Abstract
Background:
Identification of the genetic factors that underlie stimulant responsiveness in animal models has significant implications for better understanding and treating stimulant addiction in humans.
Methods:
F2 progeny derived from parental rat strains F344/NHsd and LEW/NHsd, which differ in responses to drugs of abuse, were used in quantitative trait locus (QTL) analyses to identify genomic regions associated with amphetamine-induced locomotion (AIL) and G-protein levels in the nucleus accumbens (NAc).
Results:
The most robust QTLs were observed on chromosome 3 (maximal log ratio statistic score (LRSmax)=21.3) for AIL and on chromosome 2 (LRSmax=22.0) for Gαi3. A “suggestive” QTL (LRSmax=12.5) was observed for AIL in a region of chromosome 2 that overlaps the Gαi3 QTL. Novelty-induced locomotion showed different QTL patterns from AIL, with the most robust QTL on chromosome 13 (LRSmax=12.2).
Conclusions:
Specific unique and overlapping genomic regions influence AIL, novelty-induced locomotion, and inhibitory G-protein levels in the NAc. These findings suggest that common genetic mechanisms influence certain biochemical and behavioral aspects of stimulant responsiveness.
Keywords: Addiction, F344 rats, Lewis rats, genetics, QTL, amphetamine, novelty-induced locomotion, nucleus accumbens, G proteins
Introduction
Differences in individual responses to stimulants exist in humans and other species. Some exposed individuals become dependent on amphetamine, methamphetamine, or cocaine, while others do not (Barr et al, 2006; Sofuoglu and Kosten, 2006). Genetic influences on psychiatric disorders associated with stimulant usage are also significant, with the genetic contributions to drug dependence (including to stimulants) estimated at 30%-60% (Tsuang et al, 1998). Divergent responses to stimulants are also important in the clinical arena, in which some individuals with attention deficit hyperactivity disorder (ADHD) are treated effectively with amphetamine or methylphenidate (Biederman et al, 2004). Cocaine dependence risk loci have been mapped in humans via genetic linkage (Gelernter et al, 2005); animal studies, however, provide the opportunity to identify risk loci for specific traits associated with pharmacological responses and other phenotypes that cannot be readily ascertained in human subjects.
Amphetamine-induced locomotion (AIL) is a well-studied phenomenon in animal models of psychiatric disorders, including stimulant addiction and psychoses (Blackburn and Szumlinski 1997; Ikemoto and Witkin 2003; Rajakumar et al, 2005). AIL, regarded as a measure of mesolimbic dopamine function (Dellu-Hagedorn, 2005), is mediated via dopaminergic transmission in the nucleus accumbens (NAc) (Kim and Vezina, 1998; Chausmer and Ettenberg, 1999; Millan et al, 1999; Ikemoto and Witkin, 2003). AIL differs from other types of locomotion, such as novelty-induced locomotion (NIL), which has been associated with stress responses. A complex relationship exists between drug-induced behaviors and novelty responses in rodents (Hiroi and Agatsuma, 2005; Agatsuma et al, 2006). AIL is differentially regulated in rats with high or low levels of spontaneous exploratory behavior (Corda et al, 2005; Alttoa et al, 2007). While AIL and NIL correlate with one another in some studies, genetically distinct animal strains show differences in AIL and NIL, and these animals provide an opportunity for identifying genetic factors influencing stimulant responsiveness (Brodkin et al, 1998; Marley et al, 1998; Stohr et al, 1998; Conversi et al, 2006). For example, Fisher (F344/NHsd) as compared with Lewis (LEW/NHsd), rats show more robust AIL, and roughly equivalent NIL (Brodkin et al, 1998; Stohr et al, 1998).
