Abstract
Plasmodium falciparum causes the virulent form of malaria and disease manifestations are linked to growth inside infected erythrocytes. In order to survive and evade host responses the parasite remodels the erythrocyte by exporting several hundred effector proteins beyond the surrounding parasitophorous vacuole membrane. A feature of exported proteins is a pentameric motif (RxLxE/Q/D) that is a substrate for an unknown protease. Here, we show the protein responsible for cleavage of this motif is Plasmepsin V, an aspartic acid protease located in the endoplasmic reticulum. Plasmepsin V cleavage reveals the export signal (xE/Q/D) at the N-terminus of cargo proteins. Expression of an identical mature protein with xQ at the N-terminus generated by signal peptidase was not exported demonstrating Plasmepsin V activity is essential and linked with other key export events. Identification of the protease responsible for export into erythrocytes provides a novel target for therapeutic intervention against this devastating disease.
Malaria is a disease of global importance causing significant morbidity and death 1. Plasmodium falciparum is responsible for the lethal form of malaria and morphological changes occur in infected erythrocytes altering physical properties and impairing circulation2. Parasitised erythrocytes become rigid and adhere to different cell types and this plays a pivotal role in complications of falciparum malaria3,4. As the parasite develops inside the erythrocyte over 300 effector proteins are exported to the host5,6, some of which associate with the cytoskeleton7 or are inserted into the host membrane8,9,10 and these alter mechanical and adhesive properties of infected erythrocytes 11. Trafficking of parasite proteins to erythrocytes is a multi-step process involving entry into the endoplasmic reticulum (ER) and trafficking to the cell surface followed by translocation across the parasitophorous vacuole membrane (PVM) into the erythrocyte or assembly into membranous structures (Maurer’s clefts) to reach their destinations 12 (Supplementary Fig. 1). The PTEX translocon complex is associated with the PVM and appears to be responsible for export across this barrier 13.
For export beyond the PVM of infected erythrocytes, or hepatocytes by liver stage parasites, P. falciparum employs a signal sequence for ER entry followed by a motif termed PEXEL (Plasmodium EXport ELement) or Vacuolar Translocation Signal, which is conserved across Plasmodium spp. 5,6. The motif allowed identification of exported proteins in Plasmodium spp., revealing 5–8% of parasite proteins are destined for export 14,15. The PEXEL consists of the pentameric sequence RxLxE/Q/D that is processed after the conserved leucine, and N-acetylated, in the ER of the parasite 16,17. The parasite protease responsible for cleavage of the PEXEL remains unknown, although signal peptidase has been suggested as a candidate 16. Since successful export to the host relies on the PEXEL motif 5,6,18,17, the protease responsible for processing would be a crucial part of the export machinery and its identification provides an important target for development of novel antimalarials.
Plasmepsin V is an ER resident aspartic acid protease that cleaves the PEXEL motif
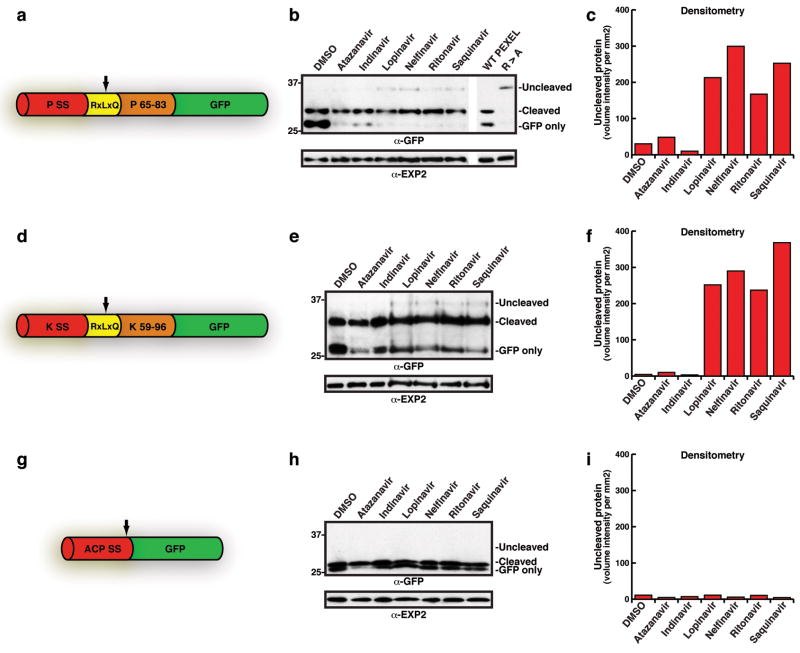
To identify candidate proteases responsible for PEXEL processing we used features of the cleavage site to predict the type of enzyme involved. We noted that HIV-1 aspartic acid protease cleaved substrate proteins within a motif similar to PEXEL 19,20, suggesting the Plasmodium enzyme shares similarity. To test this we incubated inhibitors of HIV-1 protease in culture with P. falciparum-infected erythrocytes and monitored PEXEL cleavage. PEXEL processing was reduced in the presence of lopinavir, nelfinavir, ritonavir and saquinavir for PfEMP3- and knob associated histidine-rich (KAHRP)-GFP proteins (Fig. 1a–f). Whilst the inhibition was inefficient it suggested PEXEL processing might involve a protease similar to HIV-1 aspartic acid protease. Sensitivity of signal peptidase was assessed using the inhibitors with acyl carrier protein signal sequence (ACP)-GFP 21. This transgenic protein lacks a PEXEL and is secreted into the parasitophorous vacuole 21. In contrast to the PEXEL-containing proteins, we observed no detectable reduction in processing of the signal sequence from the ACPs-GFP chimera with inhibitors (Fig. 1g–i). We conclude that the PEXEL protease is not the parasite signal peptidase but involves a unique protease belonging to the aspartic acid family.
