Summary
Mutations in the enzyme cytosolic isocitrate dehydrogenase 1 (IDH1) are a common feature of a major subset of primary human brain cancers. These mutations occur at a single amino acid residue of the IDH1 active site resulting in loss of the enzyme’s ability to catalyze conversion of isocitrate to α-ketoglutarate. However, only a single copy of the gene is mutated in tumors, raising the possibility that the mutations do not result in a simple loss of function. Here we show that cancer-associated IDH1 mutations result in a new ability of the enzyme to catalyze the NADPH-dependent reduction of α-ketoglutarate to R(−)-2-hydroxyglutarate (2HG). Structural studies demonstrate that when R132 is mutated to histidine, residues in the active site are shifted to produce structural changes consistent with reduced oxidative decarboxylation of isocitrate and acquisition of the ability to convert α-ketoglutarate to 2HG. Excess accumulation of 2HG has been shown to lead to an elevated risk of malignant brain tumors in patients with inborn errors of 2HG metabolism. Similarly, in human malignant gliomas harboring IDH1 mutations, we find dramatically elevated levels of 2HG. These data demonstrate that the IDH1 mutations result in production of the onco-metabolite 2HG, and suggest that the excess 2HG which accumulates in vivo contributes to the formation and malignant progression of gliomas.
Mutations in the enzyme cytosolic isocitrate dehydrogenase 1 (IDH1) are found in approximately 80% of grade II-III gliomas and secondary glioblastomas in humans 1–3. These mutations occur at a single amino acid residue of IDH1, arginine 132, which is most commonly mutated to histidine (R132H)1, 3, 4. Only a single copy of the gene has been found to be mutated in tumors 1–6. Many of the high grade gliomas with IDH1 mutations are secondary glioblastomas that have progressed from lower grade lesions 1–3, 5. When analyzed in relation to other genes implicated in brain tumors, the compiled evidence suggests that IDH1 is often the first mutation that occurs 2. While these findings suggest that IDH1 mutations are selected for early during tumorigenesis, why mutations in a single allele of IDH1 result in predilection for malignant progression is uncertain. It has been reported that the R132H mutation disrupts the ability of IDHl to convert isocitrate to αKG 3, 7, but the consequences of this impaired enzymatic activity on cellular metabolism have not been systematically analyzed. For example, while R132 IDH1 mutations might reduce the rate of cytosolic αKG production as suggested by others 7, whether IDH1 mutations can influence the enzyme’s ability to act on αKG as a substrate has not been explored. This latter activity may be particularly important for the tumorigenic role of IDH1 mutations since cytosolic αKG is in equilibrium via transamination with the glutamate that plays a unique role in glial cell physiology, and IDH1 mutations are especially prevalent in malignant gliomas.
Metabolite profiling identifies elevated 2HG levels in IDH1 mutant expressing cells
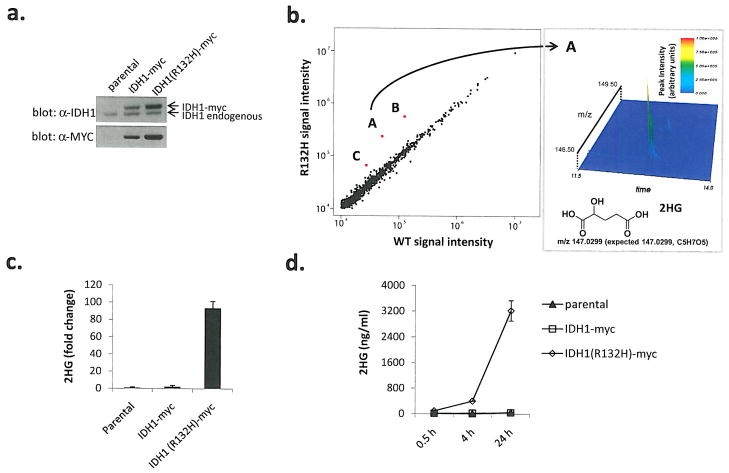
To understand the impact of IDH1 mutation on cellular metabolism, we profiled metabolites to identify changes in metabolite levels in cells expressing R132 mutant IDH1 compared with cells expressing wild-type IDH1. To initiate these studies, we stably transfected U87MG glioblastoma cells, which are wild-type for IDH1, with myc-tagged wild-type or R132H mutant IDH1. Cells expressing either myc-tagged wild-type or mutant IDH1 were used for metabolite profiling experiments (Figure 1a). Metabolites extracted from exponentially growing cells were profiled by liquid chromatography - electrospray ionization - mass spectrometry (LC-MS). In initial survey analyses, full-scan LC-MS in negative ion mode (exact mass) was used to examine differences in metabolite species with an m/z between 110 and 1000. Relative quantitative data were collected for approximately 850 ions and identities were proposed by comparison with known human metabolites. Identities of > 100 species identified by a combination of exact mass and retention time match to purified standards were assigned 8. There were no significant differences between cells expressing wild-type IDH1 when compared with parental cells. The levels of most observed ions were also similar between wild type and R132H mutant IDH1 expressing cells (Figure 1b), with no significant changes found in canonical TCA cycle species (P > 0.05 for citrate, isocitrate, αKG, succinate, fumarate, and malate). However, three species were significantly more abundant in R132H mutant IDH1 expressing cells (P < 0.001 for each). The mass of the one of these ions matched precisely to 2HG (expected m/z 147.0299, measured 147.0299). The other ions co-eluted with 2HG, and had masses consistent with the sodium adduct and a dehydrated form of 2HG. Subsequent injection of 2HG standard confirmed a retention time match to the biological peak, and that it forms all three of the observed ions during LC-MS ionization (data not shown). The cellular accumulation of 2HG was quantified by targeted triple-quadrupole LC-MS-MS analysis of cell extracts (Figure 1c). The structure of 2HG is close to αKG, the product of the IDH1 enzyme. Thus, the sole metabolite identified by untargeted metabolite profiling to be markedly altered by R132H mutant IDH1 expression was also implicated by its structure to be IDHl-related.
Figure 1. Cells expressing human R132H IDH1 contain dramatically elevated levels of 2HG.
