Abstract
We demonstrated that a yeast deletion mutant in IPT1 and SKN1, encoding proteins involved in biosynthesis of mannosyldiinositolphosphoryl ceramides, is characterized by increased autophagy and DNA fragmentation upon nitrogen starvation as compared to the single deletion mutants or wild type (WT). Apoptotic features were not significantly different between single and double deletion mutants upon nitrogen starvation, pointing to increased autophagy in the double Δipt1 Δskn1 deletion mutant independent of apoptosis. We observed increased basal levels of phytosphingosine in membranes of the double Δipt1 Δskn1 deletion mutant as compared to the single deletion mutants or WT. These data point to a negative regulation of autophagy by both Ipt1 and Skn1 in yeast, with a putative involvement of phytosphingosine in this process.
Keywords: autophagy, DNA fragmentation, apoptosis, sphingolipid
Introduction
We previously demonstrated that biosynthesis of the sphingolipid class of mannosyldiinositolphosphoryl ceramides (M(IP)2C) in yeast depends on nutrient conditions (Im et al., 2003; Thevissen et al., 2005). Skn1 and Ipt1 in yeast are both involved in the biosynthesis of M(IP)2C (Dickson et al., 1997; Thevissen et al., 2005). When grown in nutrient rich media, Δipt1 and Δskn1 single and double deletion mutants are characterized by membranes devoid of M(IP)2C (Dickson et al., 1997; Thevissen et al., 2005). However, when grown under nutrient limitation in half strength potato dextrose broth (PDB), the single deletion mutants Δipt1 and Δskn1 show reappearance of M(IP)2C in their membranes, whereas M(IP)2C is completely absent in membranes of the double Δipt1 Δskn1 deletion mutant grown under these conditions (Im et al., 2003; Thevissen et al., 2005). These data point to SKN1- or IPT1- dependent M(IP)2C biosynthesis in a Δipt1 or Δskn1 deletion mutant, respectively, upon nutrient depletion, whereas in the double Δipt1 Δskn1 deletion mutant, M(IP)2C biosynthesis is blocked independent of medium composition. Hence, nutrient conditions influence biosynthesis of M(IP)2C in yeast.
Autophagy is a catabolic membrane-trafficking phenomenon that occurs in response to dramatic changes in the nutrients available to yeast cells, for example during starvation for nitrogen or carbon (Abeliovich and Klionsky, 2001). Although autophagy and M(IP)2C content of yeast membranes seem both responsive to nutritional stress, a direct link between these processes has not been investigated in yeast to date. Hence, the question arises whether Δipt1 or Δskn1 single and double deletion mutants are characterized by an altered autophagic response as compared to the corresponding wild type (WT). Therefore, in this study, we used nitrogen (N) starvation to assess differences in the autophagic response of the different Δipt1 and/or Δskn1 deletion mutants as compared to WT, as well as their sphingolipid profiles and putative induction of apoptosis, which has previously been linked to autophagy (Maiuri et al., 2007; Scott et al., 2007). Since overexpression of autophagy-related protein 1, Atg1, in Drosophila was previously shown to induce autophagy and to cause cell death accompanied by increased DNA fragmentation (Scott et al., 2007), we further assessed DNA fragmentation upon N starvation in all mutants and WT.
Materials and methods
Materials and Microorganisms
Yeast strains used are S. cerevisiae BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and the corresponding Δipt1, Δskn1 (Invitrogen, Carlsbad, CA) mutants and the double Δipt1 Δskn1 deletion mutant (Thevissen et al., 2005), the pho8Δ60::pho8 pho13Δ::kan-lox strain (wild type, YTS158) (Noda et al., 1995) and the corresponding Δatg1, Δipt1, Δskn1, and Δipt1 Δskn1 mutants.
