling of δM1 in the fast and slow desensitized states
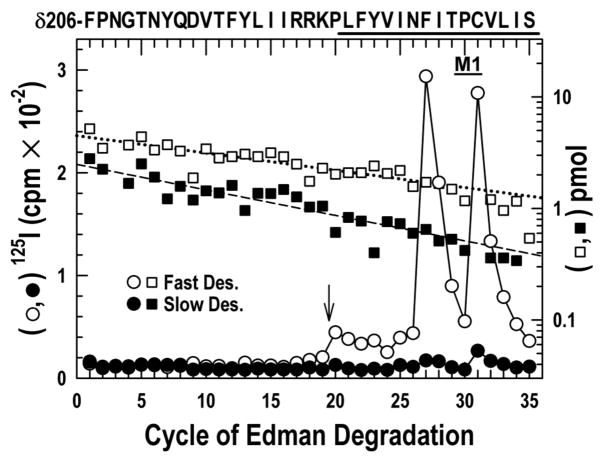
Our previous work established that two residues in the δM1 domain, δPhe-232 and δCys-236, were photolabeled in the open but not the resting state (28). To determine if these residues behave synchronously with δM2–18′ Thr-274, δM2–22′ Leu-278 and Ile-288 in δM2–M3 loop, we took advantage of the fact that the fragment beginning at δPhe-206 before δM1 can be isolated from the same HPLC fractionation (Fig. S1C) of the Endo-Lys-C digest of the δ-subunit that yielded δM2 (28). A single peptide beginning at δPhe-206 in the N-terminal domain was sequenced for 19 cycles without 125I release (Fig. 3). Sequencing was stopped before δPro-225, two residues before the beginning of δM1, for OPA treatment, followed by 16 additional cycles of Edman degradation. Strong release of 125I was observed in cycles 27 and 31 in the fast but not the slow desensitized state sample. The photoincorporation in the fast vs. the slow desensitized state respectively was: δPhe-232, 29 vs. 2, and δCys-236, 30 vs. 7 cpm/pmol. In a separate experiment (not shown), we confirmed that there is photoincorporation into δPhe-232 in the open state (24 cpm/pmol), as was observed previously (28).
Figure 3. Photoincorporation of [125I]TID in the δM1 helix in the fast desensitized and slow desensitized states of nAChR.
125I (○, ●) and PTH-amino acids (□, ■) released during sequencing of the fragment beginning at δPhe-206 and extending through δM1, which was isolated by the reversed phase HPLC fractionation of the EndoLys-C digests of the δ subunit described in Fig. 1 and Fig. S1 from nAChRs photolabeled in the fast desensitized (○, □) and slow desensitized (○, ■) states. For sequencing the fast desensitized and slow desensitized state samples, 7,600 and 840 cpm of 125I were loaded onto filters respectively. To confirm that the 125I release after cycle 20 was attributable to the δM1, both samples were treated with OPA after 19 cycles of Edman degradation (i.e. when δPro-225 was the N-terminal residue) (↓) to prevent further sequencing of any other peptides in the samples not containing an N-terminal proline at that cycle. Even before OPA treatment however, the only sequence detected began at δPhe-206 (□, Io = 4.5 pmol, R = 97 %;■, Io = 2.5 pmol, R = 95 %). Prominent labeling at cycles 27 and 31 was only seen in the fast desensitized state.
Comparison of photolabeling of αM2 in the resting and open states
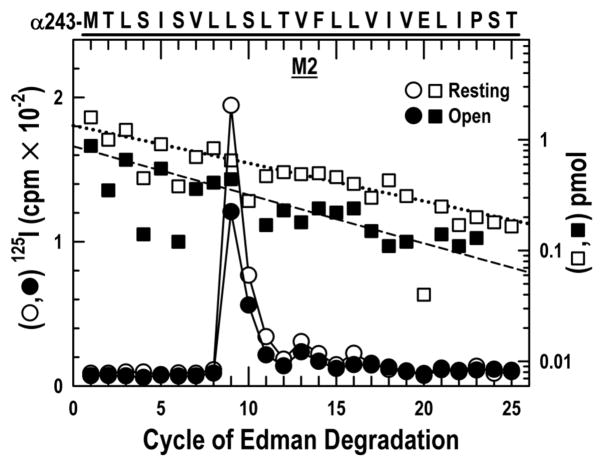
Next we asked whether the α-subunit contained residues equivalent to those in the δM2 that are only labeled after activation. We therefore isolated for sequence analysis a fragment beginning at the N-terminus of αM2 (αMet-243, αM2–1′) by HPLC fractionation of an EndoLys-C digest of an ~20kDa fragment beginning at αSer-173 that is produced when the α-subunit is digested with V8 protease (Fig. S3) (See Methods). Sequencing (Fig. 4) revealed a peptide beginning at αMet-243. Under both labeling conditions there was a single robust peak of 125I release at cycle 9 followed by two smaller ones at cycles 13 and 16, with photoincorporation efficiencies similar in the resting and the open state respectively: αM2–9′ Leu-251, 57 vs. 64; αM2–13′ Val-255, 5 vs. 8 and αM2–16′ Leu-258, 4 vs. 3 cpm/pmol. Thus, photoincorporation in the channel lumen of αM2 is not strongly affected by channel opening. There was no release of 125I at cycle 18 or 22 in either sample.