(a) Western blots for myc-tagged human isocitrate dehydrogenase 1 (IDH1-myc) or R132H mutant (R132H-myc) in stably transfected U87MG human glioblastoma cells, (b) Metabolite profiles from cells expressing R132H IDH1 or WT IDH1 detected by LC-MS scanning for species between 110–1000 m/z (M-H+). Red spots labeled A, B, and C represent species assigned to 2HG, dehydro-2HG, and 2HG-sodium adduct respectively. Spectrometric details supporting the identification of species “A” as 2HG are shown in the right panel, (c) Cells expressing R132H IDH1 contain elevated levels of 2HG. Data were normalized by cell number and expressed as fold difference relative to parental values. Error bars depict one standard deviation (SD) from the mean of 3 independent experiments, (d) Cells expressing R132H IDH1 display time-dependent accumulation of 2HG in cell culture media, normalization was as described in (c). Errors bars depict one SD from the mean of four independent experiments.
The accumulation of 2HG was not restricted to cell extracts as 2HG was found to rapidly accumulate in the medium of cells expressing R132H mutant IDH1 (Figure 1d). No appreciable 2HG could be found in the medium of wild-type cells or cells transfected with wild-type IDH1 (Figure 1d). Isotope-labeling experiments on whole cells using uniformly labeled 13C-glutamine as a culture media nutrient demonstrated that the carbons in 2HG are derived from glutamine, with reasonably high overall pathway flux from glutamine through glutamate and αKG to 2HG. Moreover, labeling experiments did not demonstrate any other major alterations in central carbon metabolic flux in cells expressing R132H mutant 2HG (data not shown). The presence of a myc epitope tag did not alter activity of R132H mutant IDH1. Despite being expressed at lower levels than the myc-tagged R132H IDH1, cells transfected with untagged R132H IDH1 demonstrated a comparable increase in 2HG production (Supplementary Figure 1). To determine whether 2HG production in cells expressing R132H mutant IDH1 is unique to U87MG cells, we stably expressed wild-type and R132H mutant IDH1 in wild-type IDHl-expressing LN-18 glioblastoma cells (Supplementary Figure 2a). Similar to results obtained with U87MG cells, the major difference in metabolite levels observed in LN-18 cells expressing R132H mutant IDH1 was an increased level of 2HG (Supplementary Figure 2b).
Mutant IDH1 directly converts αKG to 2HG
The R132H mutation has been reported to result in loss of function for enzyme activity3, 7. However, in these studies only the NADP+-dependent oxidative decarboxylation of isocitrate to αKG was assessed. To understand how IDH1 activity is altered in cells by the presence of R132H mutant IDH1, we expressed increasing amounts of wild-type and R132H mutant IDH1 separately or in combination and assessed isocitrate-dependent NADPH production and αKG-dependent NADPH consumption in cell lysates. Consistent with published results, expression of R132H mutant IDH1 resulted in no measurable production of NADPH from isocitrate, and isocitrate-dependent NADPH production increased with increasing amounts of wild-type enzyme (Supplementary Figure 3a, b). The ability of the wild-type enzyme to generate NADPH was decreased slightly by co-expression of the R132H mutant IDH1. Opposite results were obtained, however, when NADPH consumption was measured in the presence of αKG. NADPH consumption by wild-type enzyme was not observed, while R132H mutant IDH1 expression resulted in αKG-dependent NADPH consumption (Supplementary Figure 3c). Although the overall consumption of NADPH was slow, if anything co-expression of wild-type IDH1 with R132H mutant IDH1 facilitated the αKG-dependent consumption of NADPH. These findings demonstrate that in contrast to wild-type IDH1, R132H mutant IDH1 promotes an NADPH-dependent reduction of αKG. Furthermore, as this reduction was not inhibited by co-expression of wild-type IDH1, these data suggest that the novel activity of mutant IDH1 can persist even in the presence of a wild-type IDH1 allele. In fact, it is possible that in the case of a heterodimer of wild-type and mutant IDH1, the αKG and NADPH produced locally by the wild-type subunit could be used as substrates for the mutant subunit explaining the decrease in NADPH production observed in lysates when wild-type and mutant IDH1 are co-expressed.
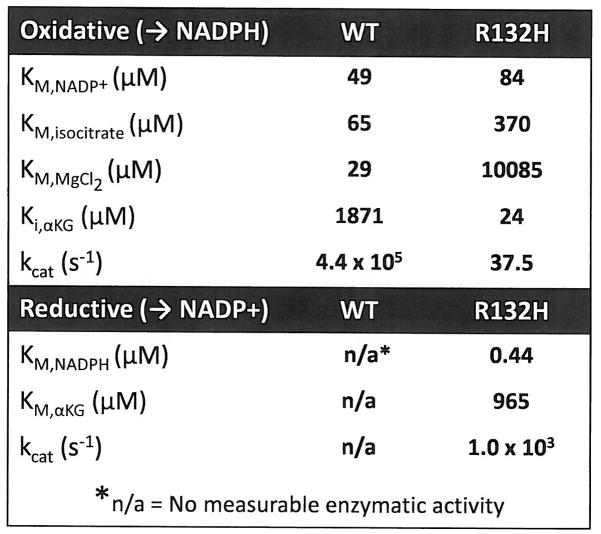
To understand how R132 mutations alter the enzymatic properties of IDH1, wild-type and R132H mutant IDH1 proteins were produced and purified from E coli. When NADP+-dependent oxidative decarboxylation of isocitrate was measured using purified wild-type or R132H mutant IDH1 protein, it was confirmed that R132H mutation impairs the ability of IDH1 to catalyze this reaction3, 7, as evident by the loss in binding affinity for both isocitrate and MgCl2 along with a 1000-fold decrease in catalytic turnover (Figure 2, Supplementary Figure 4a). In contrast, when NADPH-dependent reduction of αKG was assessed using either wild-type or R132H mutant IDH1 protein, only R132H mutant could catalyze this reaction (Figure 2, Supplementary Figure 4b). Part of this increased rate of αKG reduction results from an apparent increase in affinity for both the cofactor NADPH and substrate αKG in the R132H mutant IDH1 (Figure 2). Taken together, these data demonstrate that while the R132H mutation leads to a loss of enzymatic function for oxidative decarboxylation of isocitrate, this mutation also results in a gain of enzyme function for the NADPH-dependent reduction of αKG.
Figure 2. R132H mutation alters the enzymatic properties of IDH1.
