Summary
Mixed Lineage Leukemia (MLL) and its metazoan orthologs have been linked with the epigenetic maintenance of transcriptional activity. To identify mechanisms by which MLL might perpetuate active transcription in dividing cells, we investigated its role during M-phase of the cell cycle. Unlike other histone methyltransferases examined, MLL remained globally embedded within condensed mitotic chromosomes. Genome-wide location analysis revealed a rearranged pattern of MLL occupancy in mitosis compared with interphase, characterized by a strong preference for MLL to occupy genes in mitosis possessing the highest levels of interphase transcription. Knockdown experiments revealed that MLL is required for rapid post-mitotic reactivation of its mitotic target genes, suggesting a bookmarking function. MLL tethers Menin, RbBP5, and ASH2L to genes during mitosis, but is dispensable for preserving H3K4 methylation. These findings implicate mitotic retention as a novel component of MLL-based gene regulation which may facilitate inheritance of active gene expression states during cell division.
MLL is a transcriptional regulator that maintains gene activity through the preservation of active chromatin structure. Gene knockout studies in mice have found Mll to be required for embryonic development as well as for self-renewal of several adult stem cell lineages (Jude et al., 2007; Lim et al., 2009; Yu et al., 1995). MLL maintains expression of a host of downstream target genes, most notably, members of the HOX transcription factor gene clusters. MLL-deficient tissues are able to initiate proper Hox gene activation during early embryogenesis, however, expression fails to be maintained throughout later development (Ernst et al., 2004; Yu et al., 1995). MLL is also a proto-oncogene that can be mutated via chromosomal translocation to form leukemogenic fusion proteins that hyperactivate normal MLL target genes and block hematopoietic cell differentiation (reviewed in Krivtsov and Armstrong, 2007).
MLL is a human ortholog of the Trithorax gene in Drosophila, which is likewise implicated in maintaining cellular identity during development through the maintenance of gene activity (Ringrose and Paro, 2007). Trithorax, as well as its silencing counterpart, Polycomb, are described as ‘epigenetic memory’ pathways, due to their ability to maintain expression states that propagate through extensive rounds of cell division despite the lack of continued expression of the DNA-binding factors that originally initiated the respective “ON” or “OFF” state (Cavalli and Paro, 1998; Maurange and Paro, 2002). It has been proposed that Trithorax and Polycomb proteins maintain heritable chromatin structure via specialized molecular interactions that resist disruption by DNA replication and mitosis (Francis and Kingston, 2001; Ringrose and Paro, 2007). Indeed, recent evidence suggests that Polycomb complexes propagate a repressed expression state via an inability of DNA replication to dissociate PRC1 from chromatin or through self-perpetuating cycles of histone H3K27 methylation performed by PRC2 (Francis et al., 2009; Hansen et al., 2008; Margueron et al., 2009). However, mechanisms have yet to be identified for how MLL/Trithorax proteins enable active chromatin states to withstand the disruptive forces generated during mitotic chromosome condensation.
MLL possesses histone H3K4 methyltransferase activity, which could provide a mechanism for maintaining heritable chromatin states by labeling histones with stable chromatin marks (Milne et al., 2002). However, the H3K4 methyltransferase activity of MLL is not essential in vivo, as mice harboring a homozygous deletion of only the MLL catalytic domain are born at normal Mendelian ratios with a relatively mild phenotype. In sharp contrast, MLL-null mice fail to survive past E10.5 (Terranova et al., 2006; Yu et al., 1995). Therefore, MLL is likely to serve methyltransferase-independent functions to propagate active chromatin. MLL has been shown to interact with several proteins, such as Menin, RbBP5, and ASH2L, that likely contribute to the regulation of transcription by MLL (Nakamura et al., 2002; Yokoyama et al., 2004). While MLL associates with a large number of active transcription units in vivo (Guenther et al., 2005; Scacheri et al., 2006), it remains uncertain whether the association of MLL with chromatin endures through DNA replication or mitosis. Some prior studies noted co-localization of MLL with mitotic chromosomes (Caslini et al., 2000; Ennas et al., 1997), however, these findings have been challenged by a recent study that observed MLL displacement from chromatin during mitosis (Mishra et al., 2009). Furthermore, the fluctuation of MLL levels throughout the cell cycle controlled by targeted degradation points towards a temporal regulation of MLL in cycling cells (Liu et al., 2007). Nevertheless, it remains uncertain whether MLL has evolved specific mechanisms to avoid disturbance by DNA replication or mitosis which might underlie a role in epigenetic inheritance of gene expression.
M-phase of the cell cycle is associated with a global condensation of chromosomes into compact structures, genome-wide silencing of transcription, and dissociation of the vast majority of transcriptional regulators from their target genes. The mass-removal of transcription factors from mitotic chromatin may destabilize existing transcriptional programs and allow ‘cellular reprogramming’ opportunities during development or during in vitro manipulation of cell fates (reviewed in Egli et al., 2008; Martinez-Balbas et al., 1995). We speculated that MLL/Trithorax might exert an epigenetic inheritance function via retention within mitotic chromatin. This would allow MLL to mark active genes during their transiently silenced state in mitosis. Here, we have found that MLL displays a unique pattern of chromatin occupancy during mitosis that contributes to the rapid restoration of target gene activity within daughter cell chromatin.
Results
MLL N- and C-fragments colocalize with condensed mitotic chromosomes
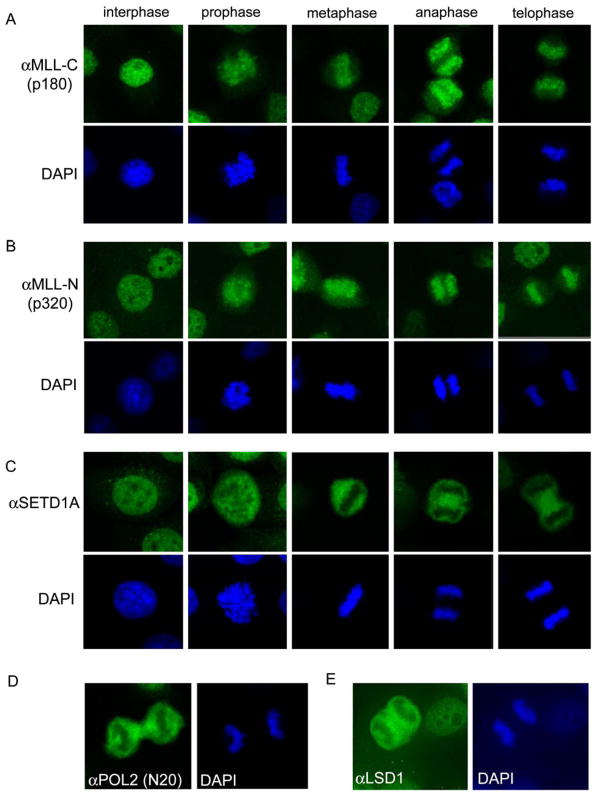
It has been noted previously that overexpressed fragments of MLL can colocalize with topoisomerase II on mitotic scaffolds (Caslini et al., 2000), however the functional relevance of this localization was never determined. As an alternative to scaffold binding, we considered whether MLL might instead associate with gene promoters in condensed mitotic chromosomes. To evaluate this, we first confirmed the localization pattern of endogenous MLL by confocal immunofluorescence (IF) microscopy in HeLa using antibodies against both Taspase1 cleavage products of MLL (N320 and C180) (Hsieh et al., 2003). Both MLL fragments colocalized with chromatin throughout the mitotic interval, spanning from prophase to telophase (Figure 1A, B). In contrast, SETD1A, the major H3K4 methyltransferase in mammalian cells (Wu et al., 2008), was dissociated from mitotic chromosomes, similar to RNA polymerase II and the H3K4 demethylase LSD1 (Figure 1C, D, E). The same MLL localization pattern was found in U2OS and in IMR90 primary human fibroblasts (Supplemental Figure 1). Our observations disagree with a recent report that found MLL to be displaced from mitotic HeLa chromosomes (Mishra et al., 2009). To resolve this issue, we performed additional controls to verify the specificity of our antibodies. First, shRNA knockdown of MLL resulted in a complete removal of IF and ChIP signals from mitotic chromosomes (Supplemental Figure 1C, Figure 4). Second, mitotic chromosome localization was confirmed in living cells using a GFP-MLL fusion protein (Supplemental Figure 1B). Together, our findings support a widespread and specific association of MLL with mitotic chromosomes in diverse cell types.