Challenge with N starvation medium
Overnight cultures in YPD medium (1% yeast extract; 2% peptone, 2% glucose) were transferred to SD medium (0.8 g/l CSM, complete amino acid supplement mixture, Bio 101 Systems; 6.5 g/l YNB, yeast nitrogen base; 20 g/l glucose) at a start OD600 = 0.2, grown to exponential phase till OD600 = 0.8, washed twice with SD-N medium (0.17 % YNB w/o ammonium sulfate and amino acids, 2% glucose), and shifted to SD-N medium for 4 h. As a control, cells were also shifted to SD medium after reaching exponential phase.
Assay for monitoring autophagy
For monitoring bulk autophagy, the alkaline phosphatase activity of Pho8Δ60 was carried out as described previously (Klionsky, 2007; Noda et al., 1995). The percentage of autophagy of the different mutants was relative to the WT autophagy level in the different conditions.
Assay for ROS induction and phosphatidylserine externalization
After challenge with SD-N medium, cell numbers were measured (using CASY cell counter), ROS levels were determined (via staining with DHE (dihydroethidium)), and phosphatidylserine externalization of the yeast cultures (via staining with FITC-labeled annexin V in combination with propidium iodide) was quantified using flow cytometry and BD FACSDiva software (Büttner et al., 2007; Madeo et al., 1997). Experiments were repeated at least three times.
Assay for DNA fragmentation
DNA fragmentation following shift to SD-N medium was quantified using flow cytometryand BD FACSDiva software after TUNEL-staining (terminal deoxynucleotidyl transferase biotin-dUTP nick-end labeling) (Büttner et al., 2007; Madeo et al., 1997).
Sphingolipid analysis
For sphingolipid labeling, yeast cultures were incubated in SD-N-inositol containing [3H]myo-inositol (1μCi/mL; American Radiolabelled Chemicals (St. Louis, MO, US)) whereafter sphingolipids were extracted and analysed (Thevissen et al., 2005). Ceramide and sphingoid base analysis was performed as described previously using a sphingolipidomics approach (Aerts et al., 2008; Bielawski et al., 2006). For each condition, experiments were done twice at least in duplicate.
Statistical analysis
Statistical analysis was performed using paired T-test.
Results
Δipt1 Δskn1 mutant is characterized by increased autophagy under N starvation
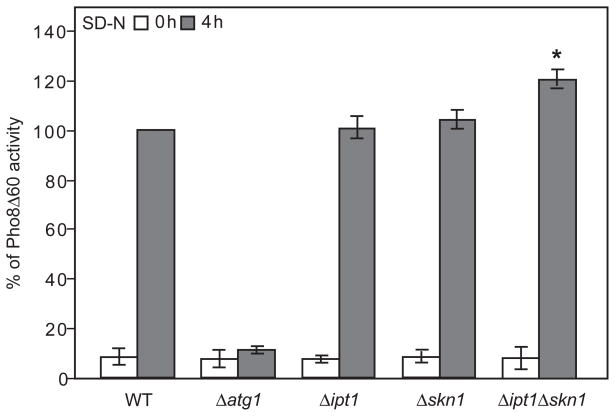
To determine whether induction of autophagy is affected in Δipt1 Δskn1 mutant as compared to the single deletion mutants and WT, we used the Pho8Δ60 assay (Klionsky, 2007). Pho8Δ60 is a truncated variant of the vacuolar alkaline phosphatase Pho8, which lacks the N-terminal transmembrane region that normally allows entry into the endoplasmic reticulum (ER), resulting in accumulation of the mutant protein in the cytosol (Noda et al., 1995). Cytosolic Pho8Δ60 is sequestered as a nonspecific cargo within autophagosomes upon induction of autophagy and delivered into the vacuole, where it is processed into an enzymatically active form due to removal of a C-terminal propeptide. Therefore, the alkaline phosphatase activity of Pho8Δ60 reflects the magnitude of autophagic cargo delivery. To this end, SKN1 and/or IPT1 were deleted in a Pho8Δ60 yeast strain background. Upon challenge with N starvation medium, Δipt1 Δskn1 cells showed a significant 10–20% increase in Pho8Δ60 activity as compared to the single deletion mutants or the corresponding Pho8Δ60 WT (Fig. 1), indicating increased autophagy upon deletion of both IPT1 and SKN1. This is in contrast to the single deletion mutants in IPT1 or SKN1, which did not show significantly increased autophagy as compared to the corresponding WT under conditions of N starvation (Fig. 1). Deletion of ATG1, encoding a protein serine/threonine kinase required for autophagy (Matsuura et al., 1997) served as a negative control in this experiment and showed essentially no increase in Pho8Δ60-dependent alkaline phosphatase activity upon starvation.