Figure 1. PEXEL processing is sensitive to HIV protease inhibitors.
a, Structure of PfEMP3-GFP. PSS, signal sequence; RxLxQ PEXEL; residues 65-83 of PfEMP3. Arrow, PEXEL cleavage. b, PfEMP3-GFP processing with HIV-1 inhibitors. c, Densitometry of uncleaved bands in b. d, Structure of KAHRP-GFP. KSS, signal sequence; RxLxQ PEXEL, residues 59-96 of KAHRP. e, KAHRP-GFP PEXEL processing with inhibitors. f, Densitometry of uncleaved bands in e. g, Structure of ACPs-GFP. ACP SS, signal sequence-GFP. Arrow, signal sequence cleavage. h, ACPs-GFP signal sequence processing is not inhibited. ‘Cleaved’, signal sequence cleavage. i, Densitometry of the uncleaved region of immunoblot. (see Supplementary for details)
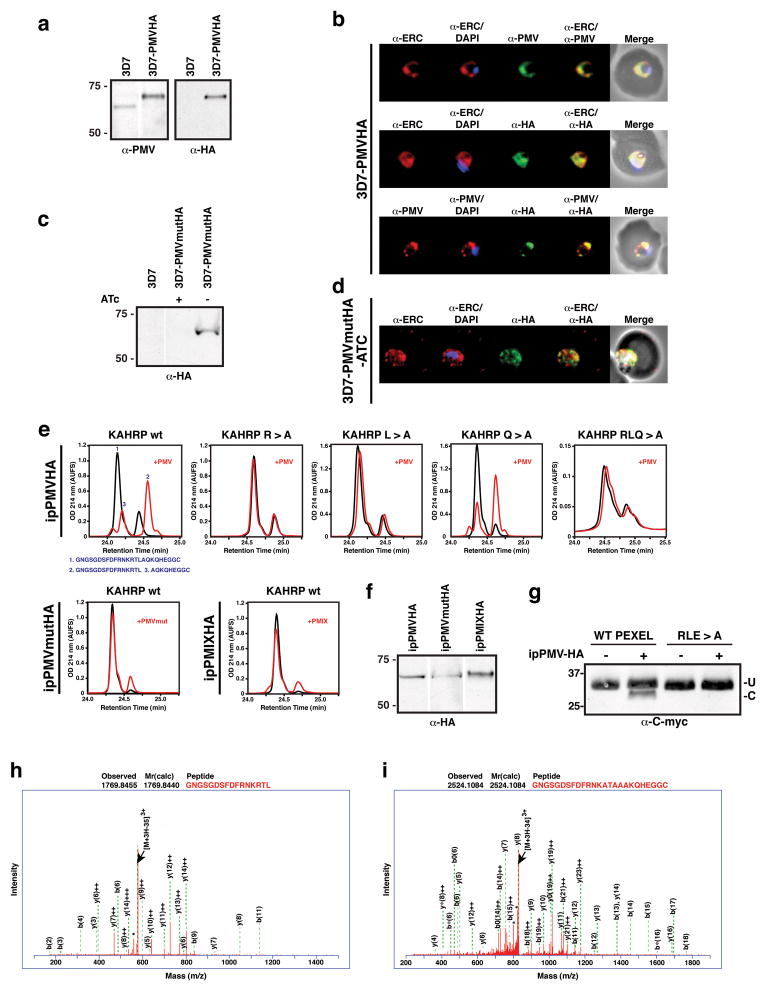
A list of candidate aspartic acid proteases was generated from the P. falciparum genome based on; firstly, expression in blood and liver stages of the parasite 22,23, secondly, presence of a signal sequence for ER entry; and thirdly, specificity to Plasmodia spp. because the PEXEL is absent from closely related apicomplexans 14. Plasmepsin V (PF13_0133), a putative aspartic acid protease localising to the ER that is functionally divergent to other members of the Plasmepsin family, satisfied our criteria 24. To investigate the function of Plasmepsin V, we introduced a hemagglutinin (HA) tag at the C-terminus (3D7-PMVHA) (Fig 2a). HA-tagged Plasmepsin V localised to the ER 25, as has been shown previously for the endogenous protein 24 (Fig. 2b).
Figure 2. Plasmepsin V cleaves PEXEL.
a, Plasmepsin V HA tagged (3D7-PMVHA). b, Plasmepsin V localises to ER. c, Inactive HA-tagged Plasmepsin V (3D7-PMVmutHA). d, Mutant Plasmepsin V (3D7-PMVmutHA) localises to ER. e, HA-tagged Plasmepsin V (ipPMVHA) with KAHRP peptides; unmutated, R>A, L>A, Q>A, RLQ>A. ipPMVmutHA (inactive) or ipPMIXHA (Plasmepsin IX) with unmutated peptide. f, Recovered enzyme. g, Recombinant GBP130 cleavage by Plasmepsin V. Last lanes are GBP130 mutant (RLE>A). U, uncleaved; C, cleaved. h, Identification of cleaved peptide GNGSGDSFDFRNKRTL. Identification of uncleaved RLQ>A mutant peptide. * refers to multiple H20 and NH3 losses. (see Supplementary for details)
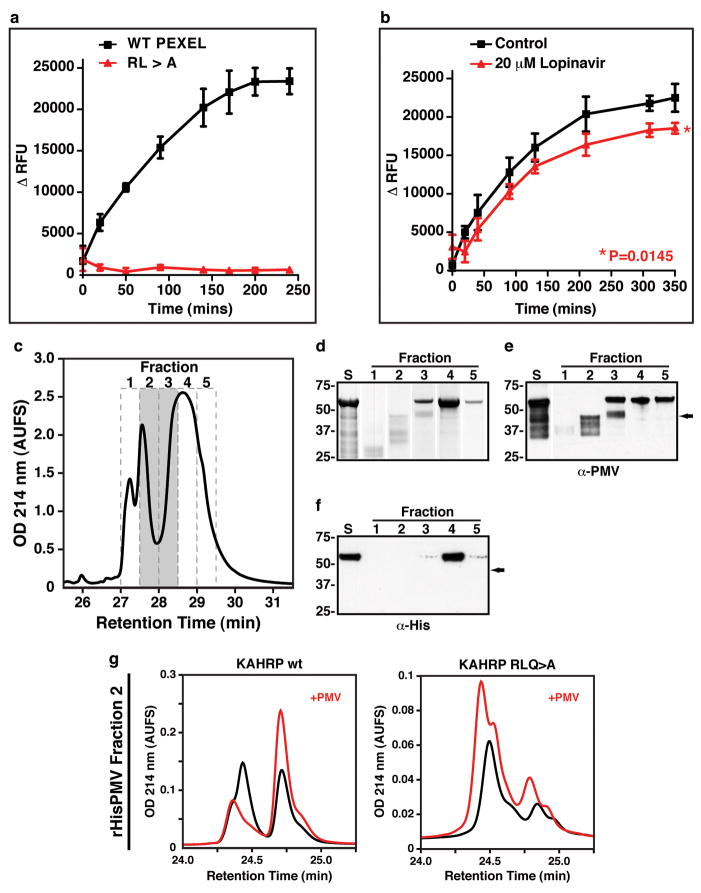
To determine if Plasmepsin V could cleave PEXEL substrates we immuno-precipitated the HA-tagged protease (ipPMVHA) and incubated KAHRP peptides containing either a wild-type PEXEL or mutations that interfere with cleavage (Fig. 2e) 17. The resulting peptides were separated by reversed-phase high-pressure liquid chromatography (RP-HPLC) and analysed by nano liquid chromatography tandem mass spectrometry (LC-MS/MS) (Fig. 2e,h,i). Incubation of the KAHRP peptide with ipPMVHA produced two major peptide species (Fig 2e; first panel) derived from cleavage within the PEXEL after leucine (Fig 2e,g). In contrast, mutation of the conserved amino acids R>A, L>A or RLQ>A resulted in no cleavage (Fig. 2e,h), consistent with the properties of the PEXEL cleaving protease in vivo 17. Furthermore, mutation of the conserved fifth amino acid (Q>A) did not inhibit cleavage (Fig. 2e), corroborating our previous data that this residue is not required for PEXEL processing 17. To verify the PEXEL cleavage was due to Plasmepsin V, and not other co-precipitated proteases, we constructed P. falciparum transgenic parasites that either conditionally, or