Figure 4. Photoincorporation of [125I]TID in the αM2 helix in the resting and open states.
125I (○, ●) and PTH-amino acids (□, ■) released while sequencing the fragment beginning at αMet-243 (αM2–1′), which was isolated from the α-subunits photolabeled in the experiment described in Fig. 1A by HPLC purification of an EndoLys-C digest (Fig. S3) of a 20 kDa fragment produced by digestion with V8 protease. For sequencing the resting and open state samples, 2,217 and 1,874 cpm of 125I were loaded onto filters respectively. Sequencing revealed a single peptide (resting, □, Io = 1.3 pmol, R = 92%; open state, ■, Io = 0.9 pmol, R = 90%). A major peak of 125I release was observed in cycle 9 (αLeu-251), with minor peaks at cycles 13 (αVal-255) and 16 (αLeu-258).
Comparison of photolabeling of βM2 in the resting and open states
To identify residues photolabeled by [125I]TID, β-subunits of nAChR were digested with trypsin and a band of ~10 kDa was isolated and purified by HPLC (Fig. S4A). A single peptide beginning with βMet-249 at the N terminus of βM2 was detected with a single peak of 125I release in cycle 9, corresponding to βM2–9′ Leu-257. Photoincorporation in this residue was 109 vs. 72 cpm/pmol for the resting and open states respectively (Fig. 5A). There was no release at cycle 18 and 22 in either sample.
Figure 5. Photoincorporation of [125I]TID in the βM2 helix in the resting, open and fast desensitized states.
125I (○, ●) and PTH-amino acids (□, ■) released while sequencing the fragment beginning at βMet-249 (βM2–1′), which was isolated from the β-subunits photolabeled in the experiments described in Figs. 1A & B by HPLC purification (Fig. S4) of an ~10 kDa fragment isolated by Tricine SDS-PAGE from trypsin digests.
A, nAChRs were photolabeled in the resting (○, □) and open (●, ■) states. For sequencing the resting and open state samples, 4252 and 4523 cpm of 125I were loaded onto filters respectively. Sequencing revealed a single peptide beginning at βMet-249 (□: Io = 2.5 pmol, R= 93%; ■: Io = 3.6 pmol, R= 94%). There was a major peak of 125I release in both states at cycle 9, corresponding to βLeu-257.
B, nAChRs were photolabeled in the open (●, ■) and the fast desensitized (○, □) states. For sequencing the open and fast desensitized state samples, 3166 and 1060 cpm of 125I were loaded onto filters respectively. Sequencing revealed a single peptide beginning at βMet-249 (■: Io = 3.9 pmol, R= 92%; □: Io = 3.8 pmol, R= 92%). The major peak of 125I release in both states was at cycle 9 (βLeu-257), with minor release at cycles 13 (βVal-261) and 16 (βLeu-264).
Comparison of photolabeling of βM2 in the open and fast desensitized states
Using the procedure in the above paragraph, 125I release was observed in cycles 9 and 13 corresponding to the labeling of the residues βM2–9′ Leu-257 and βM2–13′ Val-261 (Fig. 5B). Photoincorporation efficiencies for the open vs. the fast desensitized state respectively for these two residues were: 55 vs. 19, and 4 vs. 5 cpm/pmol. There was no release at cycle 18 and 22 in either sample.