Kinetic parameters of oxidative and reductive reactions as measured for WT and R132H IDH1 enzymes are shown. KM and kcat values for the reductive activity of the WT enzyme were unable to be determined as no measurable enzyme activity was detectable at any substrate concentration.
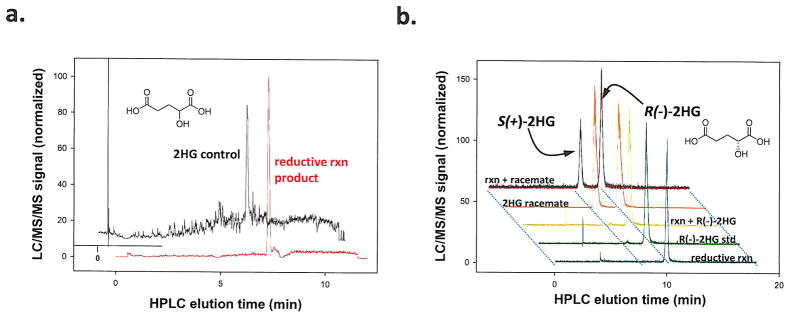
Reduction of the α-ketone in αKG can result in 2HG. To determine whether R132H mutant protein directly produced 2HG from αKG we examined the product of the mutant IDH1 reaction using negative ion mode triple quadrupole electrospray LC-MS. These experiments confirmed that 2HG was the direct product of NADPH-dependent αKG reduction by the purified R132H mutant protein through comparison with known metabolite standards (Figure 3a). Conversion of αKG to isocitrate was not observed. To determine the chirality of the 2HG produced, we derivatized the products of the R132H reaction with diacetyl-L-tartaric anhydride, which allowed us to separate the (S) and (R) enantiomers of 2HG by simple reverse-phase LC and detect the products by tandem mass spectrometry 9 (Figure 3b). The peaks corresponding to the (S) and (R) isomers of 2HG were confirmed using racemic and R(−)-2HG standards. The reaction product from R132H co-eluted with R(−)-2HG peak, demonstrating that the R(−) stereoisomer is the product produced from αKG by R132H mutant IDH1.
Figure 3. R132H mutation in IDH1 results in production of R(−)-2HG.
(a) 2HG was identified as the reductive reaction product of recombinant human R132H mutant IDH1 using LC-MS as shown, (b) The chirality of 2HG produced by R132H mutant IDH1 was assessed as by diacetyl-L-tartaric anhydride derivatization and LC-MS analysis. Normalized LC-MS signal for the reductive reaction (rxn) product alone, an R(−)-2HG standard alone, and the two together (Rxn + R(−)-2HG) are shown as is the signal for a racemic mixture of R(−) and S(+) forms (2HG Racemate) alone or with the reaction products (Rxn + Racemate).
To determine whether the altered enzyme properties resulting from R132H mutation were shared by other R132 mutations found in human gliomas, recombinant R132C, R132L and R132S mutant IDH1 proteins were generated and the enzymatic properties assessed. Similar to R132H mutant protein, R132C, R132L, and R132S mutations all result in a gain-of-function for NADPH-dependent reduction of αKG (Supplementary Figure 4). Thus, in addition to impaired oxidative decarboxylation of isocitrate, one common feature shared among the IDH1 mutations found in human gliomas is the ability to catalyze direct NADPH-dependent reduction of αKG.
Structural studies revealed a distinct active site of R132H mutant IDH1
To define how R132 mutations alter the enzymatic properties of IDH1, the crystal structure of R132H mutant IDH1 bound to αKG, NADPH, and Ca2+ was solved at 2.1 Å resolution (see Supplementary Table 1 for crystallographic data and refinement statistics). The overall quaternary structure of the homodimeric R132H mutant enzyme adopts the same closed catalytically competent conformation (shown as a monomer in Figure 4a) that has been previously described for the wild-type enzyme 10.
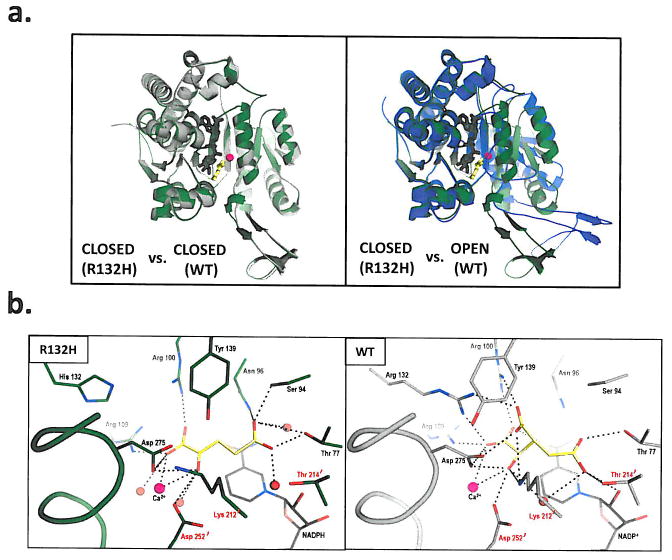
Figure 4. Structural analysis of R132H mutant IDH1.
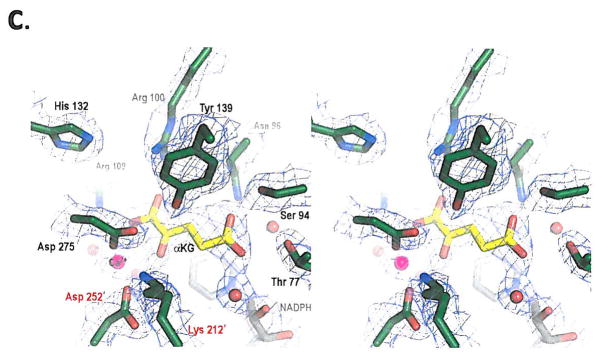
(a) On the left is shown an overlay of R132H mutant IDH1 (green) and WT IDH1 (gray) structures in the ‘closed’ conformation. On the right is shown an overlay of WT IDH1 (blue) structure in the ‘open’ conformation with mutant IDH1 (green) for comparison, (b) Close-up comparison of the R132H IDH1 active site (left) with αKG (yellow) and NADPH (gray) and the WT IDH active-site (right) with isocitrate (yellow) and NADP (gray). Residues coming from the other monomer are denoted with a prime (‘) symbol. In addition to the mutation at residue 132, the major changes are the positions of the catalytic residues Tyr 139 and Lys 212′. (c) Walleyed stereo image showing the composite omit map for αKG, NADPH, calcium ion, His 132 and other key catalytic residues in the R132H mutant active site contoured at 1σ level.