Figure 1.
MLL N- and C- fragments localize globally with mitotic chromatin.
Immunofluorescence (IF) of asynchronous HeLa cells stained with A) anti-MLL-C (468), B) anti-MLL-N (457), C) anti-SETD1A, D) anti-RNA polymerase II (N20), or E) anti-LSD1 (09–058) antibodies. All secondary antibodies were conjugated with Cy2 and counterstained with DAPI.
MLL occupancy within mitotic chromatin favors genes with high levels of interphase transcription
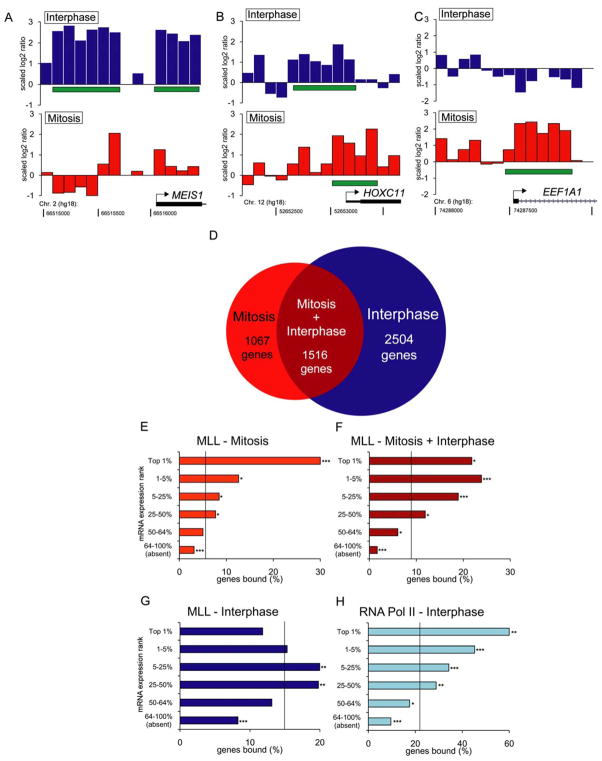
Genome-wide location analysis was performed to compare the interphase and mitotic occupancy pattern of MLL. Anti-MLL ChIP of interphase and mitotically arrested HeLa cells (Supplemental Figure 2) was analyzed using a RefSeq array representing 18,028 unique promoter regions. In interphase, MLL occupied a substantial number of genes, including many of its known targets, such as MEIS1 and HOXC11 (Figure 2A, B) (note: the HOXA cluster is not highly active in this cell type, data not shown). Gene ontology (GO) analysis revealed a strong tendency for MLL to occupy interphase genes encoding transcription factors, consistent with known MLL functions (Supplemental Figure 3). In addition, MLL binding peaks frequently encompassed sequences previously known to be linked with MLL, such as motifs for E2F, which recruits MLL via HCF-1 (Supplemental Figure 4) (Tyagi et al., 2007). The widespread occupancy of MLL (thousands of target genes) during interphase is consistent with prior studies (Guenther et al., 2005; Scacheri et al., 2006). Moreover, alignment of published data sets (derived from asynchronous HeLa S3 cells (Scacheri et al., 2006)) with ours revealed significant overlap with regard to genomic occupancy (Supplemental Figure 5). Thus, the genomic distribution pattern of MLL in interphase is consistent with its known roles in gene expression regulation.
Figure 2.
ChIP-chip analysis of MLL occupancy in interphase and mitosis. A, B, C) Scaled log2-ratio data for the MEIS1, HOXC11, and EEF1A1 promoter regions. Green horizontal bars denote regions called as peaks. D) Venn diagram evaluating the relative frequency of MLL occupancy at overlapping and unique peak locations between interphase and mitosis. E) Percentile rank of mRNA abundance for genes bound by MLL or Pol II. Vertical line represents the percentage of genes expected to be bound if no relationship existed between occupancy and expression. Chi-squared statistical test was used. * indicates p < 0.05. ** indicates p < 10−10. *** indicates p < 10−20
Unexpectedly, during mitosis MLL was only retained at a subset of its interphase target genes. For example, HOXC11 was occupied in mitosis and interphase, while MEIS1 was occupied only in interphase (Figure 2A, B). Equally surprising, a substantial number of genes were occupied by MLL exclusively in mitosis, such as EEF1A1 (Figure 2C). These cell-cycle preferences in MLL occupancy were verified by extensive qPCR analysis performed in multiple independent biological replicates at >70 genes (Supplemental Figure 6). Collectively, our comparison of interphase and mitotic occupancy identified thousands of unique, as well as overlapping MLL-occupied genes (Figure 2D), suggesting a global reorganization of occupancy coupled with cell cycle progression.
To gain further insight into the cell-cycle preferences of MLL occupancy, we compared the absolute expression levels of MLL’s interphase and mitotic target genes using published expression array data (Viegas et al., 2007) (reflecting gene activity in interphase). This revealed that genes occupied by MLL in mitosis tended to be expressed at very high levels in interphase (Figure 2E, F). In contrast, genes occupied by MLL exclusively in interphase tended to be expressed at moderate levels (Figure 2G). As a control, we performed an anti-RNA polymerase II (pol2) ChIP-chip using interphase chromatin, which, as expected, showed a strong correlation with the level of gene expression (Figure 2H). In addition, GO analysis revealed that MLL occupancy in mitosis tended to occur at genes encoding cell-cycle regulators, chaperones, and components of the translation machinery, which are all gene categories expressed at high levels (Supplemental Figure 3). Taken together, these data suggest that MLL occupancy is significantly reconfigured between interphase and mitosis toward genes possessing the highest levels of interphase transcription.