Discussion
Location of the photolabeled residues
Using time–resolved photolabeling, we have followed structural changes as the nAChR passed from the open to the fast desensitized state and from that state to the slow desensitized state. Combined with our previous study on changes occurring during agonist–induced channel opening (28), the work covers a time span from 1.5 ms to many minutes after rapid addition of agonist. Our results are summarized in graphical form in Fig. 6. TID photolabeling reported on structural changes in three distinct domains of the Torpedo nAChR. The first domain is the pseudo-centrosymmetric subunit interface site in the channel lumen, predominantly at the conserved M2–9′ leucines, αLeu-251, βLeu-257 and δLeu-265, but also more extracellular at M2–13′ & 16′. The photolabeling kinetics of the channel lumen residues are summarized in Fig 6C. The second domain is the interface between the ligand–binding domain (LBD) and the transmembrane domain. Here in the δM2–M3 loop the photolabeled δIle-288 makes contact with the conserved Cys-loop of the LBD at the highly conserved δPhe-137 and δPro-138 (Fig. S5). The third domain is the extracellular end of the δ–subunit’s transmembrane domain where we photolabeled two residues one turn apart both on M1 (δPhe-232, δCys-236) and on M2–18′ & 22′ (δThr-274, δLeu-278). On one side, these residues are bounded by the α–δ subunit interface and on the other by the space within the δ–subunit’s four helix bundle (Figs. 6A & S5). In the second domain (δM2–M3 loop), the α–carbon of δIle-288 is 13–20 Å from those of the residues on M1 and M2. Although structurally distinct, the photolabeling kinetics of the residues in the second and third domains were similar (Fig. 6B) and they may be considered together; we have previously termed them the activation–dependent residues and predicted that they all contribute to a single TID binding pocket (28).
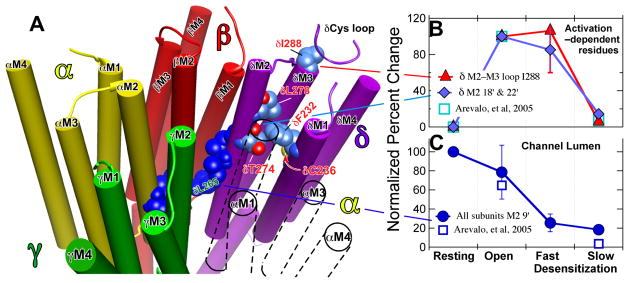
Figure 6. Location (Panel A) and contrasting state–dependence of the activation–dependent (Panel B) and the channel lumen (Panel C) groups of residues.
Panel A: A representation of the transmembrane region of the Torpedo nAChR based on the 2005 structure (2gb9.pdb: (5)). The receptor is viewed from the extracellular, N-terminal side and the structure is sliced to show the transmembrane domain (TMD) only. The subunits are color coded as follows (Alpha, yellow; Beta, red; Gamma, green; Delta, purple; the TMD of the second α–subunit that is situated between the γ– and δ–subunits is shown only in outline for clarity). The channel lumen residues are shown in dark blue; the M2–9′ residues are shown for each subunit and the M2–13′ & 16′ residues for the δ-subunit. The δM2–9′ Leu-265 is identified in green lettering. For the activation dependent–residues on the δ–subunit the carbon atoms are shown in corn blue, oxygen in red and nitrogen in mid blue; residues are identified in red lettering. In the top right near δIle-288 a portion of the δ–subunit’s Cys–loop from the LBD is shown in ribbon representation. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081) (48). Panels B & C: The normalized efficiency of photoincorporation of [125I]TID as the nAChR passes sequentially from the resting through the open and fast desensitized states to the slow desensitized state. Panel B: The biphasic state dependence of photoincorporation into the activation–dependent residus in the δ–subunit; the residues are identified in the key. The percent change in each pair of experiments was calculated. The data for δM2–18′ & 22′ (Thr-274 and Leu-278) were grouped and averaged (2 replicates per point). For the δM2–M3 loop residue, Ile-288, there were two replicates for each state transition (Fig. 1 and Fig 2.). The data were then normalized with error propagation to the open state, which was set to 100. There was no photolabeling in the resting state, equal degrees of photoincorporation in the open and fast desensitized states and only modest photolabeling in the slow desensitized state. Panel C: The state dependence of the efficiency of photoincorporation into the channel lumen M2–9′ residues on the α–, β–& δ–subunits (αM2 Leu-251; βM2 Leu-257 & δM2 Leu-265). The data were analyzed as in Panel B. The percent change in each pair of experiments was calculated. We assumed 9′ leucine photoincorporation was equal in all subunits and averaged the data, which were then normalized with error propagation to the resting state, which was set to 100. For comparison the squares show data from our previous study (28) for the mean of δM2–18 & 22′ normalized to the open state (B) and for δM2 Leu-265 normalized to the resting state (C).