constitutively, expressed an inactive form of Plasmepsin V tagged with HA (PMVmutHA and PMVmutHA2, respectively). We confirmed expression and ER localisation of PMVmutHA (Fig 2c,d) and the immuno-precipitated proteins (ipPMVmutHA or ipPMVmutHA2) were incubated with KAHRP peptides and no processing occurred (Fig 2e and Supplementary Fig. 2). Transgenic parasites were generated that expressed HA-tagged Plasmepsin IX (PF14_0281) (3D7-PMIXHA), another member of the aspartic acid protease family in P. falciparum. Immuno-precipitated Plasmepsin IX (ipPMIXHA) could not process the KAHRP peptide (Fig 2e,f). ipPMVHA also cleaved a recombinant GBP130 protein to the expected size (Fig. 2g) as well as a quenched fluorescent substrate of 9 amino acids (Fig. 3a), both of which contained a PEXEL. Mutation of conserved amino acids in the PEXEL blocked cleavage of both substrates (Fig. 2g and 3a). The HIV protease inhibitor lopinavir reduced Plasmepsin V activity by 19.1% (P=0.0145; Fig 3b) consistent with in vivo inhibition of PEXEL cleavage of PfEMP3- and KAHRP-GFP (Fig. 1). Taken together, these results show that Plasmepsin V specifically cleaves the PEXEL and has the functional specificity expected of the PEXEL protease.
Figure 3. Activity of Plasmepsin V and cleavage of PEXEL.
a, Plasmepsin V cleavage of fluorescent substrate with PEXEL (black line) and PEXEL RL>A mutant (red line). Data shown is one representative experiment done in triplicate. Data shown is the mean ± standard deviation. b, Lopinavir reduces Plasmepsin V activity (P=0.0145). Data shown is one representative experiment done in triplicate. Data shown is the mean ± standard deviation. c, N-terminal hexa-His fusion of Plasmepsin V (rHisPMV) from E. coli separated by HPLC. Elutions with activity indicated in grey. d, Starting material (S) and re-folded fractions were Coomassie stained. e, Starting material (S) and fractions probed with anti-Plasmepsin V antibodies. f, Starting material (S) and fractions probed with anti-hexaHis antibodies. g, HPLC of recombinant Plasmepsin V activity (rHisPMV) with KAHRP peptides. (see Supplementary for details)
To provide further evidence that Plasmepsin V was responsible for PEXEL cleavage we expressed amino acids 37 to 521 in Escherichia coli with an N-terminal Hexa-His fusion tag. The recombinant Plasmepsin V was extracted from inclusion bodies and purified on NiNTA under denaturing conditions and further fractionated using RP-HPLC (Fig 3c). Individual fractions were analysed using SDS-PAGE (Fig. 3d) or transferred to nitrocellulose and probed with anti-Plasmepsin V antibodies (Fig. 3e). A major band was observed at approximately 55 kDa (fraction #3, 4 and 5) and smaller processed forms of Plasmepsin V were also detected (fractions 2–3) suggesting N-terminal processing (Fig. 3f) occurred either by the protease itself or other E. coli proteases present in the purified inclusion bodies. Processed forms of ipPMVHA are also detectable in immunoblots suggesting one may be the activated form (Supplementary Fig. 3). Recombinant protein fractions was assayed for PEXEL cleavage using KAHRP peptides and PEXEL-specific activity was observed with fractions 2 and 3, which contained the processed form of Plasmepsin V (Fig. 3c–g and Supplementary Fig. 4). This data suggests that a recombinant, N-terminally truncated form of Plasmepsin V can cleave the PEXEL motif.