Fig. 1.
The Δipt1 Δskn1double mutant is characterized by increased autophagy during nitrogen starvation. Wild type (WT, YTS158), Δatg1, Δipt1, Δskn1 and Δipt1 Δskn1 strains expressing Pho8Δ60 were grown in YPD medium and shifted to SD-N for 4 hours. Samples at the indicated time points were collected, and the cell lysates were assayed for alkaline phosphatase activity. The results represent the mean of three separate experiments, and the error bars indicate the standard deviation. The value for the WT strain at 4 h of starvation was normalized to 100%. *p < 0.05.
N starvation of single and double Δipt1 Δskn1 deletion mutants leads to induction of typical cell death markers
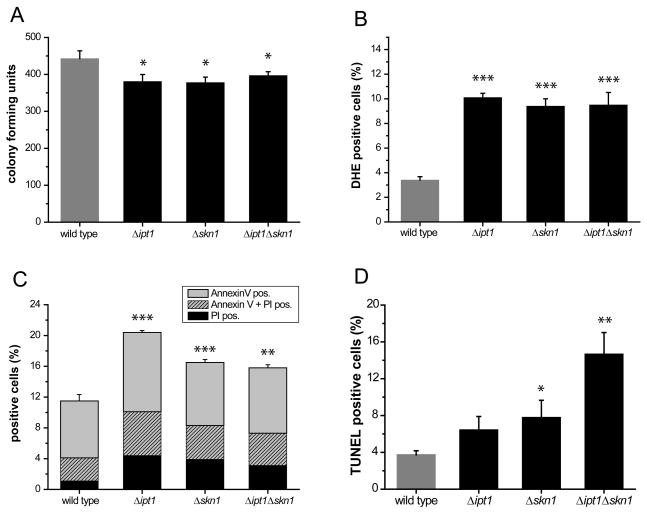
A functional cross-talk exists between autophagy and apoptosis (Maiuri et al., 2007). Upon challenge with N starvation medium, we observed a slight but significant increase in the death rate (10–15 %) of all the deletion mutant strains as compared to WT (Fig. 2A), while no difference was observed when shifting to rich medium (SD) ruling out a survival defect of the mutant strains (data not shown). Additionally, all mutant strains displayed an enhancement of reactive oxygen species (ROS) accumulation (3-fold) as compared to WT (Fig. 2B) upon challenge with N starvation medium, as well as a slight overall increase in the number of early apoptotic (AnnexinV positive, PI negative), late apoptotic (AnnexinV positive, PI positive) and necrotic (only PI positive) cells (Fig. 2C). Thus, the single and double Δipt1 Δskn1 deletion mutants show comparable death rates upon N starvation.
Fig. 2.
The double Δipt1 Δskn1 deletion mutant exhibits enhanced DNA fragmentation under N starvation. (A) survival determined by clonogenicity of WT cells, the Δipt1 and Δskn1 single mutants or the double Δipt1 Δskn1 deletion mutant after N starvation for 4 h during early exponential growth. Data represent mean ± SEM of four independent experiments. *p < 0.05. (B) quantification (FACS analysis) of ROS accumulation using DHE staining for the experiment shown in (A). ***p < 0.001. (C) quantification (FACS analysis) of phosphatidylserine externalization and loss of membrane integrity using annexin V/PI costaining for the experiment shown in (A). (D) quantification (FACS analysis) of DNA fragmentation using TUNEL staining for the experiment shown in (A). *p < 0.05; **p < 0.01. In all experiments, 30,000 cells were evaluated. Data represent mean ± SEM of four independent experiments.