An interesting spatiotemporal pattern of agonist–induced structural changes was observed to occur asynchronously over a wide time frame (Fig. 6, panels B & C). In our previous study, photoincorporation efficiency into the activation–dependent residues on the δ–subunit was similar at 1.5 ms and 10 ms following addition of agonist (28). Here we show that there is no further structural change during fast desensitization in this region. Thus, after the initial change detected 1.5 ms after addition of agonist, there is no further change until slow desensitization occurs. In contrast, in the channel lumen structural changes were detected both during opening and during fast desensitization. Thus, during fast desensitization structural changes occurring in the center of the channel lumen at M2–9′ are uncoupled from those contralateral to the extracellular end of M2 and in the δM2–M3 loop in contact with the LBD (compare Fig 6B with 6C). However, upon slow desensitization a major structural change occurs in the latter regions, while either modest or no changes are detected in the channel lumen. Finally, the slow desensitized state can be distinguished from the resting state, not only, as expected, in the centre of the channel lumen but also in the two more extracellular domains where the efficiency of photolabeling in the activation–dependent residues remains higher in the slow desensitized state than in the resting state, a result in agreement with another recent study (38).
This microscopic pattern of photolabeling is consistent with that of the intact subunits (28). The δ–subunit is the only one to experience an increase in photoincorporation upon opening because it is the only one with photoincorporation in the activation-dependent residues and this balances out the decrease in the channel lumen. During fast desensitization all subunits experience a decrease in photoincorporation originating in the channel lumen, but the level in the δ–subunit remains higher than in the other subunits. Finally, during slow desensitization only the δ–subunit photoincorporation decreases sharply because photoincorporation in the activation-dependent residues is lost.
Relevance to models of gating and desensitization
Our conclusion that the channel lumen’s structure in the center of the M2 domain is different in the resting, open and fast desensitized states is consistent with the two gate model, which proposes an activation gate and a separate desensitization gate (14, 16, 28, 39). In each of the three states we studied the model predicts a different arrangement of gates in the conduction pathway. In the resting state, the desensitization gate is open and the activation gate is closed. Activation opens the latter gate and conduction occurs. During fast desensitization, the desensitization gate now closes while the activation gate remains open.
The uncoupled model proposes that desensitization is not a concerted process, that each subunit has only one desensitized structure and that the difference between fast and slow desensitization is merely in the number of subunits in the desensitized state (17). Supporting evidence comes from rapid kinetics studies with a fluorescent agonist that suggest that the agonist site at the αδ–interface desensitizes more slowly than that at the αγ–interface (40, 41), pointing to a model in which a conformation change at the αγ–interface accompanies fast desensitization and a similar one at the αδ–interface accompanies slow desensitization. Our demonstration that the activation–dependent residues on the δ–subunit only change during slow desensitization is consistent with this conclusion. Furthermore, the conformation at the αδ–agonist site and that at the extracellular end of the transmembrane domain of the δ–subunit appear to be tightly coupled both across the αδ-subunit interface and across the LBD–TMD interface. At the same time, we observe changes in the centrosymmetric site in the channel lumen (δM2–9′ Leu-265), but it is likely that these reflect conformation changes in neighboring subunits rather than changes in the δ-subunit’s conformation, although we cannot rule out the latter possibility.
Although many studies have demonstrated the importance of the M2–M3 loop in channel gating, few have considered desensitization. Recent electrophysiological studies of chimeras of two receptors with very different desensitization rates, the nACh α7 receptor and 5HT3AR, show that mutations in the M2–M3 loop affect open-channel lifetime and rate of desensitization in parallel (18). This is consistent with our observations on the δ–subunit. In addition, our results suggest that structural changes in the δM2–M3 loop are rather rigidly coupled to those at the top of δM2 and δM1 because photolabeling of all five activation–dependent residues change in parallel. Comparison of the two prokaryotic structures suggests that gating involves a “tilting” of M2 pivoted on its central region so that the extracellular end moves anticlockwise around the pore’s axis (viewed from the LBD) and outwards towards M3, which in turn tilts outwards. The tilting of M2 during opening would move δM2 residues Thr-274 and Leu-278, which are contralateral to the channel lumen, into the pocket of the δ–subunit that is bounded by the four helix bundle and change the environment around δIle-288 on the δM2–M3 loop. It is likely that these coupled motions are driven by the interaction of the Cys-loop with the δM2–M3 loop (see below). One possibility is that in the open state δIle-288 takes up a position on the surface of the same helix–bounded pocket that has the other activation–dependent residues lining its surface (28). There are two arguments against this. First, δIle-288 is photolabeled more efficiently than the other residues, suggesting, but not proving, that they may not occupy the same pocket. Second, comparison of the two prokaryotic structures does not reveal such a re-arrangement of the M2–M3 loop in this direction, although this argument is complicated by the different lengths of that loop in the two representative structures. Further discussion must await the determination of the structure of a single protein in the resting and open state.