Plasmespin V directs export of effector proteins at the ER
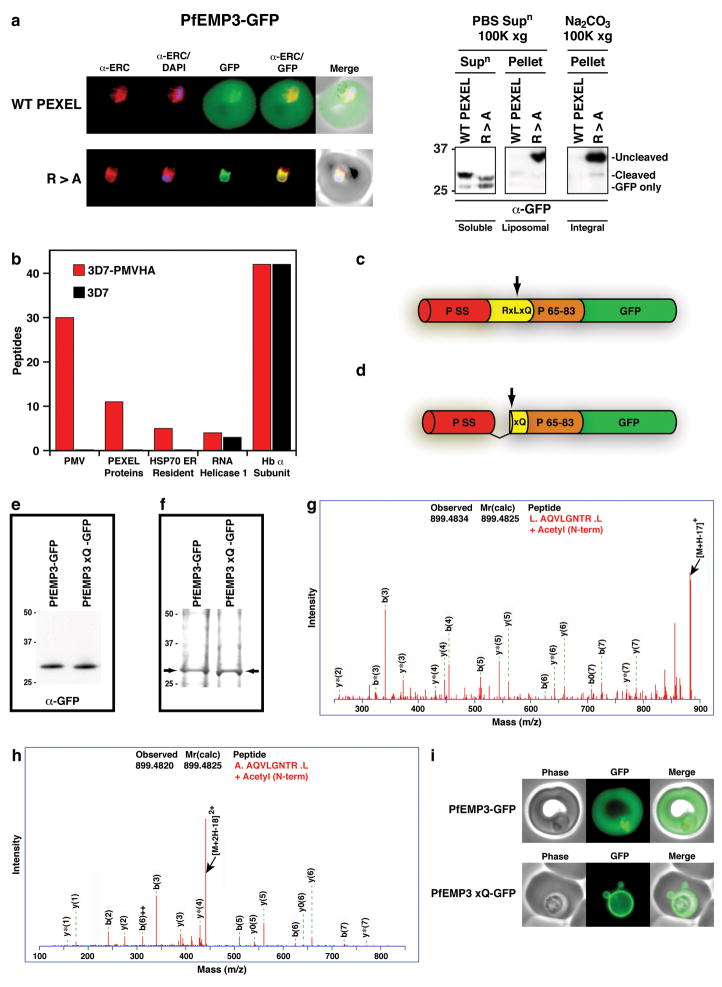
As the PEXEL motif is required for export to infected erythrocytes 5,6,18,17, we next determined the importance of Plasmepsin V for export in vivo. Firstly, we investigated the importance of signal peptidase in export by mutating the PEXEL R>A in the normally exported protein, PfEMP3-GFP, to assess if the signal sequence was processed. The mutant was retained in the ER as a full-length integral membrane protein by its signal peptide, which was not cleaved by signal peptidase (Fig. 4a), as we have shown previously for KAHRP 17 indicating that signal peptidase is not essential for trafficking. This highlights the important role Plasmepsin V plays in freeing cargo proteins from the ER membrane by processing the PEXEL (Fig. 4a). Additionally, we previously showed that the non-exported protein MSP3 is exported if the signal sequence is fused to a PEXEL 26, indicating that Plasmepsin V is the dominant protease over signal peptidase in determining the final destination of proteins. Secondly, we determined if the Plasmepsin V orthologous gene in P. berghei (PbPMV) could be disrupted, as the transfection methodology for this rodent malaria is more efficient than in P. falciparum, providing a more robust test of essentiality. Three attempts to disrupt the PbPMV gene in P. berghei were unsuccessful suggesting it is essential (Supplementary Fig. 5). Thirdly, we determined that Plasmepsin V directly binds exported proteins in vivo by immuno-precipitation of the HA-tagged enzyme and proteomic analysis of interacting proteins. This showed that Plasmepsin V was specifically immuno-precipitated and, importantly, that PEXEL-containing proteins (PF10_0159, MAL7P1.172 and PFE0060w) specifically co-precipitated with the protease (Fig. 4b, Supplementary Fig. 6). Fourthly, we constructed transgenic parasites expressing a protein that would functionally bypass Plasmepsin V. A control PfEMP3-GFP transgenic line was constructed expressing a protein that would be cleaved within the PEXEL by Plasmepsin V to reveal the acetylated export signal AcxQ (Fig 4c). The second construct was based on PfEMP3-GFP but had a modified signal peptidase cleavage site and no PEXEL RxL so it would be cleaved by signal peptidase to reveal the same mature protein (Fig 4d). We confirmed that both processed proteins had AcxQ at the N-terminus and were identical at the C-terminus (Fig. 4e–h). Additionally, mass spectrometry of trypsin cleaved peptides across regions of each protein was consistent with them being identical (Supplementary Fig. 7). The export profile of the two mature proteins was dramatically different as Plasmepsin V-processed PfEMP3-GFP was efficiently exported to the erythrocyte whilst signal peptidase-cleaved PfEMP3xQ-GFP accumulated in the parasitophorous vacuole (Fig 4i). Therefore, revealing the N-terminal signal for export (AcxE/Q/D) alone is not sufficient to reach the host cell; Plasmepsin V must cleave the PEXEL and it then directs the next step in export to the host cell.
Figure 4. Cleavage by Plasmepsin V is essential for export.
a, Exported PfEMP3-GFP (top) whilst R>A mutant accumulated in ER (bottom). Right, fractionation showing full-length R>A mutant accumulates in membrane. b, Proteomic analysis of Plasmepsin V immuno-precipitated proteins. c, Structure of PfEMP3-GFP. PSS, signal sequence; RxLxQ PEXEL, residues 65-83 of PfEMP3. Arrow, PEXEL cleavage. d, Structure of PfEMP3xQ-GFP. Arrow, signal peptidase cleavage. e, N-terminal processing confirmed with α-GFP. f, Immuno-purified PfEMP3-GFP and PfEMP3xQ-GFP analysed by Coomassie and LC-MS/MS. g, Identification of N-terminal AcAQVLGNTR of PfEMP3-GFP after Plasmepsin V processing. h, Identification of AcAQVLGNTR at N-terminus of PfEMP3xQ-GFP after signal peptidase processing. i, Imaging of exported PfEMP3-GFP (top) and non-exported PfEMP3xQ-GFP (bottom). (see Supplementary for details)
The role of Plasmepsin V in export of P. falciparum proteins
How might Plasmepsin V direct export of proteins into P. falciparum-infected erythrocytes? Plasmepsin V has an N-terminal signal sequence for ER entry and a C-terminal transmembrane sequence such that it would be inserted into the ER membrane with the protease domain protruding into the lumen 24 (Supplementary Fig. 1). The signal sequences of exported proteins would direct co-translational insertion into the ER membrane but are not efficiently cleaved 17 (Fig. 4a), suggesting the PEXEL is recognised, bound and cleaved before signal peptidase identifies its cleavage site. That AcxE/Q/D alone is insufficient for export strongly suggests that Plasmepsin V functions in a complex with another protein(s) that has exclusive access to Plasmepsin V-matured cargo. We propose that this access may assist in sorting mature cargo to a subset of vesicles for trafficking to discrete domains of the parasitophorous vacuole in a manner distinct from general secretion but dependent on xE/Q/D 13,17 (Supplementary Fig. 1). Consistent with this is localisation of the PTEX translocon on the parasitophorous vacuole membrane in discrete domains 13.
Plasmepsin V is conserved in Plasmodium spp., including P. vivax 27,28, but is absent from higher eukaryotes; its identification as a critical enzyme for protein export provides an important target for development of novel antimalarials. HIV-1 protease inhibitors have been very successful treatments in the combat against HIV and so these inhibitors may provide a platform for the design of novel antimalarial compounds intended to impede the function of this important malarial protease.