Recruitment of MLL and eviction of SETD1A, MLL2, DOT1L, ASH1L, and TBP during mitotic silencing of highly expressed genes
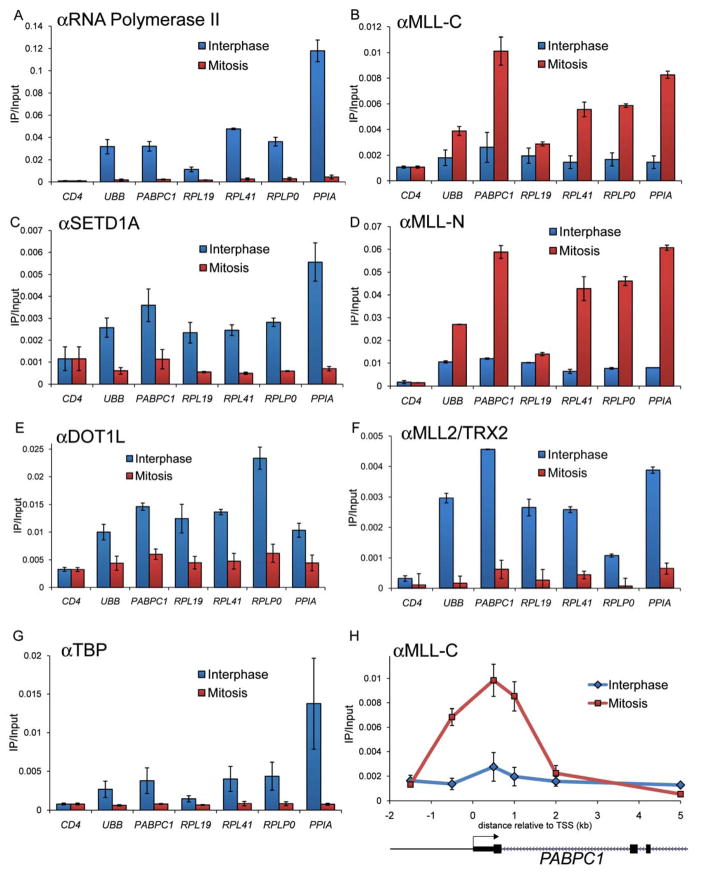
Similar to MLL, the histone methyltransferases ASH1L, MLL2/TRX2, DOT1L, and SETD1A have also been linked with actively transcribed regions, however, all prior experiments evaluating these factors have been performed in asynchronous cell populations (Demers et al., 2007; Gregory et al., 2007; Lee and Skalnik, 2008; Steger et al., 2008). Therefore, using ChIP-qPCR we next compared these enzymes to MLL with respect to their ability to occupy six genes in mitosis that were randomly chosen among the most highly expressed genes (in the top 1% of expression). As a control, we confirmed at each gene that pol2 was bound at high levels near the transcription start site (TSS) in interphase and was depleted in mitosis, consistent with a global mitotic silencing of gene transcription (Figure 3A). In striking contrast, MLL N- and C- fragments were bound at high levels in mitosis but depleted in interphase (Figure 3B, D). As expected, the constitutively silent CD4 gene lacked mitotic MLL binding. We also evaluated the spatial profile of MLL occupancy across the PABPC1 gene in mitosis and interphase and mapped its occupancy to a region extending from 0.5 kilobases (kb) upstream to 1 kb downstream of the TSS (Figure 3H). A similar spatial profile was observed at the MYC gene (Supplemental Figure 7). The occupancy pattern of MLL was also verified in HEK293T and murine embryonic stem cells as well as with additional independent antibodies raised against the C-fragment (Supplemental Figures 8, 9, and 10). Unlike MLL, the histone methyltransferases DOT1L, MLL2/TRX2, ASH1L, and SETD1A were all bound in interphase and evicted from their target genes during mitosis (Figure 3C, E, F, Supplemental Figure 11). TBP is a general transcription factor previously found to be retained at specific sites along mitotic chromosomes (Christova and Oelgeschlager, 2002; Xing et al., 2008). However, we failed to detect retention of this factor at any of the highly active genes examined here (Figure 3G). This may reflect gene-specificity in the locations where TBP is retained in mitosis, which could be distinct from MLL-occupied locations. Taken together, our findings contrast MLL recruitment with the eviction general transcription factors and certain methyltransferases during mitotic gene silencing.
Figure 3.
MLL occupancy is acquired at several highly expressed genes upon mitotic silencing. ChIP-qPCR analysis using the indicated antibodies comparing interphase and mitotic preparations of chromatin. The anti-MLL-C (468) or anti-MLL-N (456) antibody was used. (AG) Primer-pairs amplify at +0.5 kb relative to the TSS. (H) Primers amplify indicated location relative to the PABPC1 TSS. Interphase (blue) refers to cells arrested by double-thymidine block. Mitosis (red) refers to cells arrested by sequential thymidine-nocodazole treatment. CD4 is a constitutively silent control gene. Error bars denote S.E.M.
MLL promotes rapid transcriptional reactivation of its occupied genes during mitotic exit
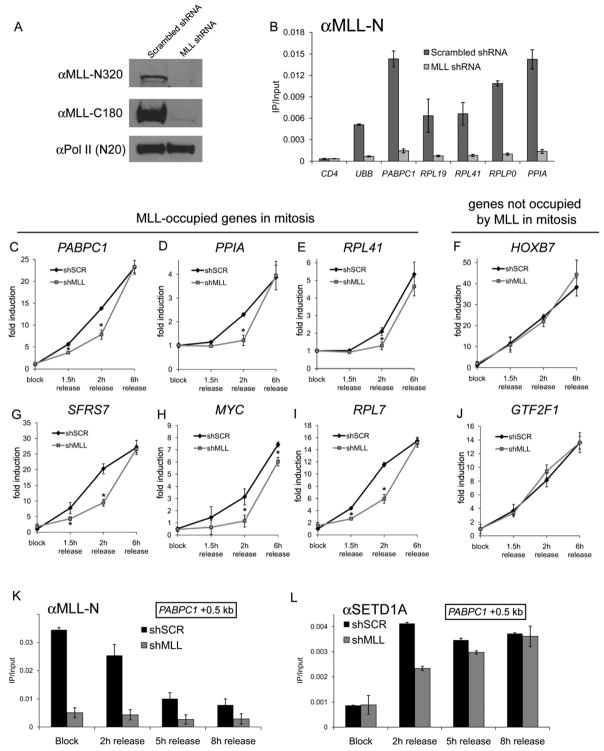
Widespread mitotic occupancy of MLL might serve to facilitate the return of transcriptional activity following mitotic exit, thereby functioning as a mitotic bookmark (Michelotti et al., 1997). To evaluate this possibility, we reduced MLL expression in HeLa cells via retroviral expression of short-hairpin RNAs (shRNAs). Western blotting of whole-cell extracts and ChIP-qPCR revealed ~90% reduction of both MLL N- and C-fragments in these cells (Figure 4A, B). Consistent with published reports, MLL-deficient cells proliferated more slowly and accumulated in M phase, suggesting a mitotic block (Supplemental Figure 12) (Liu et al., 2007; Mishra et al., 2009). Also consistent with published findings (Liu et al., 2007), we found that MLL-deficient cells exhibited a delayed transition from pro-metaphase to cytokinesis following mitotic release (Supplemental Figure 12). To measure the reacquisition of global transcriptional activity during telophase, we quantified 5-fluorouridine incorporation into nascent transcripts using flow cytometry following release from pro-metaphase arrest. When matched for their cell cycle stages using propidium iodide co-staining, MLL-deficient cells displayed a delay in the resumption of transcriptional activity (Supplemental Figure 13).
Figure 4.