We next assessed the level of DNA fragmentation, a further phenotypic marker of apoptosis in yeast (Madeo et al., 1997). All deletion mutants consistently showed enhanced DNA fragmentation as compared to WT (Fig. 2D). However, the increase in DNA fragmentation obtained for the double Δipt1 Δskn1 deletion mutant (4-fold increase) was strikingly higher than for the single deletion mutants (1.5 to 2-fold increase). This surplus DNA fragmentation may therefore be of non-apoptotic origin and points to a link between autophagy and increased DNA fragmentation, as previously demonstrated in Drosophila upon overexpression of Atg1, where autophagy is induced and causes cell death accompanied by DNA fragmentation (Scott et al., 2007).
Δipt1 Δskn1 mutant has increased levels of sphingoid bases
Nutrient conditions influence biosynthesis of M(IP)2C in yeast (Im et al., 2003; Thevissen et al., 2005). Therefore, we analyzed levels of complex sphingolipids, namely M(IP)2C, mannosylinositolphosphoryl ceramides (MIPC) and inositolphosphoryl ceramides (IPC), in membranes of the single and double Δipt1 Δskn1 deletion mutants and WT under N starvation. Unlike when grown in half strength PDB, there was no detectable M(IP)2C in any of the mutants upon challenge with N starvation medium, whereas the content of MIPC was increased in all mutants as compared to WT (data not shown), as demonstrated previously when these mutants were grown in rich medium (Thevissen et al., 2005). Hence, based on the detection limits of our system, membranes of the single and double deletion mutants were not characterized by different content of complex sphingolipids upon N starvation.
Next, we determined levels of sphingolipid metabolites including α-hydroxy-phytoceramides, dihydroceramides, phytoceramides, dihydrosphingosine and phytosphingosine and corresponding sphingoid base phosphates via sphingolipidomics approach in all mutants and WT upon N starvation (Fig. 3). Although LC/MS analysis of sphingolipid metabolites did not reveal significant differences between the double Δipt1 Δskn1 deletion mutant and the single mutants or WT upon N starvation, it was observed that higher basal levels (without starvation) of phytosphingosine were present in membranes of the double Δipt1 Δskn1 deletion mutant (Fig. 3A) as compared to the single deletion mutants or WT. In addition, the double Δipt1 Δskn1, single Δskn1 deletion mutants, and WT showed significantly increased levels of α-hydroxy-C18:1-phytoceramides upon N starvation as compared to growth without starvation (Fig. 3B) while levels of phytosphingosine-1-phosphate were decreased upon nitrogen starvation (Fig. 3C). In conclusion, membranes of the double Δipt1 Δskn1 deletion mutant are characterized by increased basal levels of phytosphingosine.
Fig. 3.