Two important features were noted by the change of R132 to histidine: the effect on conformation equilibrium and the reorganization of the active-site. Located atop a β-sheet in the relatively rigid small domain, R132 acts as a gate-keeper residue and appears to orchestrate the hinge movement between the open and closed conformations. The guanidinium moiety of R132 swings from the open to the closed conformation with a distance of nearly 8 Å. Substitution of histidine for arginine is likely to change the equilibrium in favor of the closed conformation that forms the catalytic cleft for cofactor and substrate to bind efficiently, which partly explains the high-affinity for NADPH exhibited by the R132H mutant enzyme. This feature may be advantageous for the NADPH-dependent reduction of αKG to R(−)-2HG in an environment where NADPH concentrations are low. Secondly, closer examination of the catalytic pocket of the mutant IDH1 structure in comparison to the wild-type enzyme showed not only the expected loss of key salt-bridge interactions between the guanidinium of R132 and the α/β carboxylates of isocitrate, as well as changes in the network that coordinates the metal ion, but also an unexpected reorganization of the active-site. Mutation to histidine resulted in a significant shift in position of the highly conserved residues Y139 from the A subunit and K212′ from the B subunit (Figures 4b & c), both of which are thought to be critical for catalysis by this enzyme family11. In particular, the hydroxyl moiety of Y139 now occupies the space of the β-carboxylate of isocitrate.
The electron density in the active site was not sufficient to assign αKG and its orientation unambiguously. We have modeled the substrate based on the available electron density, taking into consideration the coordination between the carbonyl oxygen of αKG and the calcium ion as well as an orientation of αKG that would produce R(−)-2HG, the experimental product. The model required a significant repositioning of αKG compared to isocitrate, such that the distal carboxylate of αKG now points upward to make new contacts with N96 and S94. Overall, this single R132H mutation results in formation of a distinct active site compared to wild-type IDH1.
2HG levels are elevated in human glioma samples
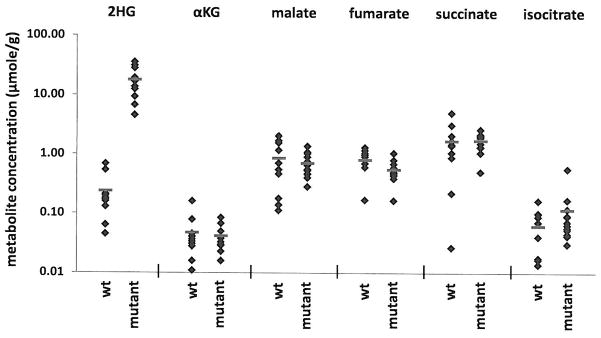
Our data demonstrate that mutation of R132 can result in the ability of IDH1 to generate R(−)-2HG from αKG. To determine if 2HG production is characteristic of tumors harboring mutations in IDH1, metabolites were extracted from human malignant gliomas that were either wild-type or mutant for IDH1 (see Supplementary Table 2 for summary of tumor characteristics). It has been suggested that αKG levels are decreased in cells transfected with mutant IDH17. We observe that the average αKG level from 12 tumor samples harboring various R132 mutations was slightly less than the average αKG level observed in 10 tumors which are wild-type for IDH1. This difference in αKG was not statistically significant, and a range of αKG levels was observed in both wild-type and mutant tumors (Figure 5). Similarly, a range of levels was observed for other proximal TCA cycle metabolites with no significant differences observed between wild-type IDH1 tumors and tumors with R132 IDH1 mutations. In contrast, increased 2HG levels were found in all tumors that contained an R132 IDH1 mutation (Figure 5, Supplementary Table 2). All R132 mutant IDH1 tumors examined had between 5 and 35 μmol of 2HG per gram of tumor, while tumors with wild-type IDH1 had over 100 fold less 2HG. This increase in 2HG in R132 mutant tumors was statistically significant (p<0.0001). We confirmed that (R)-2HG was the isomer present in tumor samples (data not shown). Together these data establish that the novel enzymatic activity associated with R132 mutations in IDH1 results in the production of 2HG in human brain tumors that harbor these mutations.
Figure 5. Human malignant gliomas containing R132 mutations in IDH1 contain increased concentrations of 2HG.
Human glioma samples obtained by surgical resection were snap frozen, genotyped to stratify as wild-type (WT) (N=10) or carrying an R132 mutant allele (Mutant) (n=12) and metabolites extracted for LC-MS analysis. Among the 12 mutant tumors, 10 carried a R132H mutation, one an R132S mutation, and one an R132G mutation. Each symbol represents the amount of the listed metabolite found in each tumor sample. Red lines indicate the group sample means. The difference in 2HG observed between WT and R132 mutant IDH1 mutant tumors was statistically significant by Student’s t-test (p<0.0001). There were no statistically significant differences in αKG, malate, fumarate, succinate, or isocitrate levels between the WT and R132 mutant IDH1 tumors.
Discussion
2HG is known to accumulate in the inherited metabolic disorder 2-hydroxyglutaric aciduria. This disease is caused by deficiency in the enzyme 2-hydroxyglutarate dehydrogenase, which converts 2HG to αKG12. Patients with 2-hydroxyglutarate dehydrogenase deficiencies accumulate 2HG in the brain as assessed by MRI and CSF analysis, develop leukoencephalopathy, and have an increased risk of developing brain tumors 13–15.