Theoretical modeling provides some insights into the conformational dynamics in the region of the activation–dependent residues. Thus, a molecular dynamics normal mode analysis study of the α7 nAChR suggested a direct coupling between a twisting motion of the LBD and dynamic changes of M2 (42). The coupling occurred at the interface between the Cys–loop and M2–M3 loop. The conserved Phe-135, (δPhe-137 in Torpedo numbering) is stabilized within a hydrophobic pocket formed by Leu-270 and Ile-271 (Torpedo δ–subunit numbering: Leu-287, Ile-288; both subunits have Pro–Leu–Ile at the M3 end of the M2–M3 loop). Downward motion of the M2–M3 loop caused the M2 helices to tilt. However, our results refer to the δ–subunit which does not bind the agonist and is a so-called complementary subunit, contributing to agonist action at the α–δ subunit interface. A theoretical study of the α4 β2 nAChR is of interest (43) because it found similar motions in the LBD–TMD interface to those in the above study and, in addition, that those in the complementary β–subunits were greater than those in the α–subunits.
Implications for drug action
This work, together with our previous study (28), has delineated the behavior of hydrophobic pockets through four conformations of the nAChR. It adds weight to the allosteric hypothesis of general anesthetic action that requires the existence of general anesthetic binding sites whose affinity varies with the protein’s conformation. Furthermore, our work shows that more than one drug binding pocket can be occupied simultaneously in the nAChR TMD and that the binding properties of these pockets may depend on the protein’s conformation in different ways, a conclusion that is supported by recent studies with TDBzl-etomidate, a photoreactive analog of the general anesthetic etomidate (44).
The kinetic behavior of the δ–subunit’s activation–dependent residues on M1 and M2, predicts that this four helix bundle binding pocket, reminiscent of that hypothesized to be responsible for volatile anesthetic action on GABAA receptors (45), has a similar structure in the open and fast desensitized states, but two different structures in the resting state and slow desensitized state. Nonetheless the much smaller general anesthetic, halothane, does label the resting state at δTyr-228 (46), one helical turn above the activation–dependent residue δPhe-232, illustrating the importance of an agent’s size; the smaller halothane has access to the pocket that is denied to the bulkier TID in this conformation. Furthermore, consistent with our conclusion that the resting and slow desensitized states are not equivalent, halothane photoincorporation is enhanced relative to the resting state by slow desensitization (46).
In the channel lumen at the level of the M2–9′ residues and in the hydrophobic patch extracellular to these regions (M2–13′ and 16′), the environment appears to be different in each of the conformations studied and an agent binding here might have a different affinity for each state. On the extracellular side of the hydrophobic patch in the channel lumen at M2–20′ (αGlu-262 & δGln-276), azietomidate, another photoactivable analog of the general anesthetic etomidate, labels about two-fold more efficiently in the open state than in the slow desensitized state, suggesting more modest changes in this region (47).
Conclusions
We have provided new information on structural changes that take place during fast desensitization. Specifically, we have been able to probe a region at the extracellular end of the TMD of the δ–subunit. The structure here changes when the channel opens (28). The open state structure persists in the fast desensitized state, only changing again when the slow desensitized state is attained. At the same time in the channel lumen at the level of the conserved M2–9′ leucines, the structures of the closed, open and fast desensitized states are all different, Thus, the structure of the ion pore in the two nonconducting states (resting and fast desensitized) connected to the open state are not equivalent.
Supplementary Material
Acknowledgments
We thank Z. Dostalova for help in preparing acetylcholine receptor rich Torpedo membranes.
This research was supported by a grant from the National Institute of Health (GM-58448), by the Department of Anesthesia and Critical Care, Massachusetts General Hospital and by an award to the Harvard Medical School from the Howard Hughes Biomedical Research Support Program.