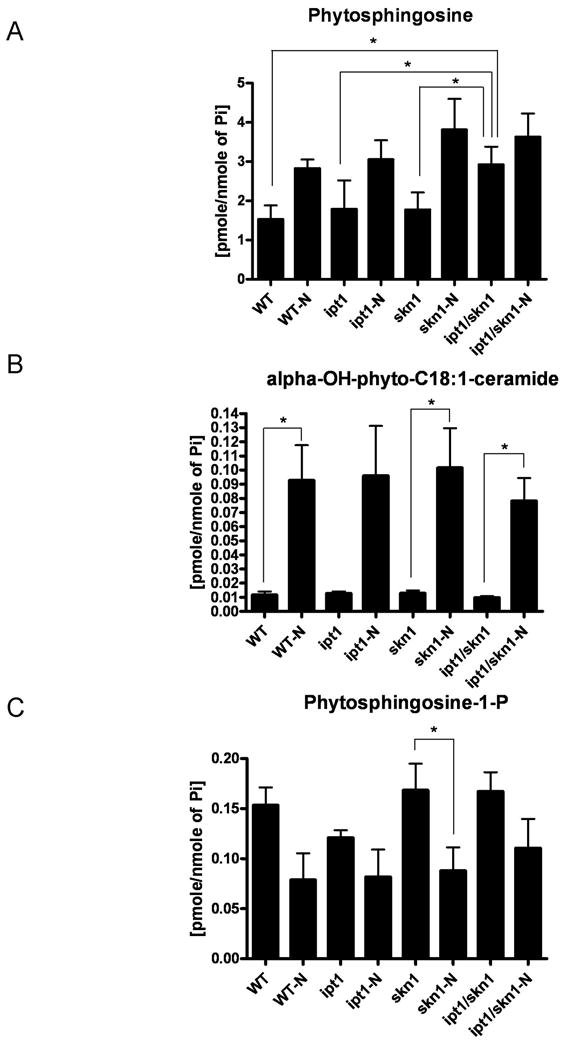
Sphingolipidomics analysis. Profiles of representative sphingolipid metabolites in WT, the single and double Δipt1 Δskn1 deletion mutants under N starvation (-N) or without N starvation. Levels of (A) phytosphingosine, (B) alpha-OH-phyto-C18:1-ceramide, (C) phytosphingosine-P were normalized to total phosphate. *p < 0.05.
Discussion
This study reports on the increased induction of autophagy upon N starvation in a double Δipt1 Δskn1 deletion mutant of yeast as compared to the single deletion mutants or WT. Apoptotic features were slightly increased in the single and double Δipt1 Δskn1 deletion mutants as compared to WT upon N starvation, but there was no significant difference between single and double deletion mutants in this regard, pointing to increased autophagy in the double Δipt1 Δskn1 deletion mutant independent of apoptosis. The double Δipt1 Δskn1 deletion mutant was further characterized by increased DNA fragmentation upon N starvation as compared to the single deletion mutants or WT. This surplus DNA fragmentation seems to be of non-apoptotic origin since apoptotic features of the double Δipt1 Δskn1 deletion mutant were not significantly different from those of single mutants upon N starvation. Hence, these data point to a link between autophagy and increased DNA fragmentation, as previously demonstrated in Drosophila upon overexpression of Atg1 (Scott et al., 2007).
To get more mechanistic insight in the increased autophagy and DNA fragmentation in the double Δipt1 Δskn1 deletion mutant as compared to the single deletion mutants and WT, we focused on putative differences in complex sphingolipids and sphingolipid metabolites in the different yeast strains upon N starvation. In contrast to previous observations for nutrient starvation in half strength PDB media, which induced the presence of M(IP)2C in Δipt1 and Δskn1 single deletion mutants (Im et al., 2003; Thevissen et al., 2005), N starvation did not lead to detectable differences in levels of complex sphingolipids or sphingolipid metabolites in the double Δipt1 Δskn1 deletion mutant as compared to the single deletion mutants or WT. Interestingly, higher basal levels of the sphingoid base phytosphingosine were observed in the double Δipt1 Δskn1 mutant as compared to the single deletion mutants or WT. Treatment of Pho8Δ60 yeast cells with the ceramide synthase inhibitor fumonisin B1, resulting in the accumulation of sphingoid bases, resulted in a slight but reproducible increase in alkaline phophatase activity under starvation conditions (data not shown). All these data point to a putative role for sphingoid bases in the induction of autophagy and/or DNA fragmentation in yeast. Up till now, there are no reports on a link between sphingolipids or sphingolipid metabolism and autophagy or DNA fragmentation in yeast. In mammals, however, few reports highlight the link between the sphingolipid rheostat and autophagy (Lavieu et al., 2007, 2008). The sphingolipid rheostat in mammals is composed of the relative levels of sphingolipids and their metabolites, namely ceramide (Cer), sphingosine (Sph), and sphingosine-1-phosphate (S1P). In mammalian cells, both ceramide and S1P stimulate autophagy (Lavieu et al., 2008), whereas in yeast, it seems that non-phosphorylated sphingoid bases might stimulate autophagy as documented in this study.