Furthermore, elevated brain levels of 2HG result in increased ROS levels16, 17, potentially contributing to an increased risk of cancer, and alterations in NADPH metabolism resulting from mutant IDH1 expression could further exacerbate this effect. The ability of 2HG to act as an NMDA receptor agonist may contribute to this effect 16. 2HG may also be toxic to cells by competitively inhibiting glutamate and/or αKG utilizing enzymes. These include transaminases which allow utilization of glutamate nitrogen for amino and nucleic acid biosynthesis, and αKG-dependent prolyl hydroxylases such as those that regulate Hif1α levels. Alterations in Hif1α have been reported to result from mutant IDH1 protein expression 7. Regardless of mechanism, it appears likely that the gain-of-function ability of cells to produce 2HG as a result of R132 mutations in IDH1 contributes to tumorigenesis. Patients with 2-hydroxyglutarate dehydrogenase deficiency have a high risk of CNS malignancy 15. The ability of mutant IDH1 to directly act on αKG may explain the prevalence of IDH1 mutations in tumors from CNS tissue, which are unique in their high level of glutamate uptake and its ready conversion to αKG in the cytosol18, thereby providing high levels of substrate for 2HG production. Myeloid cells also display a high ability to metabolize glutamine and recently R132 IDH1 mutations have also been described in a subset of AML19. The apparent co-dominance of the activity of mutant IDH1 with that of the wild-type enzyme is consistent with the genetics of the disease, in which only a single copy of the gene is mutated. As discussed above, the wild-type IDH1 could directly provide NADPH and αKG to the mutant enzyme. These data also demonstrate that mutation of R132 to histidine, serine, cysteine, glycine, or leucine share a common ability to catalyze the NADPH-dependent conversion of αKG to 2HG. These findings help clarify why mutations at other amino acid residues of IDH1, including other residues essential for catalytic activity, are not found. Finally, these findings have clinical implications in that they suggest that 2HG production will identify patients with IDH1 mutant brain tumors. This will be important for prognosis as patients with IDH1 mutations live longer than patients with gliomas characterized by other mutations5. In addition, patients with lower grade gliomas may benefit by the therapeutic inhibition of 2HG production. Inhibition of 2HG production by mutant IDH1 might slow or halt conversion of lower grade glioma into lethal secondary glioblastoma, changing the course of the disease.
Methods Summary
R132H, R132C, R132L and R132S mutations were introduced into human IDH1 by standard molecular biology techniques. 293T and human glioma U87MG and LN-18 cell lines were transfected using standard techniques. Protein expression levels were determined by Western blot. Metabolites were extracted from cultured cells and from tissue samples using 80% aqueous methanol (−80°C) as previously reported 8. Metabolite levels in samples were determined by negative mode electrospray LC-MS. For untargeted profiling, extract components were resolved using reverse phase HPLC running and metabolites were detected in ultra-high resolution mode (resolution ~100,000) by accurate mass ion-trap MS, collecting at one scan/second over an m/z range of 110–1100. For targeted evaluation of 2HG, αKG, and other TCA intermediates, extracts were resolved by reverse-phase HPLC system and metabolites detected by triple-quadrupole mass spectrometry, using multiple-reaction monitoring. Enzymatic activity in cell lysates was assessed by following a change in NADPH fluorescence over time in the presence of isocitrate and NADP, or αKG and NADPH. For enzyme assays using recombinant IDH1 enzyme, proteins were purified from E coli using Ni affinity and size-exclusion chromatography. Enzymatic activity for recombinant IDH1 protein was assessed by following a change in NADPH absorbance at 340 nm using a stop-flow spectrophotometer. Chirality of 2HG was determined as described previously9. For crystallography studies, purified R132H IDH1 was pre-incubated with NADPH, calcium chloride, and αKG. Crystals were obtained at 20°C by vapor diffusion equilibration using 3 μL drops mixed 2:1 (protein:precipitant) against a well-solution of MES pH 6.5 and PEG 6000. Patient tumor samples were obtained after informed consent as part of a UCLA IRB-approved research protocol, collected by surgical resection, snap frozen in isopentane cooled by liquid nitrogen and stored at −80°C. The IDH1 mutation status of each sample was determined as described previously3.
Supplemental Methods
Cloning, Expression, and Purification of ICDH1 wt and mutants in E coli
The open reading frame (ORF) clone of human isocitrate dehydrogenase 1 (cDNA) (IDH1; ref. ID NM_005896) was purchased from Invitrogen in pENTR221 (Carlsbad, CA) and Origene Inc. in pCMV6 (Rockville, MD). To transfect cells with wild-type or mutant IDH1, standard molecular biology mutagenesis techniques were utilized to alter the DNA sequence at base pair 395 of the ORF in pCMV6 to introduce base pair change from guanine to adenine, which resulted in a change in the amino acid code at position 132 from arginine (wt) to histidine (mutant; or R132H), and confirmed by standard DNA sequencing methods. For 293T cell transfection, wild-type and R132H mutant IDH1 were subcloned into pCMV-Sport6 with or without a carboxy-terminal Myc-DDK-tag. For stable cell line generation, constructs in pCMV6 were used. For expression in E coli, the coding region was amplified from pENTR221 by PCR using primers designed to add NDEI and XHO1 restrictions sites at the 5′ and 3′ ends respectively. The resultant fragment was cloned into vector pET41a (EMD Biosciences, Madison, Wl) to enable the E coli expression of C-terminus His8-tagged protein. Site directed mutagenesis was performed on the pET41a-ICHD1 plasmid using the QuikChange® MultiSite-Directed Mutagenesis Kit (Stratagene, La Jolla, CA)to change G395 to A, resulting in the Arg to His mutation. R132C, R132L and R132S mutants were introduced into pET41a-ICHD1 in an analogous way.
Wild-type and mutant proteins were expressed in and purified from the E coli Rosetta™ strain (Invitrogen, Carlsbad, CA) as follows. Cells were grown in LB (20 μg/ml Kanamycin) at 37°C with shaking until OD600 reaches 0.6. The temperature was changed to 18°C and protein expression was induced by adding IPTG to final concentration of 1 mM. After 12–16 hours of IPTG induction, cells were resuspended in Lysis Buffer (20mM Tris, pH7.4, 0.1% Triton X-100, 500 mM NaCl, 1 mM PMSF, 5 mM β-mercaptoethanol, 10 % glycerol) and disrupted by microfluidation. The 20,000g supernatant was loaded on metal chelate affinity resin (MCAC) equilibrated with Nickel Column Buffer A (20 mM Tris, pH7.4, 500mM NaCl, 5 mM β-mercaptoethanol, 10% glycerol) and washed for 20 column volumes. Elution from the column was effected by a 20 column-volume linear gradient of 10% to 100% Nickel Column Buffer B (20 mM Tris, pH7.4, 500 mM NaCl, 5 mM β-mercaptoethanol, 500 mM imidazole, 10% glycerol) in Nickel Column Buffer A). Fractions containing the protein of interest were identified by SDS-PAGE, pooled, and dialyzed twice against a 200-volume excess of Gel Filtration Buffer (200 mM NaCl, 50 mM Tris 7.5, 5 mM β-mercaptoethanol, 2 mM MnSO4, 10% glycerol), then concentrated to 10 ml using Centricon (Millipore, Billerica, MA) centrifugal concentrators. Purification of active dimers was achieved by applying the concentrated eluent from the MCAC column to a Sephacryl S-200 (GE Life Sciences, Piscataway, NJ) column equilibrated with Gel Filtration Buffer and eluting the column with 20 column volumes of the same buffer. Fractions corresponding to the retention time of the dimeric protein were identified by SDS-PAGE and pooled for storage at −80°C.
