Abstract
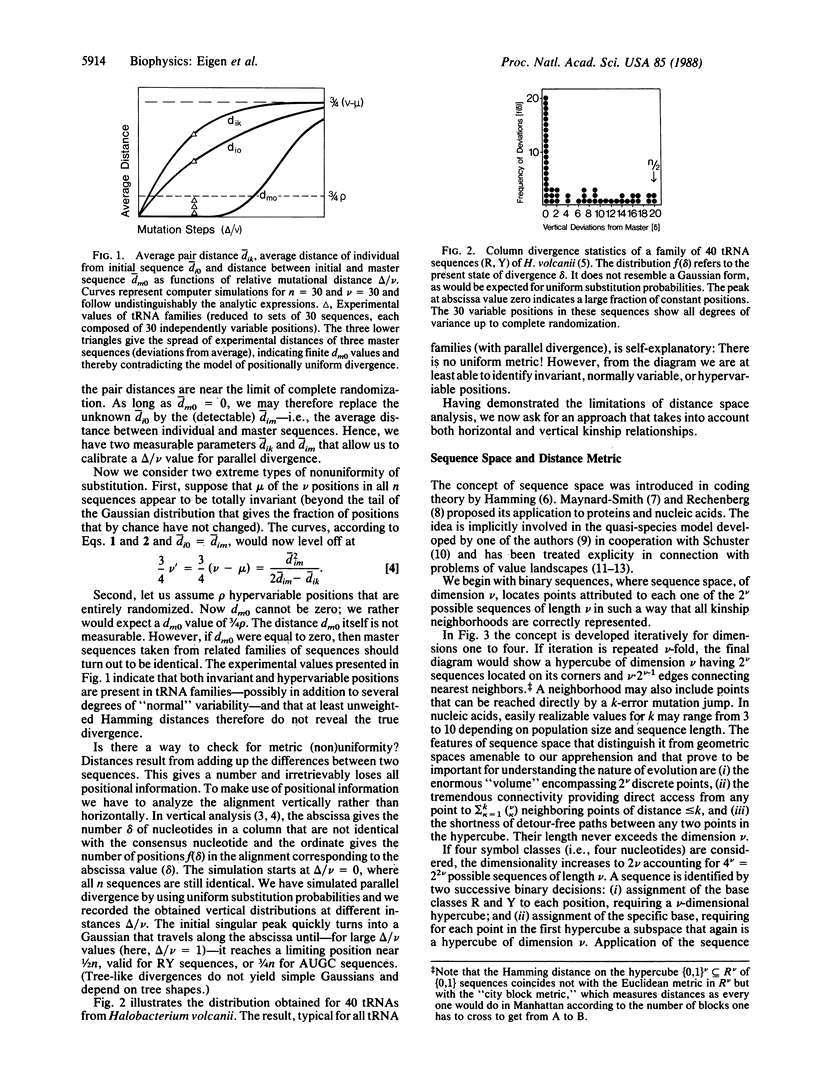
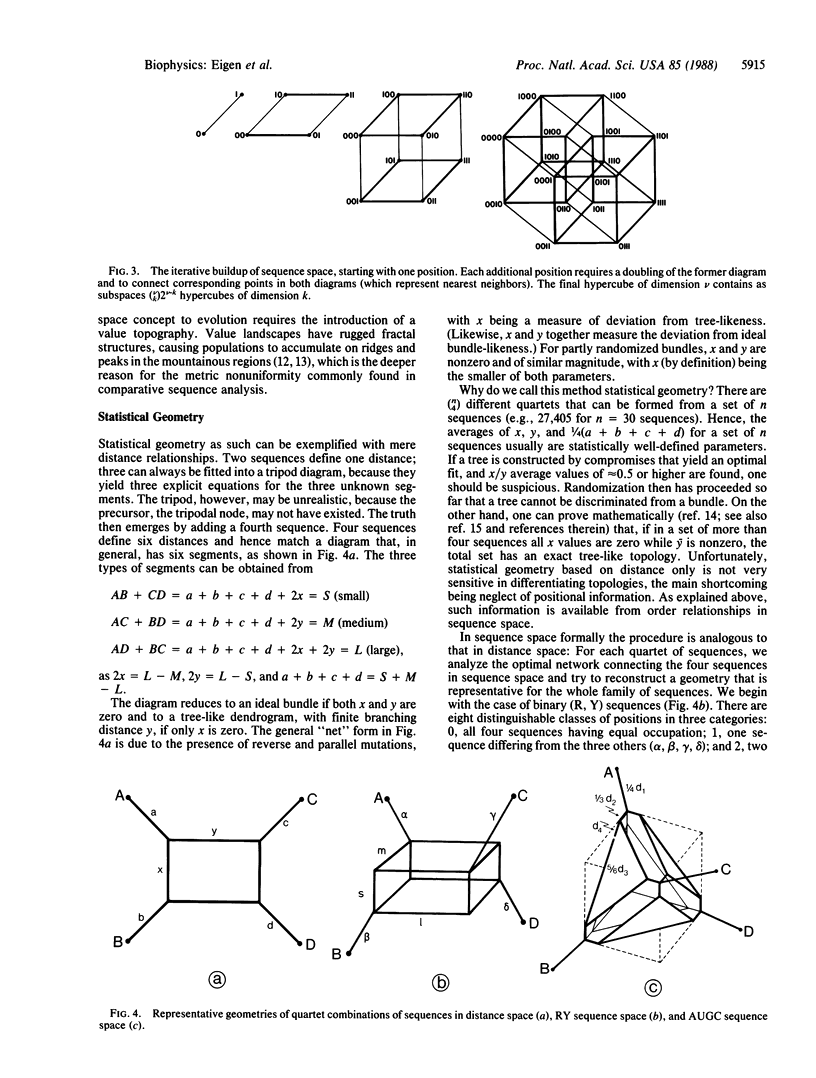
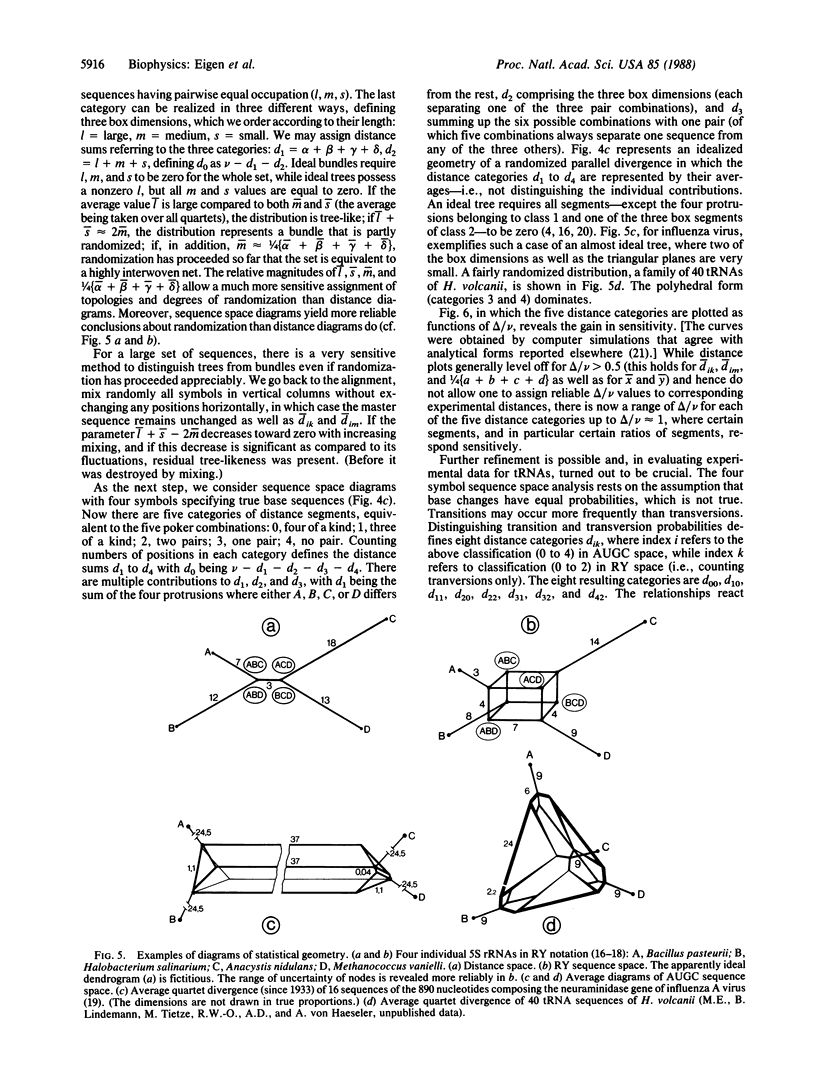
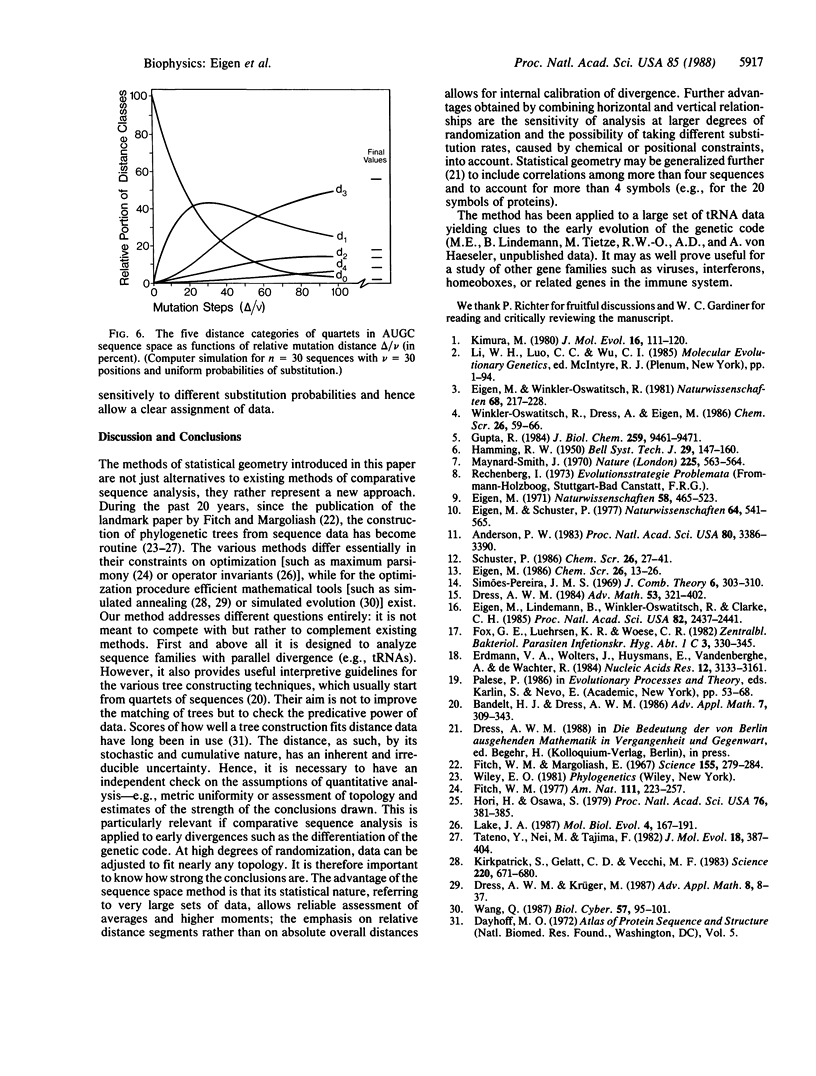
A statistical method of comparative sequence analysis that combines horizontal and vertical correlations among aligned sequences is introduced. It is based on the analysis mainly of quartet combinations of sequences considered as geometric configurations in sequence space. Numerical invariants related to relative internal segment lengths are assigned to each such configuration and statistical averages of these invariants are established. They are used for internal calibration of the topology of divergence and for quantitative determination of the noise level. Comparison of computer simulations with experimental data reveals the high sensitivity of assignment of basic topologies even if much randomized. In addition, these procedures are checked by vertical analysis of the aligned sequences to allow the study of divergences with positionally varying substitution probabilities.
Full text
PDF