MLL is required for rapid post-mitotic reactivation of its occupied genes. A) Western blot of whole-cell extracts prepared from cells expressing control or MLL-specific shRNA. B) ChIP q-PCR with anti-MLL-N (456) antibodies performed in mitotic cells. C-J) Primary transcript RT-qPCR analysis of the indicated genes in cells released from mitotic arrest at the indicated timepoints. Signals were normalized to the level of stable GAPDH mature transcript. Asterisks indicate p < 0.05 calculated using a two-tailed student’s t-test. Error bars denote S.E.M. K-L) Timecourse ChIP-qPCR performed with the indicated antibodies using primers amplifying the PABPC1 +0.5 kb region.
To better discern the direct action of MLL on transcription re-initiation from indirect effects on cell cycle progression, we determined whether the MLL-dependent onset of transcription occurs specifically at genes occupied by MLL during mitosis. To this end, MLL-deficient cells were released from mitotic arrest, and gene expression levels were measured at various time points using RT-qPCR. Primers were employed that span intron-exon junctions to detect short-lived primary transcripts that more closely reflect the transcriptional rate than more stable mRNAs. Six genes that are occupied by MLL exclusively in mitosis were selected from our ChIP-chip analysis. Two genes that lack MLL occupancy in mitosis served as controls. This analysis revealed that genes with MLL occupancy in mitotic chromatin exhibited a significant delay in their reactivation kinetics at 1.5 and/or 2 hours following mitotic release (Figure 4C, D, E, G, H, I). Eventually these genes achieved their full activity, as might be expected once the cells have moved deeper into G1. Importantly, HOXB7 and GTF2F1, genes that lacked MLL binding in mitosis, reactivated with normal kinetics following mitotic release (Figure 4F, J), suggesting that the effects of MLL are gene-specific and not simply due to a delay in cell cycle progression. These findings were further verified with multiple independent shRNAs as well as following a shorter duration knockdown with siRNA (Supplemental Figures 14, 15). In addition, expression of several genes encoding known regulators of mitotic progression were unaffected by MLL depletion (Supplemental Figure 16). As an additional control for the gene-specific requirements for MLL during the cell cycle, we verified that an alternative means of delaying mitotic progression (growth at reduced temperature) led to delays at all genes examined regardless of their association with MLL during mitosis (Supplemental Figure 17).
A direct effect of MLL on post-mitotic gene reactivation requires the continued presence of MLL at its mitotic locations until the resumption gene activity. To verify this, we first verified by western blotting that MLL protein levels remained present at early timepoints following mitotic release (Supplemental Figure 18). Next, we measured by ChIP-qPCR whether MLL continues to associate with PABPC1 as the gene reactivates following mitotic release. This analysis revealed the continued presence of detectable MLL occupancy at 2 hours following release at this gene, whereas by 8 hours of release MLL occupancy has been reduced to low levels (Figure 4K). We also observed that SETD1A recruitment was delayed at the 2 hour timepoint of release, consistent with a delayed return of interphase transcription machinery in MLL-deficient cells (Figure 4L). Taken together, these results implicate MLL occupancy of mitotic chromosomes in the restoration of gene activity in cells as they undergo mitotic exit.
MLL recruits Menin, Ash2L, and RbBP5 to mitotic chromatin
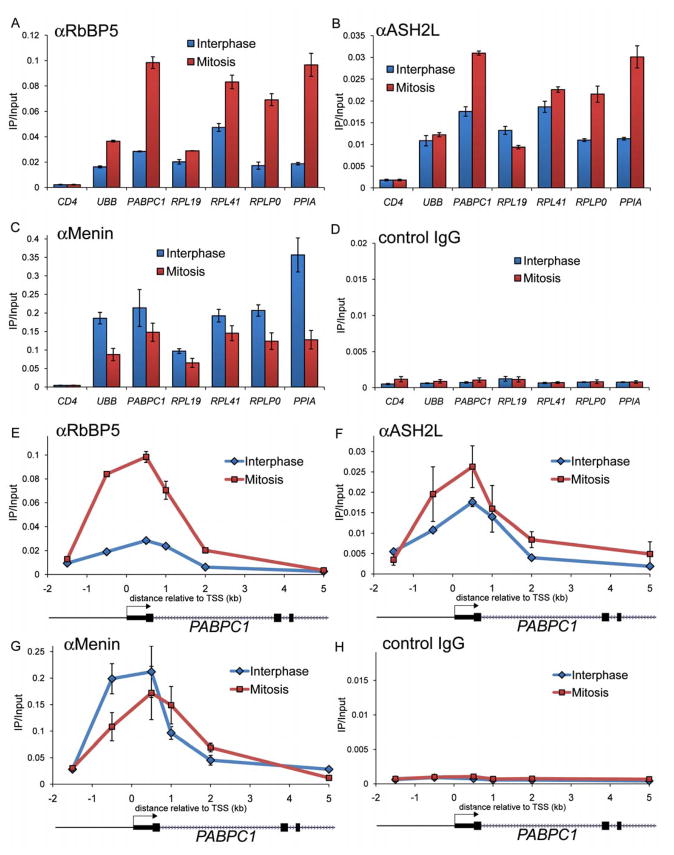
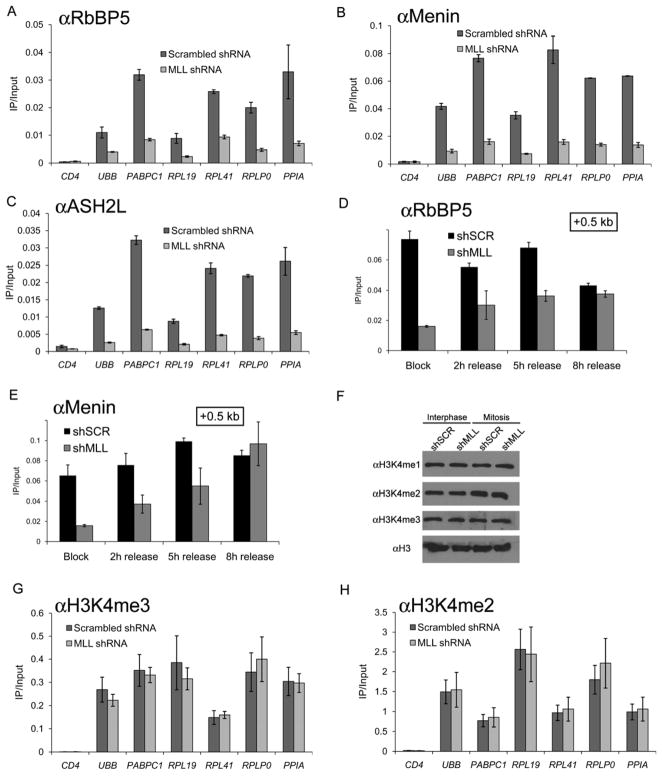
MLL interacts with numerous proteins linked with transcriptional regulation, including RbBP5, ASH2L, and the tumor suppressor protein Menin (Yokoyama et al., 2004). We evaluated by ChIP-qPCR whether MLL recruits these three factors to mitotic chromatin, which could be part of the mechanism through which MLL influences transcription following completion of mitosis. All three proteins displayed significant occupancy in both interphase and in mitosis at all six examined highly transcribed genes, with significant enrichment above control IgG and the silent CD4 region (Figure 5A–D). Importantly, all three proteins displayed a similar spatial localization as MLL around the PABPC1 TSS in mitotic chromatin (Figure 5E–H). RbBP5 and ASH2L displayed a trend towards higher occupancy in mitosis while Menin displayed a slight preference for interphase chromatin, suggesting that the stoichiometry of these proteins is different in the context of interphase and mitotic chromatin. To evaluate whether the mitotic association of these proteins requires MLL, we evaluated their occupancy in MLL-deficient cells. ChIP-qPCR revealed a substantial reduction of RbBP5, Menin, and ASH2L occupancy in mitotic chromatin at all sites analyzed, demonstrating that their association with mitotic chromatin is MLL-dependent (Figure 6A–C). In contrast, the chromatin occupancy of these proteins recovered in MLL-deficient cells following progression into G1 (Figure 6D, E), suggesting that they are recruited to interphase and mitotic chromatin via distinct mechanisms. While mitotic chromatin association is mediated by MLL, interphase chromatin association is likely to be mediated through an association with SETD1A or MLL2 (Hughes et al., 2004; Lee and Skalnik, 2008). In agreement with a published report (Mishra et al., 2009), we observed that most of RbBP5, ASH2L, and Menin localize to the cytoplasm by IF during mitosis (Supplemental Figure 19), suggesting that only the MLL-bound pool of these proteins binds to genes in mitosis, while the rest is displaced, likely in association with SETD1A and MLL2. Collectively, these findings suggest that MLL nucleates the recruitment of accessory proteins to its mitotic chromatin-occupied sites.