METHODS SUMMARY
Transgenic parasites expressed PfEMP3-GFP or PfEMP3R>A-GFP chimeras from plasmids pPfEMP3WTGlux.1 or PfEMP3R>AGlux.1, respectively. Transgenic parasites expressing KAHRP-GFP contained the plasmid pKAHRPGlux.1, comprising DNA encoding residues 1-96 of KAHRP (PFB0100c). Parasites expressing ACPs-GFP are previously described 21. Parasites expressing PfEMP3xQ-GFP contained the plasmid pPfEMP3xQGlux.1, encoding residues 1-36 of PfEMP3, followed by an alanine (to allow for signal peptidase cleavage at IYSEA-) and residues 63-82 of PfEMP3 (which include residues 63aQ64 of the PEXEL) cloned into pGlux.1. The plasmids pPMVHA1.5 pPMIXHA1.5 were integrated into either the Plasmepsin V locus (PF13_0133) or Plasmepsin IX locus (PF14_0281), respectively to append HA tags. The transgenic parasites 3D7-PMVmutHA episomally expressed plasmid pTETPMVmutHA, which contains DNA encoding Plasmepsin V with D118A, D365A and F370A substitutions fused to the same tags as in 3D7-PMVHA, cloned into the anhydrotetracycline (ATc) regulatable plasmid pTGFP 29. The same DNA encoding Plasmepsin V with D118A, D365A and F370A substitutions fused to HA, but with alternate cloning sites, was cloned into pGlux.1, removing GFP, to create pPMVmutHA2. This plasmid contains the constitutively expressed crt promoter and is non-inducible. Recombinant GBP130 or GBP130 RLE>A was expressed in E. coli harbouring plasmids pGBP130-3Cmyc18His or pGBP130-3A-3Cmyc18His, respectively, which encodes residues 66 to 196 of GBP130 (containing the wild-type or mutant PEXEL). Recombinant Plasmepsin V was expressed in E. coli harbouring plasmid pHisPMVtrunc, which contains DNA encoding residues 37-521 of Plasmepsin V cloned into pProExHTb.
Plasmepsin V activity was assayed either with anti-HA beads containing ipPMVHA or ipPMVmutHA or 3–5 μg of HPLC-eluted protein added to the reaction that contained either PEXEL peptide substrate, recombinant PEXEL substrate or fluorogenic PEXEL peptide substrate. Digest reactions were analysed by HPLC and LC-MS/MS, immunoblot and fluorescence intensity, respectively.
Methods
Parasites, plasmid construction and transfection
P. falciparum strain 3D7 and transgenic parasites were cultured in human O+ erythrocytes at 4% haematocrit as described previously 17. Transgenic parasites expressed PfEMP3-GFP or PfEMP3R>A-GFP chimeras from plasmids pPfEMP3WTGlux.1 or PfEMP3R>AGlux.1, respectively. These constructs contained DNA encoding residues 1-38 of PfEMP3 (PFB0095c), double HA followed by PfEMP3 residues 56-82, which contained either a native PEXEL or PEXEL R>A point mutation, respectively (DNA synthesised by Epoch Biolabs, Sugar Land, Texas) cloned into pGlux.1 17 using Xho I and Xma I sites to generate GFPmut2 fusions. Transgenic parasites expressing KAHRP-GFP contained the plasmid pKAHRPGlux.1, comprising DNA encoding residues 1-96 of KAHRP (PFB0100c; DNA synthesized by Epoch Biolabs) cloned into pGlux.1 with Xho I and Xma I to fuse with GFPmut2. Parasites expressing ACPs-GFP are previously described 21. Parasites expressing PfEMP3xQ-GFP contained the plasmid pPfEMP3xQGlux.1, which contained DNA encoding residues 1-36 of PfEMP3, followed by an alanine (to allow for signal peptidase cleavage at IYSEA-) and residues 63-82 of PfEMP3 (which include residues 63aQ64 of the PEXEL; DNA synthesized by Epoch Biolabs) cloned into pGlux.1 using Xho I and Xma I to fuse with GFPmut2. Transgenic parasites 3D7-PMVHA and 3D7-PMIXHA contained the plasmids pPMVHA1.5 or pPMIXHA1.5, respectively, which integrated into either the Plasmepsin V locus (PF13_0133) or Plasmepsin IX locus (PF14_0281), respectively. The pPMVHA1.5 insert was generated by amplifying the 3′ end of PF13_0133 using the oligonucleotides JB116 5′-ATCAGATCTGGGCTGTCATATGCATGAAG-3′ and JB117 5′-ATCCTGCAGGTGTTGATTCCTGTATGGGAG-3′ and the pPMIXHA1.5 insert was generated by amplifying the 3′ end of PF14_0281 using the oligonucleotides PF14_0281-HA-fwd 5′-AGATCTGTGATGATAGTAACATTGATCAG-3′ and PF14_0281-HA-rev 5′-CTGCAGCTAAATTATTTATTTTATTATGTAAGG-3′. Inserts were individually cloned into p1.5-SHA 30 using Bgl II and Pst I (underlined) to generate pPMVHA1.5 or pPMIXHA1.5, respectively. The transgenic parasites 3D7-PMVmutHA episomally expressed plasmid pTETPMVmutHA, which contains DNA encoding Plasmepsin V with D118A, D365A and F370A substitutions fused to exactly the same tags as in 3D7-PMVHA (DNA synthesised by Epoch Biolabs), cloned into the anhydrotetracycline (ATc) regulatable plasmid pTGFP 31 via Pst I and Spe I, which removed GFP. The transgenic parasites 3D7-PMVmutHA2 episomally expressed plasmid pPMVmutHA2, which contains DNA encoding Plasmepsin V with D118A, D365A and F370A substitutions fused to exactly the same tags as in 3D7-PMVHA (DNA synthesised by Epoch Biolabs), cloned into the non-inducible crt promoter-containing plasmid, pGlux.1, via Xho I and Pac I, which removed GFP. Recombinant GBP130 was expressed in E. coli harbouring plasmid pGBP130-3Cmyc18His, which contains codon optimized DNA encoding residues 66 to 196 (containing the PEXEL) of GBP130 fused to triple C-myc tags and triple hexa His tags (DNA synthesised by Epoch Biolabs) cloned into pET11a using Nde I and Bam HI. The PEXEL mutant GBP130 RLE>A protein was expressed as above but contained R84A, L86A and E88A mutations. Recombinant Plasmepsin V was expressed in E. coli harbouring plasmid pHisPMVtrunc, which contains DNA encoding residues E37 to N521 of Plasmepsin V (DNA synthesised by GENEART AG, Regensburg, Germany) cloned into pProExHTb (Invitrogen, Carlsbad, CA, USA) using Bam HI and Xho I to create an N-terminal hexa-His fusion protein.
All plasmids (100 μg) were transfected into P. falciparum 3D7 as previously described 32 and selected with WR99210 and, where appropriate, ATc.