Abbreviations
- Carbachol
carbamylcholine
- EndoLys-C
endoproteinase Lys-C
- HPLC
high-pressure liquid chromatography
- LBD
ligand–binding domain
- nAChR
nicotinic acetylcholine receptor
- OPA
o-phthalaldehyde
- PAGE
polyacrylamide gel electrophoresis
- PTH
phenylthiohydantoin
- SDS
sodium dodecyl sulfate
- TID
3-(trifluoromethyl)-3-(m-iodophenyl) diazirine
- TMD
transmembrane domain
- TPS
Torpedo physiological saline
- V8 protease
Staphylococcus aureus endopeptidase Glu-C
Footnotes
Supporting information is available showing HPLC purification of labeled peptides and a molecular model. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Cederholm JM, Schofield PR, Lewis TM. Gating mechanisms in Cys-loop receptors. Eur Biophys J. 2009 doi: 10.1007/s00249-009-0452-y. in press. [DOI] [PubMed] [Google Scholar]
- 2.Sine SM, Engel AG. Recent advances in Cys-loop receptor structure and function. Nature. 2006;440:448–455. doi: 10.1038/nature04708. [DOI] [PubMed] [Google Scholar]
- 3.Giniatullin R, Nistri A, Yakel JL. Desensitization of nicotinic ACh receptors: shaping cholinergic signaling. Trends Neurosci. 2005;28:371–378. doi: 10.1016/j.tins.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Wells GB. Structural answers and persistent questions about how nicotinic receptors work. Front Biosci. 2008;13:5479–5510. doi: 10.2741/3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. Journal of molecular biology. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 6.Hilf RJ, Dutzler R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature. 2008;452:375–379. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- 7.Hilf RJ, Dutzler R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature. 2009;457:115–118. doi: 10.1038/nature07461. [DOI] [PubMed] [Google Scholar]
- 8.Bocquet N, Nury H, Baaden M, Le Poupon C, Changeux JP, Delarue M, Corringer PJ. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457:111–114. doi: 10.1038/nature07462. [DOI] [PubMed] [Google Scholar]
- 9.Auerbach A. Life at the top: the transition state of AChR gating. Sci STKE 2003. 2003:re11. doi: 10.1126/stke.2003.188.re11. [DOI] [PubMed] [Google Scholar]
- 10.Burzomato V, Beato M, Groot-Kormelink PJ, Colquhoun D, Sivilotti LG. Single-channel behavior of heteromeric alpha1beta glycine receptors: an attempt to detect a conformational change before the channel opens. J Neurosci. 2004;24:10924–10940. doi: 10.1523/JNEUROSCI.3424-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lape R, Colquhoun D, Sivilotti LG. On the nature of partial agonism in the nicotinic receptor superfamily. Nature. 2008;454:722–727. doi: 10.1038/nature07139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beckstein O, Sansom MS. The influence of geometry, surface character, and flexibility on the permeation of ions and water through biological pores. Phys Biol. 2004;1:42–52. doi: 10.1088/1478-3967/1/1/005. [DOI] [PubMed] [Google Scholar]
- 13.Cymes GD, Grosman C. Pore-opening mechanism of the nicotinic acetylcholinereceptor evinced by proton transfer. Nature Struct Molec Biol. 2008;15:389–389. doi: 10.1038/nsmb.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auerbach A, Akk G. Desensitization of mouse nicotinic acetylcholine receptor channels. A two-gate mechanism. J Gen Physiol. 1998;112:181–197. doi: 10.1085/jgp.112.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karlin A. Emerging structure of the nicotinic acetylcholine receptors. Nature reviews. 2002;3:102–114. doi: 10.1038/nrn731. [DOI] [PubMed] [Google Scholar]
- 16.Purohit Y, Grosman C. Block of muscle nicotinic receptors by choline suggests that the activation and desensitization gates act as distinct molecular entities. J Gen Physiol. 2006;127:703–717. doi: 10.1085/jgp.200509437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prince RJ, Sine SM. Acetylcholine and epibatidine binding to muscle acetylcholine receptors distinguish between concerted and uncoupled models. The Journal of biological chemistry. 1999;274:19623–19629. doi: 10.1074/jbc.274.28.19623. [DOI] [PubMed] [Google Scholar]