Cell lines and Cell Culture
293T cells were cultured in DMEM (Dulbecco’s modified Eagles Medium) with 10% fetal bovine serum and were transfected using pCMV-6-based IDH1 constructs in six-well plates with Fugene 6 (Roche) or Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions. Parental vector pCMV6 (no insert), pCMV6-wt IDH1 or pCMV6-R132H were transfected into human glioblastoma cell lines (U87MG; LN-18 (ATCC, HTB-14 and CRL-2610; respectively) cultured in DMEM with 10 % fetal bovine serum. Approximately 24 hrs after transfection, the cell cultures were transitioned to medium containing G418 sodium salt at concentrations of either 500 ug/ml (U87MG) or 750 ug/ml (LN-18) to select stable transfectants. Pooled populations of G418 resistant cells were generated and expression of either wild-type IDH1 or R132 IDH1 was confirmed by standard Western blot analysis.
Western blot
For transient transfection experiments in 293T cells, cells were lysed 72 hours after transfection with standard RIPA buffer. Lysates were separated by SDS-PAGE, transferred to nitrocellulose and probed with goat-anti-IDHc antibody (Santa Cruz Biotechnology sc49996) or rabbit-anti-MYC tag antibody (Cell Signaling Technology #2278) and then detected with HRP-conjugated donkey anti-goat or HRP-conjugated goat-anti-rabbit antibody (Santa Cruz Biotechnology sc2004). IDH1 antibody to confirm expression of both wild-type and R132H IDH1 was obtained from Proteintech. The IDH2 mouse monoclonal antibody used was obtained from Abeam.
Metabolite extraction
48h prior to profiling the appropriate culture medium was changed to identical medium with dialyzed fetal calf serum. All cells were grown in 10 cm tissue culture dishes, and the medium was replaced again with identical medium 24 h and 1h prior to metabolite extraction. All samples were harvested at a non-confluent density (1.5 – 2.0 × 106 cells). Metabolism was quenched and metabolites extracted by aspiration of media and immediate addition of 3.6 mL 80:20 methanol:water at −80°C, and transfer to a dry-ice bed to simultaneously lyse cells and quench metabolism. Cell remnants were scraped from the tissue culture dish and transferred, along with the methanol:water, into a 15 mL conical centrifuge tube. The resulting mixture was centrifuged at 14,000 × g for 20 min, and the supernatant was moved to a new tube. A 1 mL portion of the supernatant was then dried under nitrogen gas, dissolved in 100 μL of aq. LC buffer, spun at 13,000 × g for 10 min to remove any remaining debris and analyzed by LC-MS within 24h, as described below. At the time of harvest, an equivalently treated plate to each of those harvested was used to determine the total cell count, enabling subsequent normalization of LCMS signal intensities between culture dishes.
Non-targeted LC-MS
Samples were analyzed using high resolution (~100,000 fwhm resolution) exact mass ion-trap mass spectrometry in full-scan mode (1 scan/second, set to high dynamic range), coupled to liquid chromatography via negative mode electrospray ionization (−3.0 kV). The scan range was picked to avoid abundant ions like phosphate and sulfate entering the ion trap, and was set as follows: 0–3 min: no scanning, 3–5 min: 85–800 m/z, 5.0–6.7 min: 100–800 m/z, 6.7–9.0 min: 85–800 m/z, 9–16 min: 110–1000 m/z, 16–24 min: 220–1000 m/z. For chromatography, a Synergi Hydro-RP, 100mm × 2 mm, 2.1 μm particle size column was used (Phenomonex, Torrance, CA). Solvent A was 10 mM tributylamine and 15 mM acetic acid in 97% water/3% methanol. Solvent B was methanol. The gradient was (t=0–2.5 min, 0% B; t=5–7.5 min, 20% B; t=13 min, 55% B; t=15.5–18.5 min, 95% B; 19–25 min 0% B). The flow rate was 200 μL/min.
Targeted Liquid Chromatography—Mass Spectrometry, 2HG and TCA metabolite measurements
To detect secreted 2HG in culture media, 500 μL aliquots of conditioned media were collected, mixed 80:20 with methanol, and centrifuged at 3,000 rpm for 20 minutes at 4 degrees Celsius. The resulting supernatant was collected and stored at −80°C prior to LC-MS to assess 2HG levels. To measure whole-cell associated metabolites, media was aspirated and cells were harvested as described above. Two different liquid chromatography (LC) separation methods were used, each coupled by negative electrospray ionization (ESI, −3.0 kV) to triple-quadrupole mass spectrometers operating in multiple reaction monitoring (MRM) mode, with MS parameters optimized on infused metabolite standard solutions. In both methods, metabolites were separated by reversed phase chromatography using 10 mM tributyl-amine as an ion pairing agent in the aqueous mobile phase, according to a variant of a previously reported method20. The first method allowed resolution of TCA metabolites: t = 0, 50% B; t = 5, 95% B; t= 7, 95% B; t= 8, 0% B, where B refers to an organic mobile phase of 100% methanol. The second method was specific for 2HG, running a fast linear gradient from 50%–95% B (buffers as defined above) over 5 minutes. In both methods, the column was a Synergi Hydro-RP, 100mm × 2 mm, 2.1 μm particle size (Phenomonex), as above. Metabolites were quantified by comparison of peak areas with pure metabolite standards at known concentration. Metabolite flux studies from 13C-glutamine were performed as described previously21.