HIV protease inhibitor assays
In vivo assays: Tightly synchronous, 20 hr old transgenic parasites (expressing chimeras from the crt promoter) at 15% parasitaemia were cultured with 50 μM HIV protease inhibitors solubilized in DMSO for 7 hours. Saponin-treated parasite pellets were washed twice in media containing the inhibitor and solubilized in 4x reducing SDS-PAGE sample buffer. In vitro cleavage assays: ipPMVHA activity was assessed with fluorogenic substrates (see below) in the presence of lopinavir at various concentrations solubilised in 100% ethanol, or ethanol alone (maximum 2% final ethanol concentration).
Indirect Immunofluorescence analysis (IFA)
Erythrocytes infected with transgenic parasites were either viewed live or fixed in 100% ice-cold methanol for 3 min and air dried or fixed in 4% paraformaldehyde, 0.01% gluteraldehyde as described previously 17. Cells were incubated with rabbit α-Pf ERC (1:500) 25, 3F10 rat α-HA (1:50) or mouse α-Plasmepsin V (1:5) for 2 hr followed by Alexa Fluor 488- or 594-conjugated secondary IgG antibodies (1:1000; Molecular Probes) for 1 hr and nuclei were labelled with DAPI nuclear stain (Roche) at 0.2 μg/ml in Vectorshield (Vector Labs). Cells were viewed with a Carl Zeiss Axioskop 2 microscope (Thornwood) and images collected using a PCO SensiCam (Motion Engineering Co.) and Axiovision 3 software (Carl Zeiss). Images were assembled with Photoshop CS2 v9.0.2 (Adobe).
Immunoprecipitations
PMVHA and PMVmutHA2 was affinity purified from trophozoite parasites at >5% parasitaemia by solubilizing saponin pellets in 1% (v/v) Triton X-100 in PBS by gentle sonication and rotation at 4 °C for 1 hr. The supernatant was mixed with goat anti-HA antibodies coupled to agarose beads (Sapphire BioScience) for 1 hr at 4 °C and the beads washed six times with PBS, and resuspended in the same buffer. Beads could be stored with active enzyme for 5 days (longer was not tested). PMVmutHA was affinity purified as above from late stage parasites (expressed from MSP2 promoter in pTETPMVmutHA) using magnet separation through CS columns (Miltenyi Biotech) and whole cells solubilized in 1% (v/v) Triton X-100 in PBS.
GFP-tagged chimeras were affinity purified from saponin supernatants for mass spectrometry using one litre of each parasite line and α-GFP agarose (MBL) exactly as described previously 17.
Recombinant Plasmepsin V expression and purification
Plasmepsin V was produced and purified in a manner similar to that described previously 33. Briefly, the protein was synthesised in E. coli strain BL21(DE3) for 3 hr at 37 °C and deposited into insoluble inclusion bodies. The cells were lysed by sonication and the insoluble inclusion bodies solubilised by the addition of 6 M guanidine-HCl, pH 8.0. The solubilised protein was isolated by metal-chelate chromatography using Ni-NTA agarose resin (Qiagen), and eluted in 8 M urea, 1 M imidazole, pH 8.0. The protein was then HPLC purified as described below then various fractions containing Plasmepsin V were concentrated using a vacuum concentrator (Centrivac, Labconco). The acidified Plasmepsin V fractions were then added directly to digest solutions and incubated for various periods at 37 °C.
Plasmepsin V cleavage assays
1. Cleavage assays with synthetic PEXEL peptide substrate: Plasmepsin V activity was measured in 100 μl total digest volume consisting of digest buffer (25 mM Tris hydrochloride + 25mM 4-morpholine ethane sulphonic acid monohydrate, pH 6.4), 2 nmol PEXEL peptide substrate (synthesised by GenScript USA Inc. Piscataway, New Jersey) and either 5 or 20 μl of α-HA beads containing ipPMVHA or ipPMVmutHA, respectively (< 400 μg). Digests were carried out overnight at 37 °C, with shaking. The following day, beads with bound enzyme were removed by passing the reaction through Micro Bio-Spin Chromatography Columns (Bio-Rad) and collecting the flow through, which was analysed using an Agilant 1100 modular HPLC (see below). For activity with the recombinant Plasmepsin V, approximately 3–5 μg of each fraction (1–5) of HPLC-eluted protein was added directly to the reaction, as above. 2. Cleavage assays with recombinant PEXEL substrate: Approximately 1 μg of recombinant GBP130, or the PEXEL mutant protein GBP130 RLE>A, was incubated with ipPMVHA in digest buffer (30 μl total volume) for 1 hr at 37 °C with shaking. Beads with bound enzyme were removed, as above, and the flow through was resuspended in SDS loading buffer and analysed by SDS-PAGE and immunoblot with α-C-myc antibodies. 3. Cleavage assays with fluorogenic PEXEL peptide substrate: 5 μM FRET substrate (DABCYL-G-NKRTLAQKQ-G-EDANS or PEXEL mutant DABCYL-G-NKATAAQKQ-G-EDANS; Genscript Corporation) in water was incubated with 0.5 μl of α-HA beads containing ipPMVHA or ipPMVmutHA2 (< 100 μg) in digest buffer (100 μl total volume). Reactions were prepared on ice and the fluorescence intensity (excitation 340 nm, emission 492 nm) was measured using a GENios fluorescence plate reader (Tecan Group Ltd., Switzerland) heated to 37 °C with orbital shaking between measurements.
After all cleavage assays, enzyme was eluted from the beads retained in the column in non-reducing sample buffer, reduced in 0.1 M dithiothreitol and analysed by SDS-PAGE and immunoblotted with α-HA antibodies.
Reversed-phase HPLC analysis of the refolding reaction
Reversed-phase HPLC was performed using an Agilant 1100 modular HPLC consisting of an on-line degasser, piston pump, autosampler, column oven, diode-array detector and fraction collector. Instrument control, data acquisition and evaluation were performed using Hewlett-Packard Chemstation software for LC and LC/MS systems. Buffer A was comprised 0.05% (v/v) trifluoroacetic acid (HPLC/Spectro grade, Pierce, Rockford, Ill. USA) in Milli-Q grade water (Millipore, Bedford, MA. USA), while buffer B was comprised 0.05% (v/v) trifluoroacetic acid in acetonitrile (ChromAR HPLC grade, Malinckrodt Baker, New Jersey, USA). Protein/peptide samples were centrifuged at 14000 rpm for 15 min at room temperature prior to loading onto a C8 column [2.1 mm I.D. × 100 mm (Brownlee columns, Perkin-Elmer Instruments, Norwalk, CT, USA) in the presence of buffer A. Bound proteins/peptides were eluted using alinear gradient of 0–100% B over 12 min at a flow-rate of 0.5 ml/min at 37 °C.