Figure 5.
RbBP5, ASH2L, and Menin occupy gene promoters during mitosis. ChIP-qPCR analysis using the indicated antibodies comparing occupancy between interphase and mitosis, as described in Figure 3.
Figure 6.
MLL is required for the association of Menin, RbBP5, and ASH2L with mitotic chromatin. A, B, C, G, H) ChIP-qPCR performed in control or MLL-shRNA expressing HeLa cells arrested in mitosis. D, E) Timecourse ChIP-qPCR performed in cells released from nocodazole arrest at the indicated timepoints. F) Western blot of acid extracted histones prepared from control or MLL-shRNA expressing cells. Error bars denote S.E.M.
Since MLL possesses H3K4 methyltransferase activity, we examined this modification in MLL-deficient mitotic chromatin. In control cells, H3K4 methylation was equally abundant in interphase and mitotic chromatin, consistent with published findings (Figure 6F, Supplemental Figure 20) (Kouskouti and Talianidis, 2005). Surprisingly, depletion of MLL had no effect on H3K4 methylation in bulk chromatin (Figure 6F) or specifically at MLL-occupied sites in mitosis (Figure 6G, H). Thus, despite a requirement of MLL for mitotic retention of partner proteins and for normal gene re-activation, full MLL occupancy appears dispensable for the preservation of H3K4 methylation during mitosis.
MLL-dependent HOX genes are MLL-occupied during both interphase and mitosis
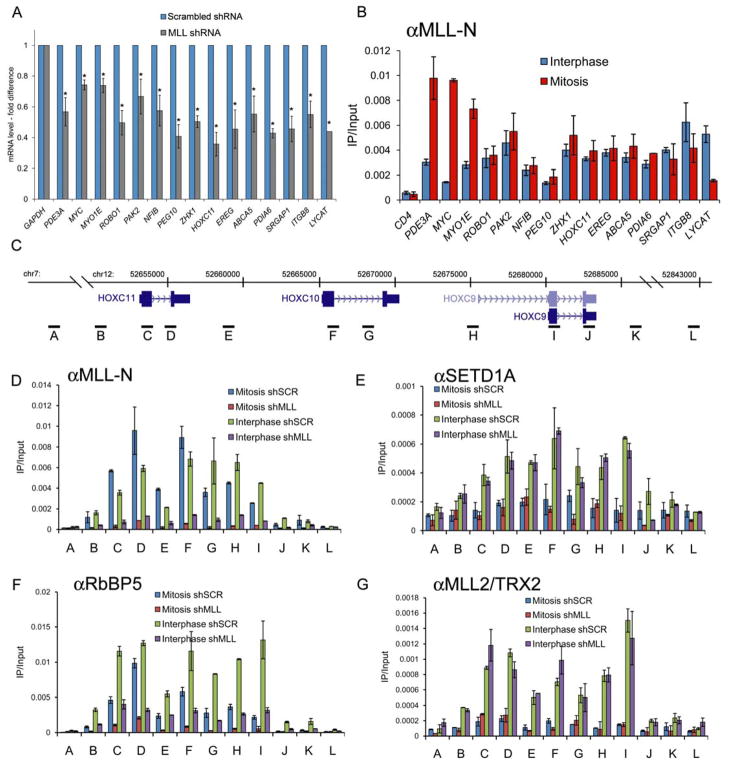
MLL/Trithorax has been classically linked with the maintenance of heritable HOX gene expression patterns. Therefore, we examined whether mitotic chromosome occupancy is a relevant feature of bona-fide MLL-dependent genes, such as HOX. First, we identified the most MLL-dependent genes in HeLa cells by performing expression microarrays comparing asynchronous MLL knockdown and control cultures, which revealed HOXC11 as among the most MLL dependent (Supplemental Figure 21). RT-qPCR verified the reduced expression of this panel of genes (Figure 7A). Notably, ChIP assays revealed that most MLL-dependent target genes were occupied by MLL during both interphase and mitosis (Figure 7B). We next performed a detailed characterization of MLL, other Trithorax-related proteins, and histone H3K4 di-methylation across 12 qPCR amplicons surrounding HOXC11 in interphase and mitotic chromatin (Figure 7C). MLL displayed a broad profile of occupancy at HOXC9, C10, and C11 in both interphase and mitotic chromatin, whereas SETD1A and MLL2 occupied the locus only during interphase (Figure 7D, E, G). In addition, we observed RbBP5 associated with the locus in both interphase and mitosis in an MLL-dependent manner (Figure 7F). It should be noted that MLL-knockdown renders a more complete eviction of RbBP5 from mitotic chromatin, whereas RbBP5 is only partially released from interphase chromatin in MLL-deficient cells, an effect likely due to the continued presence of SETD1A and MLL2 in interphase. The persistence of MLL occupancy in interphase and mitosis was also verified at the MLL-dependent HoxA genes in murine embryonic fibroblasts (Supplemental Figure 22). Thus, we observed that inheritance of HOX chromatin through mitosis occurs with the persistent association of MLL and RbBP5, whereas SETD1A and MLL2 are evicted in mitosis and resume their association only during interphase. Together, these data support MLL employing a dual interphase and mitotic occupancy pattern at its developmentally relevant target loci within HOX clusters.
Figure 7.
MLL-dependent HOX genes display MLL occupancy in both interphase and mitosis. A) RT-qPCR performed in control or MLL-shRNA expressing HeLa cells. B) ChIP-qPCR with anti-MLL-N antibodies (456) using the indicated primers, which amplify at +0.5 kb relative to the transcription start site. C) UCSC genome browser track (Kent et al., 2002) showing the HOXC cluster. qPCR amplicons are denoted by labeled horizontal bars. D-G) ChIP qPCR with the indicated antibodies comparing interphase, mitotic preparations of chromatin in cells expressing either control hairpin (shSCR) or MLL-specific hairpins (shMLL). Error bars denote S.E.M.