- 18.Bouzat C, Bartos M, Corradi J, Sine SM. The interface between extracellular and transmembrane domains of homomeric Cys-loop receptors governs open-channel lifetime and rate of desensitization. J Neurosci. 2008;28:7808–7819. doi: 10.1523/JNEUROSCI.0448-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heidmann T, Changeux JP. Fast kinetic studies on the allosteric interactions between acetylcholine receptor and local anesthetic binding sites. European Journal Of Biochemistry. 1979;94:281–296. doi: 10.1111/j.1432-1033.1979.tb12894.x. [DOI] [PubMed] [Google Scholar]
- 20.Quast U, Schimerlik MI, Raftery MA. Ligand-induced changes in membrane-bound acetylcholine receptor observed by ethidium fluorescence. 2. Stopped-flow studies with agonists and antagonists. Biochemistry. 1979;18:1891–1901. doi: 10.1021/bi00577a007. [DOI] [PubMed] [Google Scholar]
- 21.Steinbach JH, Sine SM. Function of nicotinic acetylcholine receptors. Society of General Physiologists series. 1987;41:19–42. [PubMed] [Google Scholar]
- 22.Forman SA, Miller KW. High acetylcholine concentrations cause rapid inactivation before fast desensitization in nicotinic acetylcholine receptors from Torpedo. Biophysical journal. 1988;54:149–158. doi: 10.1016/S0006-3495(88)82939-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edelstein SJ, Schaad O, Henry E, Bertrand D, Changeux JP. A kinetic mechanism for nicotinic acetylcholine receptors based on multiple allosteric transitions. Biological cybernetics. 1996;75:361–379. doi: 10.1007/s004220050302. [DOI] [PubMed] [Google Scholar]
- 24.Muhn P, Fahr A, Hucho F. Rapid laser flash photoaffinity labeling of binding sites for a noncompetitive inhibitor of the acetylcholine receptor. Biochemistry. 1984;23:2725–2730. doi: 10.1021/bi00307a029. [DOI] [PubMed] [Google Scholar]
- 25.Cox RN, Kaldany RR, DiPaola M, Karlin A. Time-resolved photolabeling by quinacrine azide of a noncompetitive inhibitor site of the nicotinic acetylcholine receptor in a transient, agonist-induced state. The Journal of biological chemistry. 1985;260:7186–7193. [PubMed] [Google Scholar]
- 26.Heidmann T, Changeux JP. Characterization of the transient agonist-triggered state of the acetylcholine receptor rapidly labeled by the noncompetitive blocker [3H]chlorpromazine: additional evidence for the open channel conformation. Biochemistry. 1986;25:6109–6113. doi: 10.1021/bi00368a041. [DOI] [PubMed] [Google Scholar]
- 27.Chiara DC, Kloczewiak MA, Addona GH, Yu JA, Cohen JB, Miller KW. Site of resting state inhibition of the nicotinic acetylcholine receptor by a hydrophobic inhibitor. Biochemistry. 2001;40:296–304. doi: 10.1021/bi0021481. [DOI] [PubMed] [Google Scholar]
- 28.Arevalo E, Chiara DC, Forman SA, Cohen JB, Miller KW. Gating-enhanced accessibility of hydrophobic sites within the transmembrane region of the nicotinic acetylcholine receptor’s {delta}-subunit. A time-resolved photolabeling study. The Journal of biological chemistry. 2005;280:13631–13640. doi: 10.1074/jbc.M413911200. [DOI] [PubMed] [Google Scholar]
- 29.Pedersen SE, Sharp SD, Liu WS, Cohen JB. Structure of the noncompetitive antagonist-binding site of the Torpedo nicotinic acetylcholine receptor. [3H]meproadifen mustard reacts selectively with alpha-subunit Glu-262. The Journal of biological chemistry. 1992;267:10489–10499. [PubMed] [Google Scholar]
- 30.White BH, Cohen JB. Photolabeling of membrane-bound Torpedo nicotinic acetylcholine receptor with the hydrophobic probe 3-trifluoromethyl-3-(m-[125I]iodophenyl)diazirine. Biochemistry. 1988;27:8741–8751. doi: 10.1021/bi00424a009. [DOI] [PubMed] [Google Scholar]
- 31.Brauer AW, Oman CL, Margolies MN. Use of o-phthalaldehyde to reduce background during automated Edman degradation. Anal Biochem. 1984;137:134–142. doi: 10.1016/0003-2697(84)90359-2. [DOI] [PubMed] [Google Scholar]
- 32.Middleton RE, Cohen JB. Mapping of the acetylcholine binding site of the nicotinic acetylcholine receptor: [3H]nicotine as an agonist photoaffinity label. Biochemistry. 1991;30:6987–6997. doi: 10.1021/bi00242a026. [DOI] [PubMed] [Google Scholar]