An important signaling pathway involved in the regulation of autophagy is the Ras/PKA pathway (Budovskaya et al., 2004). Inactivation of the Ras/PKA pathway, by overexpression of a dominant negative allele of RAS2, known as RAS2ala22, resulted in increased induction of autophagy as compared to WT. However, additional inactivation of the genes encoding the PKA catalytic subunits, TPK1, TPK2 and TPK3, in the double Δipt1 Δskn1 deletion mutant did not result in an enhanced autophagy phenotype (data not shown) as compared to the double Δipt1 Δskn1 deletion mutant, indicating that Skn1 together with Ipt1 might act in the same pathway as Ras/PKA regarding induction/regulation of autophagy. Moreover, PKA and Sch9 signaling pathways are known to regulate autophagy cooperatively in yeast (Yorimitsu et al., 2007). Long-chain bases including phytosphingosine are recognized as regulators of AGC-type protein kinase (where AGC stands for protein kinases A, G and C) Pkh1 and Pkh2, which are homologues of mammalian phosphoinositide-dependent protein kinase 1 (Sun et al., 2000). Based on in vitro data, Liu and coworkers demonstrated that phytosphingosine stimulates Pkh1 to activate additional downstream kinases including Ypk1, Ypk2 and Sch9, and additionally, that phytosphingosine can directly activate Ypk1, Ypk2 and Sch9 (Liu et al., 2005a, 2005b). In conclusion, it could be that the higher basal levels of phytosphingosine, which we observed in the double Δipt1 Δskn1 mutant, affect Sch9 function directly or indirectly, and concommitantly, the authophagy response. Hence, future research will be directed to determine if Sch9 or other kinases are part of the link between sphingolipids and autophagy in yeast.
In conclusion, all the data obtained in this study point to a negative regulation of autophagy by both Ipt1 and Skn1 in yeast, which could be mediated by sphingoid bases and might act in the same pathway as Ras/PKA signaling pathway. Apparently, Ipt1 and Skn1 can functionally complement each other under nutrient limitation, not only regarding synthesis of the complex sphingolipid M(IP)2C upon nutrient limitation in half strength PDB (Thevissen et al., 2005), but also regarding the negative regulation of autophagy under N starvation, as demonstrated in this study.
Acknowledgments
This work was supported by a grant from FWO-Vlaanderen (research project G.0440.07) to B.P.A.C. Postdoctoral fellowships to A.M.A. (Research Council) and to K.T. (Industrial Research Found), both from K.U. Leuven, are gratefully acknowledged. F.M. and D.C.G. are grateful to the FWF for SFB “Lipotox” and NRN S-9304-B05. Lipidomics CORE at the Medical University of South Carolina (USA) is supported by NIH Grant No. C06 RR018823. D.J.K. is supported by National Institutes of Health Public Health Service grant GM53396.