Discussion
In this study, we investigated the mechanism by which MLL, a conserved component of the Trithorax-Polycomb system, perpetuates active chromatin states during cell division. While MLL is best known for influencing transcription through catalysis of H3K4 methylation, our findings revealed an additional epigenetic mechanism employed by this factor: a pervasive occupancy of promoter regions packaged into condensed mitotic chromosomes as they are inherited from mother to daughter cells. Our functional studies suggest that mitotic retention allows MLL to accelerate the kinetics of gene reactivation following the exit from mitosis. Surprisingly, this activity appears largely independent of H3K4 methylation on mitotic chromosomes. The precise mechanism through which MLL serves as a mitotic bookmark remains unclear, but might involve the action of key accessory proteins such as RbBP5, ASH2L, and Menin, which are tethered by MLL to select sites in mitotic chromatin.
An unexpected observation in this study was the vast reorganization of MLL occupancy observed during the cell-cycle. In contrast to the situation in interphase, MLL occupancy during mitosis is heavily biased towards genes with the highest levels of pre-mitotic activity. One explanation for this binding behavior is that highly-expressed genes undergo the profoundest transition from silent to active as cells exit mitosis. Such a rapid switch in transcriptional rate may require auxiliary factors like MLL to faithfully restore pre-mitotic expression levels. It is also interesting to note that a prior study demonstrated that MLL can be degraded and resynthesized at two stages of the cell cycle (M/G1 transition and G1/S transition), thus raising the possibility that the reorganization of MLL occupancy described here could be mediated via regulated proteolysis (Liu et al., 2007). Importantly, the natural uncoupling of MLL’s interphase and mitotic occupancy allowed us to interrogate the functional relevance of its mitotic association independently of its interphase binding. Thus, our findings emphasize the extent to which the transcription factor landscape can be restructured during the cell cycle to facilitate perpetuation of transcriptional states.
A provocative question raised by our observations is regarding the relevant physical interactions utilized by MLL to remain associated with mitotic chromosomes. MLL contains several conserved domains that have been implicated in DNA or chromatin binding, such as several AT hooks, a CxxC domain, several PHD domains, and a BROMO domain. Menin has also been shown to cooperate with the PWWP protein, LEDGF, to target MLL to certain genomic regions (Yokoyama and Cleary, 2008). MLL occupancy of mitotic chromatin correlates with the level of gene activity in interphase, thus, MLL retention could be mediated through a physical association with histone modifications or histone variants which are deposited co-transcriptionally during interphase. Since MLL is known to serve as a coactivator for sequence-specific DNA-binding proteins (Tyagi et al., 2007), we speculate that the genome-wide return of DNA-binding transcriptional activators to their target sites following mitotic exit triggers the redistribution of MLL away from its mitotic locations to specific cis elements in interphase. Future studies will be aimed at identifying the differential recruitment mechanisms utilized by MLL in interphase and mitosis and their relevance to mutant forms of MLL found in leukemia.
Surprisingly, the effects of MLL on post-mitotic gene reactivation seem to occur without a detectable influence on levels of H3K4 methylation. While prior studies found normal levels of global H3K4 methylation in MLL-deficient cells (Milne et al., 2002; Wu et al., 2008), it is notable that during mitosis H3K4 methylation is unaffected by MLL depletion, especially in light of the widespread occupancy pattern present in mitosis. This suggests that the mitotic stability of H3K4 methylation could be mediated by an alternative mechanism, such as eviction of H3K4 demethylases. As an example, we found that the H3K4 demethylase LSD1 is globally evicted from chromatin during mitosis. Nevertheless, the ostensible uncoupling of MLL’s bookmarking function from H3K4 methylation levels indicates that MLL carries out important functions independently of its enzymatic activity. This is supported by the observation that MLL’s H3K4 methyltransferase domain is largely dispensable for its essential functions in vivo (Terranova et al., 2006). Non-enzymatic mechanisms utilized by MLL might involve tethering RbBP5, Menin, and ASH2L to its binding sites in mitotic chromatin. During the transition into interphase, these components might be “handed-off” from MLL to the SETD1A/MLL2 complexes as they re-associate in G1. Alternatively, MLL-interacting proteins might engage in physical interactions that return the transcription machinery to a post-mitotic active state. Indeed, Menin has been shown to associate with the Rpb2 subunit of RNA polymerase II (Hughes et al., 2004). MLL also contains a strong acidic activation domain at residues 2829–2883 as well as a demonstrated physical association with the C-terminal domain of RNA polymerase II (Milne et al., 2005; Prasad et al., 1995). These domains/interactions may provide a non-catalytic means by which MLL and its associated proteins deliver the transcriptional machinery back to genes as they reactivate at the end of mitosis.
Our study also highlights several important technical considerations when evaluating mitotic chromosome occupancy. First, it should be emphasized that IF visualizes the ratio of protein localized in the cytoplasm to the amount of protein localized to the chromosome. Thus, findings based on this assay alone can lead to misleading findings when the proportion of protein bound to mitotic chromosomes is low relative to the proportion present in the cytoplasm, as we observed for RbBP5, ASH2L, and Menin. In such instances, ChIP assays can more clearly ascertain whether a protein remains associated or not with mitotic chromatin. In addition, knockdown controls or use of multiple independent anti-sera is essential to verify antibody specificity.
Evidence in Drosophila implicates Trithorax and Polycomb proteins in the maintenance of mitotically heritable “ON” and “OFF” states, respectively (Cavalli and Paro, 1998; Maurange and Paro, 2002). Both Polycomb and Trithorax proteins employ an assortment of self-reinforcing mechanisms to render an expression state immune to stochastic fluctuation or subversion by the opposing pathway. For example, Polycomb complexes include several chromatin-binding proteins and histone-modifying enzymes that establish a cooperative network of interactions that can stabilize a repressed expression state during the cell cycle (Francis et al., 2009; Hansen et al., 2008; Lee et al., 2007; Wang et al., 2004). In the case of Trithorax-related gene regulation in mammalian cells, heritable gene activity might also be mediated via the interplay among diverse mechanisms employed among multiple Trithorax-like and SET1 family proteins. While deposition of histone H3K4 methylation might play a role in this process, it alone is insufficient to account for all inheritance functions in vivo (Terranova et al., 2006). Thus, we wish to propose that mitotic retention of Trithorax-like proteins, when coupled with other self-perpetuating mechanisms functioning in interphase, contributes to a robust inheritance system for transcriptional programs in dividing metazoan tissues.
Experimental Procedures
Cell culture
HeLa, HEK293T, IMR90, and U2OS cells were grown in DMEM with 10% FBS (Gemini). ES cells (CCE) were grown in DMEM with 10% ES-serum (Gemini) supplemented with LIF-conditioned medium.