- 33.Rankin SE, Addona GH, Kloczewiak MA, Bugge B, Miller KW. The cholesterol dependence of activation and fast desensitization of the nicotinic acetylcholine receptor. Biophysical journal. 1997;73:2446–2455. doi: 10.1016/S0006-3495(97)78273-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maconochie DJ, Steinbach JH. The channel opening rate of adult- and fetal-type mouse muscle nicotinic receptors activated by acetylcholine. The Journal of physiology. 1998;506(Pt 1):53–72. doi: 10.1111/j.1469-7793.1998.053bx.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu G, Raines DE, Miller KW. A hydrophobic inhibitor of the nicotinic acetylcholine receptor acts on the resting state. Biochemistry. 1994;33:15375–15381. doi: 10.1021/bi00255a018. [DOI] [PubMed] [Google Scholar]
- 36.Garcia G, 3rd, Chiara DC, Nirthanan S, Hamouda AK, Stewart DS, Cohen JB. [3H]Benzophenone photolabeling identifies state-dependent changes in nicotinic acetylcholine receptor structure. Biochemistry. 2007;46:10296–10307. doi: 10.1021/bi7008163. [DOI] [PubMed] [Google Scholar]
- 37.Blanton MP, Cohen JB. Identifying the lipid-protein interface of the Torpedo nicotinic acetylcholine receptor: secondary structure implications. Biochemistry. 1994;33:2859–2872. doi: 10.1021/bi00176a016. [DOI] [PubMed] [Google Scholar]
- 38.Hamouda AK, Sanghvi M, Chiara DC, Cohen JB, Blanton MP. Identifying the lipid-protein interface of the alpha4beta2 neuronal nicotinic acetylcholine receptor: hydrophobic photolabeling studies with 3-(trifluoromethyl)-3-(m-[125I]iodophenyl)diazirine. Biochemistry. 2007;46:13837–13846. doi: 10.1021/bi701705r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson G, Karlin A. Acetylcholine receptor channel structure in the resting, open, and desensitized states probed with the substituted-cysteine-accessibility method. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:1241–1248. doi: 10.1073/pnas.031567798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andreeva IE, Pedersen SE. Conformational asymmetry of nicotinic acetylcholine receptor (AChR) desensitization. Biophysics Journal. 2005;89:3052–Pos. [Google Scholar]
- 41.Andreeva IE, Nirthanan S, Cohen JB, Pedersen SE. Site specificity of agonist-induced opening and desensitization of the Torpedo californica nicotinic acetylcholine receptor. Biochemistry. 2006;45:195–204. doi: 10.1021/bi0516024. [DOI] [PubMed] [Google Scholar]
- 42.Cheng X, Ivanov I, Wang H, Sine SM, McCammon JA. Nanosecond-timescale conformational dynamics of the human alpha7 nicotinic acetylcholine receptor. Biophysical journal. 2007;93:2622–2634. doi: 10.1529/biophysj.107.109843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haddadian EJ, Cheng MH, Coalson RD, Xu Y, Tang P. In silico models for the human alpha4beta2 nicotinic acetylcholine receptor. The journal of physical chemistry. 2008;112:13981–13990. doi: 10.1021/jp804868s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nirthanan S, Garcia G, 3rd, Chiara DC, Husain SS, Cohen JB. Identification of binding sites in the nicotinic acetylcholine receptor for TDBzl-etomidate, a photoreactive positive allosteric modulator. The Journal of biological chemistry. 2008;283:22051–22062. doi: 10.1074/jbc.M801332200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mihic SJ, Ye Q, Wick MJ, Koltchine VV, Krasowski MD, Finn SE, Mascia MP, Valenzuela CF, Hanson KK, Greenblatt EP, Harris RA, Harrison NL. Sites of alcohol and volatile anaesthetic action on GABA(A) and glycine receptors. Nature. 1997;389:385–389. doi: 10.1038/38738. [DOI] [PubMed] [Google Scholar]
- 46.Chiara DC, Dangott LJ, Eckenhoff RG, Cohen JB. Identification of nicotinic acetylcholine receptor amino acids photolabeled by the volatile anesthetic halothane. Biochemistry. 2003;42:13457–13467. doi: 10.1021/bi0351561. [DOI] [PubMed] [Google Scholar]
- 47.Chiara DC, Hong FH, Arevalo E, Husain SS, Miller KW, Forman SA, Cohen JB. Time-resolved photolabeling of the nicotinic acetylcholine receptor by [3H]azietomidate, an open-state inhibitor. Molecular pharmacology. 2009;75:1084–1095. doi: 10.1124/mol.108.054353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera - A Visualization System for Exploratory Research and Analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.