Detection of isocitrate, αKG, and 2HG in purified enzyme reactions by LC-MS
Enzyme reactions performed as described in the text were run to completion as judged by measurement of the oxidation state of NADPH at 340 nm. Reactions were extracted with eight volumes of methanol, and centrifuged to remove precipitated protein. The supernatant was dried under a stream of nitrogen and resuspended in H2O. Analysis was conducted on an API2000 LC-MS-MS (Applied Biosystems, Foster City, CA). Sample separation and analysis was performed on a 150 × 2 mm, 4 uM Synergi Hydro-RP 80 A column, using a gradient of Buffer A (10 mM tributylamine, 15 mM acetic acid, 3% (v/v) methanol, in water) and Buffer B (methanol) using MRM transitions.
Cell lysates based enzyme assays
293T cell lysates for measuring enzymatic activity were obtained 48 hours after transfection with M-PER lysis buffer supplemented with protease and phosphatase inhibitors. After lysates were sonicated and centrifuged at 12,000g, supernatants were collected and normalized for total protein concentration. To measure IDH oxidative activity, 3 μg of lysate protein was added to 200 μl of an assay solution containing 33 mM Tris-acetate buffer (pH 7.4), 1.3 mM MgCl2, 0.33 mM EDTA, 100 μM β-NADP, and varying concentrations of D-(+)-threo-isocitrate. Absorbance at 340 nm, reflecting NADPH production, was measured every 20 seconds for 30 min on a SpectraMax 190 spectrophotometer (Molecular Devices, Sunnyvale, CA), Data points represent the mean activity of 3 replicates per lysate, averaged among 5 time points centered at every 5 min. To measure IDH reductive activity, 3 μg of lysate protein was added to 200 μl of an assay solution which contained 33 mM Tris-acetate pH 7.4, 1.3 mM MgCl2,25 μM β-NADPH, 40 mM NaHCO3, and 0.6 mM αKG. The decrease in 340 nm absorbance over time was measured to assess NADPH consumption, with 3 replicates per lysate.
Recombinant IDH1 Enzyme Assays
All reactions were performed in standard enzyme reaction buffer (150 mM NaCl, 20 mM Tris-Cl, pH 7.5,10% glycerol, 5 mM MgCl2 and 0.03% (w/v) bovine serum albumin). For determination of kinetic parameters, sufficient enzyme was added to give a linear reaction for 1 to 5 seconds. Reaction progress was monitored by observation of the reduction state of the cofactor at 340 nm in an SFM-400 stopped-flow spectrophotometer (BioLogic, Knoxville, TN). Enzymatic constants were determined using curve fitting algorithms to standard kinetic models with the Sigmaplot software package (Systat Software, San Jose, CA).
Determination of chirality of reaction products from enzyme reactions and tumors
Enzyme reactions were run to completion and extracted with methanol as described above, then derivatized with enantiomerically pure tartaric acid before resolution and analysis by LC-MS. After being thoroughly dried, samples were resuspended in freshly prepared 50 mg/ml (2R,3R)-(+)-Tartaric acid in dichloromethane:acetic acid (4:1) and incubated for 30 minutes at 75°C. After cooling to room temperature, samples were briefly centrifuged at 14,000g, dried under a stream of nitrogen, and resuspended in H2O Analysis was conducted on an API200 LC-MS-MS (Applied Biosystems, Foster City, CA), using an isocratic flow of 90:10 (2 mM ammonium formate, pH 3.6:MeOH) on a Luna C18(2) 150 × 2 mm, 5 uM column. Tartaric-acid derivatized 2HG was detected using the 362.9/146.6 MRM transition and the following instrument settings: DP -1, FP -310, EP -4, CE-12, CXP-26. Analysis of the (R)-2HG standard, 2HG racemic mixture, and methanol-extracted tumor biomass (q.v.) was similarly performed. (R)-2HG standard was obtained from Sigma and synthesized internally.
Crystallography
For crystallography studies, purified R132H IDH1 was pre-incubated with NADPH, calcium chloride, and αKG. Crystals were obtained at 20°C by vapor diffusion equilibration of 3 μL drops of mixed 2:1 (protein:precipitant) against a well solution of MES pH 6.5 and PEG 6000. A diffraction dataset with 92% completeness was collected under cryo conditions with 1.08 Å wavelength radiation on an ADSC Quantum 315R detector at Beamline X29A of the NSLS at Brookhaven National Laboratories. The dataset was integrated using HKL2000 and scaled using SCALEPACK22. A single molecular replacement solution was obtained with PHASER23 using published coordinates from wild type IDH1 in the ‘closed’ conformation as search model (PDB accession code 1T0L). Model building was performed using COOT24 and the final model was refined by the CCP4 program REFMAC525; graphics were produced using PyMOL26. Water molecules were selectively modeled into the structure based upon strong spherical electron density difference peaks, having proper coordination geometry and distance, located within the first solvent shell layer, and having B-factors <80 and electron density sigma level >1.0. After the final refinement, 92.4% of residues fit in the most favored regions in the Ramachandran plot, with 6.8% in the additional allowed regions and 0.7% in general allowed regions. The final structure model in the closed form conformation at a resolution of 2.1 Å contains three polypeptide chains of IDH1R132H each chain consisting of 425 amino acids (414 residues plus a C-term His-tag, with residues 4–410 placed into the model), three NADPH molecules, and 301 total waters.
Clinical Specimens, metabolite extraction and analysis
The collection of human brain tumor samples was approved by the UCLA IRB. Human brain tumors were obtained during surgical resection, snap frozen in isopentane cooled by liquid nitrogen, and stored at −80°C. Clinical classification and grading of the tissue was performed using standard clinical histopathology as established by the WHO. Genomic sequence analysis was deployed to identify brain tumor samples containing either wild-type isocitrate dehydrogenase (IDH1) or mutations altering amino acid 132. Genomic DNA was isolated from 50–100 mgs of brain tumor tissue using standard methods. A polymerase chain reaction on the isolated genomic DNA was used to amplify a 295 base pair fragment of the genomic DNA that contains both the intron and 2nd exon sequences of human IDH1 and mutation status assessed by standard molecular biology techniques.