Abbreviations
- PDB
potato dextrose broth
- M(IP)2C
mannosyldiinositolphosphoryl ceramides
- WT
wild type
- N
nitrogen
References
- Abeliovich H, Klionsky DJ. Autophagy in yeast: mechanistic insights and physiological function. Microbiol Mol Biol Rev. 2001;65:463–479. doi: 10.1128/MMBR.65.3.463-479.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aerts AM, Zabrocki P, François IE, et al. Ydc1p ceramidase triggers organelle fragmentation, apoptosis and accelerated ageing in yeast. Cell Mol Life Sci. 2008;65:1933–1942. doi: 10.1007/s00018-008-8129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielawski J, Szulc ZM, Hannun YA, Bielawska A. Simultaneous quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods. 2006;39:82–91. doi: 10.1016/j.ymeth.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Budovskaya YV, Stephan JS, Reggiori F, Klionsky DJ, Herman PK. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J Biol Chem. 2004;279:20663–20671. doi: 10.1074/jbc.M400272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner S, Eisenberg T, Carmona-Gutierrez D, et al. Endonuclease G regulates budding yeast life and death. Mol Cell. 2007;25:233–246. doi: 10.1016/j.molcel.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Dickson RC, Nagiec EE, Wells GB, Nagiec MM, Lester RL. Synthesis of mannose-(inositol-P)2-ceramide, the major sphingolipid in Saccharomyces cerevisiae, requires the IPT1 (YDR072c) gene. J Biol Chem. 1997;272:29620–29625. doi: 10.1074/jbc.272.47.29620. [DOI] [PubMed] [Google Scholar]
- Im YJ, Idkowiak-Baldys J, Thevissen K, Cammue BP, Takemoto JY. IPT1-independent sphingolipid biosynthesis and yeast inhibition by syringomycin E and plant defensin DmAMP1. FEMS Microbiol Lett. 2003;223:199–203. doi: 10.1016/S0378-1097(03)00375-6. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ. Monitoring Autophagy in Yeast: The Pho8Delta60 Assay. Methods Mol Biol. 2007;390:363–372. doi: 10.1007/978-1-59745-466-7_24. [DOI] [PubMed] [Google Scholar]
- Lavieu G, Scarlatti F, Sala G, Carpentier S, Levade T, Ghidoni R, Botti J, Codogno P. Sphingolipids in macroautophagy. Methods Mol Biol. 2008;445:159–173. doi: 10.1007/978-1-59745-157-4_11. [DOI] [PubMed] [Google Scholar]
- Lavieu G, Scarlatti F, Sala G, Levade T, Ghidoni R, Botti J, Codogno P. Is autophagy the key mechanism by which the sphingolipid rheostat controls the cell fate decision? Autophagy. 2007;3:45–47. doi: 10.4161/auto.3416. [DOI] [PubMed] [Google Scholar]
- Liu K, Zhang X, Lester RL, Dickson RC. The sphingoid long chain base phytosphingosine activates AGC-type protein kinases in Saccharomyces cerevisiae including Ypk1, Ypk2, and Sch9. J Biol Chem. 2005;280:22679–22687. doi: 10.1074/jbc.M502972200. [DOI] [PubMed] [Google Scholar]
- Liu K, Zhang X, Sumanasekera C, Lester RL, Dickson RC. Signalling functions for sphingolipid long-chain bases in Saccharomyces cerevisiae. Biochem Soc Trans. 2005;33:1170–1173. doi: 10.1042/BST20051170. [DOI] [PubMed] [Google Scholar]
- Madeo F, Fröhlich E, Fröhlich KU. A yeast mutant showing diagnostic markers of early and late apoptosis. J Cell Biol. 1997;139:729–734. doi: 10.1083/jcb.139.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- Scott RC, Juhász G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Taniguchi R, Tanoue D, Yamaji T, Takematsu H, Mori K, Fujita T, Kawasaki T, Kozutsumi Y. Sli2 (Ypk1), a homologue of mammalian protein kinase SGK, is a downstream kinase in the sphingolipid-mediated signaling pathway of yeast. Mol Cell Biol. 2000;20:4411–4419. doi: 10.1128/mcb.20.12.4411-4419.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevissen K, Idkowiak-Baldys J, Im YJ, et al. SKN1, a novel plant defensin-sensitivity gene in Saccharomyces cerevisiae, is implicated in sphingolipid biosynthesis. FEBS Lett. 2005;579:1973–1977. doi: 10.1016/j.febslet.2005.02.043. [DOI] [PubMed] [Google Scholar]
- Yorimitsu T, Zaman S, Broach JR, Klionsky DJ. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:4180–4189. doi: 10.1091/mbc.E07-05-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]