Immunofluorescence
Cells plated on coverslips were rinsed in PBS and then fixed in 3.7% formaldehyde/10 mM HEPES 7.5/100 mM NaCl for 12 minutes. Fixation was quenched with 50 mM glycine in PBS. Cells were permeabilized with PBS containing 0.2% Triton X-100 (PBS-T) for 5 min and blocked with PBS-T containing 2% BSA for 10 minutes. Primary antibody was diluted in blocking buffer and incubated with cells for 1 hour at room temperature. Slides were washed 3 times for 3 minutes in PBS-T. Goat Anti-rabbit Cy2 antibody (Jackson Immunoresearch) was diluted in PBS-T and added for 1 hour. Slides were washed again 3 times. The last wash contained 4,6-diamidino-2-phenylindole (DAPI). Cells were mounted in 50% glycerol. Samples were viewed on a Zeiss 510 laser scanning confocal microscope with a C-Apochromat 63×1.2Wcorr objective.
Cell synchronization
For HeLa mitotic arrest, cells were first treated with 2 mM thymidine (Sigma) for 24 hours followed by 2 washes with phosphate-buffered saline (PBS) and released into fresh medium for 3 hours. Nocodazole (Sigma) was then added at a concentration of 200 ng/ml for 12 hours. Mitotic cells were then harvested by gentle pipetting (shake-off). Interphase cell populations were prepared by double thymdine arrest: cells were treated with 2 mM thymidine for 17 hours, washed twice in PBS, released into fresh medium for 8 hours, followed by an additional 18 hour treatment with 2 mM thymidine. Release experiments were performed by washing nocodazole-arrested cells with PBS three times, following by replating in fresh media. For HEK293T mitotic arrest, cells were treated with nocodazole (200 ng/ml) for 16 hours followed by gentle pipetting for shake-off. For ES cell mitotic arrest, cells were treated with nocodazole (100 ng/ml) for 12 hours, followed by shake-off by gentle pipetting.
For mitosis-release experiments, arrested cells were washed twice in PBS followed by replating into fresh media. Cells were harvested at indicated time points and analyzed by FACS, ChIP-qPCR, or RT-qPCR.
Chromatin Immunoprecipitation
ChIP assays were performed exactly as described (Steger et al., 2008: Supplemental Methods). Crosslinking was performed with sequential EGS (Pierce)/formaldehyde (Zeng et al., 2006). All results were quantified by qPCR performed using SYBR green (ABI) on an ABI 7900HT. Each IP signal was referenced to an input standard curve dilution series (IP/Input) to normalize for differences in starting cell number and for primer amplification efficiency. All experiments were performed with three independent biological replicates
ChIP-chip promoter array
Input and immunoprecipitated DNA samples from three pooled biological replicates were amplified using Whole Genome Amplification Kit (Sigma: WGA-2). Accurate amplification of enrichment sites was verified by qPCR analysis comparing pre- and post-amplification DNA samples. Nimblegen-Roche array service was used for DNA labeling of IP (Cy5) and Input (Cy3) DNA as well as for hybridization and scanning. The 385K MM8 RefSeq Promoter microarray is covered by 50–75mer probes spaced ~100bp apart, tiling regions ~2000bp upstream to 500bp downstream of 18,028 transcription start sites. Nimblescan software was used to convert raw signal intensity data into scaled log2-ratio data sets. Scaling was performed by subtracting the bi-weight mean for values of all features on the array from each individual value. Peak calling algorithms took into account log2-ratio signals from four adjacent features, which needed to be above a specified cutoff. The ratio data was then randomized 20 times to evaluate the probability of false positives. For this study, a false discovery rate cutoff of 10% was implemented. Since a single technical replicate of the array was performed, we empirically verified the accuracy of peak calling an extensive qPCR analysis within independent biological replicates (Supplemental Figure 5).
Data analysis was performed using a custom computational pipeline with R and Matlab statistical software. For the Venn diagram (Figure 2D), all MLL peaks were categorized according to whether they were found in the mitosis, interphase, or both datasets. To assess the relationship between MLL occupancy and mRNA abundance (Figure 2E), the ChIP-chip data was overlayed with expression information derived affymetrix arrays performed in asynchronous HeLa cell cultures, adapted from (Viegas et al., 2007). mRNA levels for each gene were categorized based on percentile rank into six groups: the top 1%, the top 1–5%, the top 5 to 25%, the top 25 to 50%, the top 50 to 64%, and the bottom 36% of genes (which had undetectable transcript levels). For each MLL or RNA pol II ChIP-chip dataset, the percentage of genes occupied in each expression category was graphed horizontally. The percentage of all genes occupied for each dataset (6% for MLL Mitosis, 9% for MLL Mitosis + Interphase, 16% for MLL Interphase, and 22% for RNA pol II in interphase) was plotted as a vertical line in each graph. This line corresponds to the expected percentage of genes occupied if no relationship existed between expression and occupancy (null hypothesis). Deviation from the null hypothesis was tested statistically using a chi-squared analysis. Motif analysis (Supplemental Figure 4) was performed using cis-regulatory element annotation system (CEAS) of peak coordinates from MLL interphase data (Ji et al., 2006). Gene Ontology analysis was performed using Database for Annotation, Visualization, and Integrated Discovery (DAVID) (Dennis et al., 2003). Data were plotted as the inverse log of the p value for enriched gene sets, calculated from Fisher’s exact test.
MLL knockdown
MLL-specific short hairpin sequences were adapted from (Liu et al., 2007) and cloned into a pSUPER.retro.puro (Oligoengine) expression vector according to manufacturer’s protocol. MLL hairpin sequence: GTGCCAAGCACTGTCGAAA. Scrambled hairpin sequence: GCGCGCTTTGTAGGATTCG. Retrovirus was packaged by cotransfection with VSVg and pCPG plasmids into 293T cells. HeLa cells were spin-fected with viral supernatant at 2100 g four times over two days. Puromycin was added to infected cells 48 hours later at 2 micrograms/ml. Selection was performed for 1 week before analysis. All analyses of knockdown cells were performed in selected pools. All results were verified in multiple experimental replicates performed in individual pools of knockdown cells. Additional confirmation was performed in at least three independently generated batches of pooled cells. Whole cell lysates were prepared using RIPA buffer (25 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, protease inhibitors, 1 mM DTT) and resolved using Tris-acetate NuPAGE gel electorphoresis (Invitrogen) followed by western blotting, according to manufacturer’s protocol.
Antibodies
Santa Cruz: anti-RNA pol II (sc-899), anti-TBP (sc-204, sc-273). Bethyl Labs: anti-SETD1A (A300-289A), anti-Menin (A300-105A), anti-ASH2L (A300-112A), anti-RbBP5 (A300-109A), anti-DOT1L (A300-954A), anti-MLL2/TRX2 (A300-113A). Millipore: anti-H3K4me3 (07-473), anti-H3K4me2 (07-030), anti-H3S10Phos (05-817), anti-LSD1 (09-058). Abcam: anti-H3K4me1 (ab8895) and anti-H3 (ab1791). anti-MLL (456, 457, 468, and 473) and anti-ASH1L (296) antibodies were kindly provided by Eli Canaani (Weizmann Institute). Sigma: anti-BrdU (B8434).
Supplementary Material
Acknowledgments
We thank members of the Blobel and Stillman laboratories for insightful discussions; Eli Canaani for kindly providing MLL and ASH1L antibodies; David Spector, Alea Mills, Vincenzo Pirrota, Steve Reiner, Shelley Berger, and Ken Zaret for helpful advice; James Hsieh for providing the GFP-MLL PCDNA3 plasmid. Michael Huebner for assistance with live-cell imaging; Jay Hess for providing MLL −/− MEFs. Anthony Mazurek for assistance with siRNA experiments. We also thank Peter Schacheri and Francis Collins for providing their published ChIP-chip data sets.