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson P. W. Suggested model for prebiotic evolution: the use of chaos. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3386–3390. doi: 10.1073/pnas.80.11.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigen M., Lindemann B., Winkler-Oswatitsch R., Clarke C. H. Pattern analysis of 5S rRNA. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2437–2441. doi: 10.1073/pnas.82.8.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigen M., Schuster P. The hypercycle. A principle of natural self-organization. Part A: Emergence of the hypercycle. Naturwissenschaften. 1977 Nov;64(11):541–565. doi: 10.1007/BF00450633. [DOI] [PubMed] [Google Scholar]
- Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971 Oct;58(10):465–523. doi: 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- Eigen M., Winkler-Oswatitsch R. Transfer-RNA: the early adaptor. Naturwissenschaften. 1981 May;68(5):217–228. doi: 10.1007/BF01047323. [DOI] [PubMed] [Google Scholar]
- Fitch W. M., Margoliash E. Construction of phylogenetic trees. Science. 1967 Jan 20;155(3760):279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- Gupta R. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J Biol Chem. 1984 Aug 10;259(15):9461–9471. [PubMed] [Google Scholar]
- Hori H., Osawa S. Evolutionary change in 5S RNA secondary structure and a phylogenic tree of 54 5S RNA species. Proc Natl Acad Sci U S A. 1979 Jan;76(1):381–385. doi: 10.1073/pnas.76.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980 Dec;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick S., Gelatt C. D., Jr, Vecchi M. P. Optimization by simulated annealing. Science. 1983 May 13;220(4598):671–680. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- Lake J. A. A rate-independent technique for analysis of nucleic acid sequences: evolutionary parsimony. Mol Biol Evol. 1987 Mar;4(2):167–191. doi: 10.1093/oxfordjournals.molbev.a040433. [DOI] [PubMed] [Google Scholar]
- Smith J. M. Natural selection and the concept of a protein space. Nature. 1970 Feb 7;225(5232):563–564. doi: 10.1038/225563a0. [DOI] [PubMed] [Google Scholar]
- Tateno Y., Nei M., Tajima F. Accuracy of estimated phylogenetic trees from molecular data. I. Distantly related species. J Mol Evol. 1982;18(6):387–404. doi: 10.1007/BF01840887. [DOI] [PubMed] [Google Scholar]