Subcellular fractionation, SDS-PAGE and immunoblots
To assess subcellular localisation of PfEMP3-GFP and PfEMP3xQ-GFP, infected erythrocytes were lysed in PBS by gentle sonication and centrifuged at 10,000 xg for 15 minutes to obtain PBS supernatant and PBS pellet fractions. The PBS supernatant was centrifuged at 100,000 xg for 1 hr to separate soluble proteins (supernatant) and liposomal membranes (pellet). The PBS pellet was resuspended in 0.2 M Na2CO3 by sonication and incubated on ice for 1 hr and centrifuged at 100,000 xg for 1 hr to obtain integral membrane proteins (pellet). Fractions were solubilized in SDS-PAGE loading buffer. For SDS-PAGE, samples were boiled and electrophoresed in 4–12% or 10% Bis/Tris precast polyacrylamide gels (Invitrogen) using MOPS or MES running buffer and transferred to nitrocellulose membrane using an iBlot (Invitrogen). Membranes were blocked for 1 h in 5% skim milk (Diploma) in PBS containing 0.1% Tween 20 (Sigma). Membranes were probed with either mouse α-GFP antibodies (Roche; 1:1000), mouse α-Plasmepsin V antibodies (1:300), 3F10 rat α-HA antibodies (Roche; 1:1000), mouse α-cMyc antibodies (Sigma; 1:1000) or rabbit α-His probe antibodies (H-15: sc-803; Santa Cruz Biotechnology Inc.; 1:1000) followed by horseradish peroxidase-conjugated secondary antibodies (Silenius; 1:2000) and visualised using enhanced chemiluminescence (ECL; Amersham). BioRad prestained precision markers were used throughout.
Gel excision, digestion and Nano-LC-MS/MS
For protein samples in gels, bands (~2 mm) were excised from the 1-D gel lane and subjected to automated in-gel reduction, alkylation, and tryptic digestion using the MassPREP Station (Micromass, UK) as previously described 17. Digested peptides were subjected to manual in-gel digestion. The gel bands were reduced with 10mM DTT (Calbiochem) for 30 min, alkykated for 30 min with 50mM iodoacetic acid (Fluka) and digested with 375ng trypsin (Worthington) for 16hrs at 37C. The extracted peptide solutions (0.1% formic acid) were then concentrated to approximately 10ul by centrifugal lyophilisation using a SpeedVac AES 1010 (Savant). Digested peptides were subjected to nano liquid chromatography in conjunction with collisional tandem mass spectrometry (nano-LC-MS/MS) as previously described 17. Briefly, extracted peptides were injected and fractionated by nanoflow reversed-phase liquid chromatography on a nano LC system (1200 series, Agilent, USA) using a nanoAcquity C18 150 mm × 0.15 mm I.D. column (Waters, USA) developed with a linear 60-min gradient with a flow rate of 0.5 μl/min at 45 °C from 100% solvent A (0.1% Formic acid in Milli-Q water) to 100 % solvent B (0.1% Formic acid, 60% acetonitrile, (Mallinckrodt Baker, New Jersey, USA) 40% Milli-Q water). The nano HPLC was coupled on-line to an LTQ-Orbitrap mass spectrometer equipped with a nanoelectrospray ion source (Thermo Fisher Scientific) for automated MS/MS. Up to five most intense ions per cycle were fragmented and analysed in the linear trap, with target ions already selected for MS/MS being dynamically excluded for 3 min.
Mass spectra database searching, protein identification, bioinformatic and statistical analysis
Mass spectra peak lists were extracted using extract-msn as part of Bioworks 3.3.1 (Thermo Fisher Scientific) linked into Mascot Daemon (Matrix Science, UK). The parameters used to generate the peak lists for the Orbitrap were as follows: minimum mass 400; maximum mass 5000; grouping tolerance 0.01 Da; intermediate scans 1; minimum group count 1; 10 peaks minimum and total ion current of 100. Peak lists for each nano-LC-MS/MS run were used to search MASCOT v2.2.04 search algorithm (Matrix Science, UK) provided by the Australian Proteomics Computational Facility (www.apcf.edu.au). Automatic charge state recognition was used because of the high-resolution survey scan (30000). LC-MS/MS files were searched against a PEXEL sequence database (552306 entries) comprising sequences from the latest version of Swiss-Prot (all species), Trembl (Plasmodium entries), plasmoDB (all entries), synthetic peptides and the PEXEL constructs. The search parameters consisted of carboxymethylation of cysteine as a fixed modification (+58 Da) with variable modifications set for NH2-terminal acetylation (+42 Da) and oxidation of methionine (+16 Da). A peptide mass tolerance of ±20 ppm, #13C defined as 1, fragment ion mass tolerance of ±0.8 Da, and an allowance for up to three missed cleavages for tryptic searches where trypsin was used as the digest reagent. No-enzyme (unrestricted) searches were also carried out on all Orbitrap-LTQ data and manual validation of all peptide spectral matches was done irrespective of peptide scores or expectation values (E-values) to ensure that all major fragment ions were annotated in accordance with known rules of peptide fragmentation.
The signal sequence-processing site of PfEMP3-GFP and PfEMP3xQ-GFP was predicted by SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) with both neural networks and hidden Markov models trained on eukaryotes 34. Statistical analyses were performed with GraphPad Prism 5 for Windows.
Supplementary Material
Acknowledgments
We thank the Australian Red Cross Blood Bank for the provision of human blood and serum, the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH for providing HIV protease inhibitors and MR4 (ATCC) for Plasmepsin V antibodies, contributed by D. Goldberg. This work was supported by the National Health and Medical Research Council, and a grant from the National Institutes of Health (RO1 AI44008). J.A.B. is an NHMRC Peter Doherty postdoctoral Fellow and A.F.C. is an International Research Scholar of the Howard Hughes Medical Institute and an Australia Fellow.
Footnotes
Author Contributions J.A.B. performed the biochemical and cell biological experiments. A.N.H. performed HPLC, biochemical analysis and purification of recombinant Plasmepsin V. S.G. made 3D7-PMIXHA, purified recombinant GBP130 proteins and analysed their processing. H.P. and E.A.K performed the proteomics analysis. T.F.dK-W. attempted the knockouts of Plasmepsin V in P. berghei and analysed the results. All authors contributed to the study design and to writing the manuscript.