This work was supported by a P30 Cancer Center Support Grant CA01520 from the Abramson Family Cancer Center at the University of Pennsylvania. This work was also supported by NIH grants DK58044 and DK54937 (G.A.B). C.R.V was supported by NIH training grant T32 HL007150 and by the Cold Spring Harbor President’s Council.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Caslini C, Alarcon AS, Hess JL, Tanaka R, Murti KG, Biondi A. The amino terminus targets the mixed lineage leukemia (MLL) protein to the nucleolus, nuclear matrix and mitotic chromosomal scaffolds. Leukemia. 2000;14:1898–1908. doi: 10.1038/sj.leu.2401933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G, Paro R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell. 1998;93:505–518. doi: 10.1016/s0092-8674(00)81181-2. [DOI] [PubMed] [Google Scholar]
- Christova R, Oelgeschlager T. Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat Cell Biol. 2002;4:79–82. doi: 10.1038/ncb733. [DOI] [PubMed] [Google Scholar]
- Demers C, Chaturvedi CP, Ranish JA, Juban G, Lai P, Morle F, Aebersold R, Dilworth FJ, Groudine M, Brand M. Activator-mediated recruitment of the MLL2 methyltransferase complex to the beta-globin locus. Mol Cell. 2007;27:573–584. doi: 10.1016/j.molcel.2007.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Egli D, Birkhoff G, Eggan K. Mediators of reprogramming: transcription factors and transitions through mitosis. Nat Rev Mol Cell Biol. 2008;9:505–516. doi: 10.1038/nrm2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennas MG, Sorio C, Greim R, Nieddu M, Scarpa A, Orlandini S, Croce CM, Fey GH, Marschalek R. The human ALL-1/MLL/HRX antigen is predominantly localized in the nucleus of resting and proliferating peripheral blood mononuclear cells. Cancer Res. 1997;57:2035–2041. [PubMed] [Google Scholar]
- Ernst P, Mabon M, Davidson AJ, Zon LI, Korsmeyer SJ. An Mll-dependent Hox program drives hematopoietic progenitor expansion. Curr Biol. 2004;14:2063–2069. doi: 10.1016/j.cub.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Francis NJ, Follmer NE, Simon MD, Aghia G, Butler JD. Polycomb Proteins Remain Bound to Chromatin and DNA during DNA Replication In Vitro. Cell. 2009 doi: 10.1016/j.cell.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE. Mechanisms of transcriptional memory. Nat Rev Mol Cell Biol. 2001;2:409–421. doi: 10.1038/35073039. [DOI] [PubMed] [Google Scholar]
- Gregory GD, Vakoc CR, Rozovskaia T, Zheng X, Patel S, Nakamura T, Canaani E, Blobel GA. Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes. Mol Cell Biol. 2007;27:8466–8479. doi: 10.1128/MCB.00993-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, Young RA. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci U S A. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- Hsieh JJ, Cheng EH, Korsmeyer SJ. Taspase1: a threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell. 2003;115:293–303. doi: 10.1016/s0092-8674(03)00816-x. [DOI] [PubMed] [Google Scholar]
- Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Hayes DN, Shanmugam KS, Bhattacharjee A, Biondi CA, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- Ji X, Li W, Song J, Wei L, Liu XS. CEAS: cis-regulatory element annotation system. Nucleic Acids Res. 2006;34:W551–554. doi: 10.1093/nar/gkl322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jude CD, Climer L, Xu D, Artinger E, Fisher JK, Ernst P. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1:324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouskouti A, Talianidis I. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. Embo J. 2005;24:347–357. doi: 10.1038/sj.emboj.7600516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Lee JH, Skalnik DG. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A Histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol Cell Biol. 2008;28:609–618. doi: 10.1128/MCB.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Norman J, Shilatifard A, Shiekhattar R. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell. 2007;128:877–887. doi: 10.1016/j.cell.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Lim DA, Huang YC, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, Alvarez-Buylla A. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;458:529–533. doi: 10.1038/nature07726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Cheng EH, Hsieh JJ. Bimodal degradation of MLL by SCFSkp2 and APCCdc20 assures cell cycle execution: a critical regulatory circuit lost in leukemogenic MLL fusions. Genes Dev. 2007;21:2385–2398. doi: 10.1101/gad.1574507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balbas MA, Dey A, Rabindran SK, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- Maurange C, Paro R. A cellular memory module conveys epigenetic inheritance of hedgehog expression during Drosophila wing imaginal disc development. Genes Dev. 2002;16:2672–2683. doi: 10.1101/gad.242702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelotti EF, Sanford S, Levens D. Marking of active genes on mitotic chromosomes. Nature. 1997;388:895–899. doi: 10.1038/42282. [DOI] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- Milne TA, Dou Y, Martin ME, Brock HW, Roeder RG, Hess JL. MLL associates specifically with a subset of transcriptionally active target genes. Proc Natl Acad Sci U S A. 2005;102:14765–14770. doi: 10.1073/pnas.0503630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra BP, Ansari KI, Mandal SS. Dynamic association of MLL1, H3K4 trimethylation with chromatin and Hox gene expression during the cell cycle. Febs J. 2009;276:1629–1640. doi: 10.1111/j.1742-4658.2009.06895.x. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- Prasad R, Yano T, Sorio C, Nakamura T, Rallapalli R, Gu Y, Leshkowitz D, Croce CM, Canaani E. Domains with transcriptional regulatory activity within the ALL1 and AF4 proteins involved in acute leukemia. Proc Natl Acad Sci U S A. 1995;92:12160–12164. doi: 10.1073/pnas.92.26.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- Scacheri PC, Davis S, Odom DT, Crawford GE, Perkins S, Halawi MJ, Agarwal SK, Marx SJ, Spiegel AM, Meltzer PS, Collins FS. Genome-wide analysis of menin binding provides insights into MEN1 tumorigenesis. PLoS Genet. 2006;2:e51. doi: 10.1371/journal.pgen.0020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28:2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terranova R, Agherbi H, Boned A, Meresse S, Djabali M. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc Natl Acad Sci U S A. 2006;103:6629–6634. doi: 10.1073/pnas.0507425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S, Chabes AL, Wysocka J, Herr W. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol Cell. 2007;27:107–119. doi: 10.1016/j.molcel.2007.05.030. [DOI] [PubMed] [Google Scholar]
- Viegas MH, Gehring NH, Breit S, Hentze MW, Kulozik AE. The abundance of RNPS1, a protein component of the exon junction complex, can determine the variability in efficiency of the Nonsense Mediated Decay pathway. Nucleic Acids Res. 2007;35:4542–4551. doi: 10.1093/nar/gkm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol. 2008;28:7337–7344. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H, Vanderford NL, Sarge KD. The TBP-PP2A mitotic complex bookmarks genes by preventing condensin action. Nat Cell Biol. 2008;10:1318–1323. doi: 10.1038/ncb1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14:36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- Zeng PY, Vakoc CR, Chen ZC, Blobel GA, Berger SL. In vivo dual cross-linking for identification of indirect DNA-associated proteins by chromatin immunoprecipitation. Biotechniques 41, 694. 2006;696:698. doi: 10.2144/000112297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.