Metabolite extraction was accomplished by adding a 10× volume (m/v ratio) of −80 C methanol:water mix (80%:20%) to the brain tissue (approximately 100mgs) followed by 30 s homogenization at 4 C. These chilled, methanol extracted homogenized tissues were then centrifuged at 14,000 rpm for 30 minutes to sediment the cellular and tissue debris and the cleared tissue supernatants were transferred to a screw-cap freezer vial and stored at −80°C. For analysis, a 2× volume of tributylamine (10 mM) acetic acid (10 mM) pH 5.5 was added to the samples and analyzed by LCMS as follows. Sample extracts were filtered using a Millex-FG 0.20 micron disk and 10 μl were injected onto a reverse-phase HPLC column (Synergi 150mm × 2 mm, Phenomenex Inc.) and eluted using a linear gradient LC-MS-grade methanol (50%) with 10 mM tributylamine and 10 mM acetic acid) ramping to 80 % methanohl:10 mM tributylamine: 10 mM acetic acid over 6 minutes at 200 μL/min. Eluted metabolite ions were detected using a triple-quadrupole mass spectrometer, tuned to detect in negative mode with multiple-reaction-monitoring mode transition set according to the molecular weights and fragmentation patterns for 8 known central metabolites, including 2-hydroxyglutarate as described above. Data was processed using Analyst Software (Applied Biosystems, Inc.) and metabolite signal intensities were obtained by standard peak integration methods.
Supplementary Material
Acknowledgments
We thank Robert K. Suto, Raymond S. Brown, and Eric Fontano at Xtal BioStructures, Inc., Natick, MA for performing crystallographic studies and Shaohui Wang at ChemPartner for assistance with biochemical experiments. We thank Gregory Petsko for his review of the structure data. We also thank Tak Mak, Ning Wu, Lou Tartaglia, Jeff Saunders, Francisco Salituro and David Schenkein for helpful discussions and/or comments on the manuscript. Asterand, PLC provided some of the glioma specimens and SeqWright Inc. assisted with genomic DNA SNP analysis. JDR is supported by NIH R21 CA128620.
Footnotes
Author Information
R132H mutant IDH1 structure files are deposited in the Protein Data Bank under accession code 3INM.
Author Contributions
L.D., D.W.W, S.G., B.D.B., M.A.B., E.M.D., V.R.F., H.G.J., S.J., M.C.K., K.M.M., R.M.P., P.S.W., K.E.Y., J.D.R., L.M.L., S.M.S., contributed extensively to the work presented in this paper. L.C.C., C.B.T., M.G.V.H., S.M.S. provided support and conceptual advice. L.D., M.G.V.H., S.M.S. wrote the manuscript.
References
- 1.Balss J, et al. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 2.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174:1149–53. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–73. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartmann C, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009 doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 5.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bleeker FE, et al. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat. 2009;30:7–11. doi: 10.1002/humu.20937. [DOI] [PubMed] [Google Scholar]
- 7.Zhao S, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–5. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu W, Kimball E, Rabinowitz JD. A high-performance liquid chromatography-tandem mass spectrometry method for quantitation of nitrogen-containing intracellular metabolites. J Am Soc Mass Spectrom. 2006;17:37–50. doi: 10.1016/j.jasms.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Struys EA, Jansen EE, Verhoeven NM, Jakobs C. Measurement of urinary D- and L-2-hydroxyglutarate enantiomers by stable-isotope-dilution liquid chromatography-tandem mass spectrometry after derivatization with diacetyl-L-tartaric anhydride. Clin Chem. 2004;50:1391–5. doi: 10.1373/clinchem.2004.033399. [DOI] [PubMed] [Google Scholar]
- 10.Xu X, et al. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J Biol Chem. 2004;279:33946–57. doi: 10.1074/jbc.M404298200. [DOI] [PubMed] [Google Scholar]
- 11.Aktas DF, Cook PF. A lysine-tyrosine pair carries out acid-base chemistry in the metal ion-dependent pyridine dinucleotide-linked beta-hydroxyacid oxidative decarboxylases. Biochemistry. 2009;48:3565–77. doi: 10.1021/bi8022976. [DOI] [PubMed] [Google Scholar]
- 12.Struys EA, et al. Mutations in the D-2-hydroxyglutarate dehydrogenase gene cause D-2-hydroxyglutaric aciduria. Am J Hum Genet. 2005;76:358–60. doi: 10.1086/427890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolker S, Mayatepek E, Hoffmann GF. White matter disease in cerebral organic acid disorders: clinical implications and suggested pathomechanisms. Neuropediatrics. 2002;33:225–31. doi: 10.1055/s-2002-36741. [DOI] [PubMed] [Google Scholar]
- 14.Wajner M, Latini A, Wyse AT, Dutra-Filho CS. The role of oxidative damage in the neuropathology of organic acidurias: insights from animal studies. J Inherit Metab Dis. 2004;27:427–48. doi: 10.1023/B:BOLI.0000037353.13085.e2. [DOI] [PubMed] [Google Scholar]
- 15.Aghili M, Zahedi F, Rafiee E. Hydroxyglutaric aciduria and malignant brain tumor: a case report and literature review. J Neurooncol. 2009;91:233–6. doi: 10.1007/s11060-008-9706-2. [DOI] [PubMed] [Google Scholar]
- 16.Kolker S, et al. NMDA receptor activation and respiratory chain complex V inhibition contribute to neurodegeneration in d-2-hydroxyglutaric aciduria. Eur J Neurosci. 2002;16:21–8. doi: 10.1046/j.1460-9568.2002.02055.x. [DOI] [PubMed] [Google Scholar]
- 17.Latini A, et al. D-2-hydroxyglutaric acid induces oxidative stress in cerebral cortex of young rats. Eur J Neurosci. 2003;17:2017–22. doi: 10.1046/j.1460-9568.2003.02639.x. [DOI] [PubMed] [Google Scholar]
- 18.Tsacopoulos M. Metabolic signaling between neurons and glial cells: a short review. J Physiol Paris. 2002;96:283–8. doi: 10.1016/s0928-4257(02)00017-7. [DOI] [PubMed] [Google Scholar]
- 19.Mardis ER, et al. Recurring Mutations Found by Sequencing an Acute Myeloid Leukemia Genome. N Engl J Med. 2009:1533–4406. doi: 10.1056/NEJMoa0903840. (Electronic) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo B, Groenke K, Takors R, Wandrey C, Oldiges M. Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and tricarboxylic acid cycle by liquid chromatography-mass spectrometry. J Chromatogr A. 2007;1147:153–64. doi: 10.1016/j.chroma.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 21.Munger J, et al. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol. 2008;26:1179–86. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode (HKL2000) Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. 2007. Copyright © International Union of Crystallography. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emsley P, Cowtan K. COOT: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 25.Collaborative Computational Project Number 4. The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr. 1994;D 50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 26.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.