References
- 1.Snow RW, et al. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller LH, Good MF, Milon G. Malaria pathogenesis. Science. 1994;264:1878–1883. doi: 10.1126/science.8009217. [DOI] [PubMed] [Google Scholar]
- 3.Raventos Suarez C, Kaul DK, Macaluso F, Nagel RL. Membrane knobs are required for the microcirculatory obstruction induced by Plasmodium falciparum-infected erythrocytes. Proc Natl Acad Sci USA. 1985;82:3829–3833. doi: 10.1073/pnas.82.11.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnwell JW, et al. A human 88-kD membrane glycoprotein (CD36) functions in vitro as a receptor for a cytoadherence ligand on Plasmodium falciparum-infected erythrocytes. J Clin Invest. 1989;84:765–772. doi: 10.1172/JCI114234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marti M, et al. Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science. 2004;306:1930–1933. doi: 10.1126/science.1102452. [DOI] [PubMed] [Google Scholar]
- 6.Hiller NL, et al. A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science. 2004;306:1934–1937. doi: 10.1126/science.1102737. [DOI] [PubMed] [Google Scholar]
- 7.Pei X, et al. Structural and Functional Studies of Interaction between Plasmodium falciparum Knob-associated Histidine-rich Protein (KAHRP) and Erythrocyte Spectrin. J Biol Chem. 2005;280:31166–31171. doi: 10.1074/jbc.M505298200. [DOI] [PubMed] [Google Scholar]
- 8.Baruch DI, et al. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- 9.Su XZ, et al. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 10.Smith JD, et al. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 1995;82:101–110. doi: 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maier AG, et al. Exported proteins required for virulence and rigidity of Plasmodium falciparum-infected human erythrocytes. Cell. 2008;134:48–61. doi: 10.1016/j.cell.2008.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maier AG, Cooke BM, Cowman AF, Tilley L. Malaria parasite proteins that remodel the host erythrocyte. Nat Rev Microbiol. 2009;7:341–354. doi: 10.1038/nrmicro2110. [DOI] [PubMed] [Google Scholar]
- 13.de Koning-Ward TF, et al. A newly discovered protein export machine in malaria parasites. Nature. 2009;459:945–949. doi: 10.1038/nature08104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sargeant TJ, et al. Lineage-specific expansion of proteins exported to erythrocytes in malaria parasites. Genome Biol. 2006;7:R12. doi: 10.1186/gb-2006-7-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Ooij C, et al. The malaria secretome: from algorithms to essential function in blood stage infection. PLoS Pathog. 2008;4:e1000084. doi: 10.1371/journal.ppat.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang HH, et al. N-terminal processing of proteins exported by malaria parasites. Mol Biochem Parasitol. 2008;160:107–115. doi: 10.1016/j.molbiopara.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boddey JA, Moritz RL, Simpson RJ, Cowman AF. Role of the Plasmodium export element in trafficking parasite proteins to the infected erythrocyte. Traffic. 2009;10:285–299. doi: 10.1111/j.1600-0854.2008.00864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Przyborski JM, et al. Trafficking of STEVOR to the Maurer’s clefts in Plasmodium falciparum-infected erythrocytes. EMBO J. 2005;24:2306–2317. doi: 10.1038/sj.emboj.7600720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber IT, et al. Molecular modeling of the HIV-1 protease and its substrate binding site. Science. 1989;243:928–931. doi: 10.1126/science.2537531. [DOI] [PubMed] [Google Scholar]
- 20.Miller M, et al. Structure of complex of synthetic HIV-1 protease with a substrate-based inhibitor at 2.3 A resolution. Science. 1989;246:1149–1152. doi: 10.1126/science.2686029. [DOI] [PubMed] [Google Scholar]
- 21.Waller RF, Reed MB, Cowman AF, McFadden GI. Protein trafficking to the plastid of Plasmodium falciparum is via the secretory pathway. EMBO J. 2000;19:1794–1802. doi: 10.1093/emboj/19.8.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bozdech Z, et al. The Transcriptome of the Intraerythrocytic Developmental Cycle of Plasmodium falciparum. PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Roch KG, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 24.Klemba M, Goldberg DE. Characterization of plasmepsin V, a membrane-bound aspartic protease homolog in the endoplasmic reticulum of Plasmodium falciparum. Mol Biochem Parasitol. 2005;143:183–191. doi: 10.1016/j.molbiopara.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 25.La Greca N, et al. Identification of an endoplasmic reticulum-resident calcium-binding protein with multiple EF-hand motifs in asexual stages of Plasmodium falciparum. Mol Biochem Parasitol. 1997;89:283–293. doi: 10.1016/s0166-6851(97)00134-5. [DOI] [PubMed] [Google Scholar]
- 26.Knuepfer E, Rug M, Cowman AF. Function of the Plasmodium export element can be blocked by green fluorescent protein. Mol Biochem Parasitol. 2005;142:258–262. doi: 10.1016/j.molbiopara.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 27.Carlton JM, et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pain A, et al. The genome of the simian and human malaria parasite Plasmodium knowlesi. Nature. 2008;455:799–803. doi: 10.1038/nature07306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meissner M, et al. Tetracycline analogue-regulated transgene expression in Plasmodium falciparum blood stages using Toxoplasma gondii transactivators. Proc Natl Acad Sci U S A. 2005;102:2980–2985. doi: 10.1073/pnas.0500112102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baum J, et al. A malaria parasite formin regulates actin polymerization and localizes to the parasite-erythrocyte moving junction during invasion. Cell Host Microbe. 2008;3:188–198. doi: 10.1016/j.chom.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 31.Gilson PR, et al. MSP1(19) miniproteins can serve as targets for invasion inhibitory antibodies in Plasmodium falciparum provided they contain the correct domains for cell surface trafficking. Mol Microbiol. 2008;68:124–138. doi: 10.1111/j.1365-2958.2008.06140.x. [DOI] [PubMed] [Google Scholar]
- 32.Duraisingh MT, Triglia T, Cowman AF. Negative selection of Plasmodium falciparum reveals targeted gene deletion by double crossover recombination. Int J Parasitol. 2002;32:81–89. doi: 10.1016/s0020-7519(01)00345-9. [DOI] [PubMed] [Google Scholar]
- 33.Hodder AN, et al. Structural insights into the protease-like antigen Plasmodium falciparum SERA5 and its noncanonical active-site serine. J Mol Biol. 2009;392:154–165. doi: 10.1016/j.jmb.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.