F344/NHsd and LEW/NHsd (referred to hereafter as F344 and LEW, respectively) rat lines have been used as models for multiple psychiatric disorders including addiction (Nestler et al, 1996; Kosten and Ambrosio, 2002), schizophrenia (Lipska and Weinberger, 1996), and depression (Lahmame et al, 1997). Previously, we reported differences in AIL and related phenotypes in these strains and their F2 progeny that were generated in preparation for quantitative trait locus (QTL) analysis (Brodkin et al, 1998). Multiple biochemical phenotypes related to striatal dopamine function, and implicated in stimulant addiction, were measured by western blotting specifically in the NAc, including inhibitory G protein subunits (Gαi1,2 and Gαi3), the dopamine transporter, the transcription factor ΔFosB, and the protein phosphatase inhibitor DARPP-32. Among the phenotypes investigated, levels of Gαi subunits appeared particularly promising for further study in QTL analyses (Brodkin et al, 1998). QTL analysis has been used previously to identify genomic regions contributing to a variety of quantitative traits, for example, stress-responsiveness, alcohol or morphine consumption, aggressive behaviors, and behavioral reactivity and emotionality (Moisan et al, 1996; Remmers et al, 1996; Bice et al, 1998; Ramos et al, 1999; Brodkin et al, 2002; Potenza et al, 2004; Ferraro et al, 2005). QTL analysis appears particularly applicable to the study of phenotypes related to psychiatric disorders, given its ability to identify genomic contributions to phenotypes determined by multiple genes. However, the genomic regions identified are usually large and contain multiple candidate genes, and additional investigation is typically required to identify specific genes.
Previously, we used QTL analysis to identify regions on chromosomes 4 and 10, which contribute to differences in peak corticosterone levels in F344 and LEW rats (Potenza et al, 2004). Here, we use a similar approach to investigate genetic contributions to AIL and levels of Gαi1,2 and Gαi3 in the NAc, using NIL as a behavioral control condition for AIL. We hypothesized that: 1) we would identify genomic regions contributing to AIL, NIL, Gαi1,2 levels, and Gαi3 levels; 2) genomic regions contributing to AIL would be distinct from those contributing to NIL; and 3) genomic regions contributing to AIL would partially overlap with those contributing to levels of Gαi1,2 and Gαi3 in the NAc.
Methods and Materials
Animal Procedures
The animal care and use committee at Yale University approved the study. The research was performed in strict accordance with the NIH Guide for the Care and Use of Laboratory Animals. F344 and LEW rats were maintained and characterized as described previously (Brodkin et al, 1998; Potenza et al, 2004). F344 and LEW rats were obtained from Harlan-Sprague Dawley (Indianapolis, IN) at 35-45 days of age. F1 progeny were generated by both F344 (female) X LEW (male) and LEW (female) X F344 (male) crosses and F2 intercross progeny were derived from mating of both (F344 X LEW)F1 X (F344 X LEW)F1 and (LEW X F344)F1 X (LEW X F344)F1 pairs. F2 progeny were weaned at 21 days of age. Animals were housed in groups of two to four with food (Purina chow) and tap water ad libitum in a temperature-controlled colony with a 12-hour light/dark cycle (lights on at 0700 hour). Only males were included in this study, to limit variation in measures associated with the estrus cycle in females, and given known sex differences in F344/NHsd and LEW/NHsd rats in AIL and NIL (Stohr et al, 1998).
Locomotor Activity Measures
Rats were assessed for locomotor activity between 8 and 10 am and between the ages of 50 and 60 days using a concentric circular device as described previously (Brodkin et al, 1998). Activity was recorded for 60 min. Data for the first ten min of activity in the novel environment were used to assess NIL, as prior studies showed that differences among rats in NIL were most pronounced during this time (Brodkin et al, 1998). Immediately following the 60 min test period, animals received injections of DL-amphetamine hemi-sulfate (2.0 mg/kg s.c.), and locomotor activity was measured for an additional 60 min. Amphetamine was supplied by the National Institute on Drug Abuse (Baltimore, MD), made in isotonic saline and administered in a volume of 1 ml/kg. AIL data were calculated as cumulative activity over the 60-min test period.
Serum Levels of Amphetamine
F344 and LEW rats were given amphetamine (2 mg/kg, intraperitoneally) and trunk blood was obtained 40 min later, at the peak of behavioral effects of amphetamine. Samples were centrifuged for 10 min at 3000 g, the serum removed and stored at −20°C until analysis. Before analysis, amphetamine-d3 (50ng/ml) was added to each sample as an internal standard. Serum amphetamine levels were detected using a Micromass Ultima liquid chromatography/tandem mass spectrometry system (Waters Corp; Milford, MA) in positive ion mode and a BDS-C18 column (ThermoFischer Scientific, Inc.; West Palm Beach, FL) with a 0.8 ml/min flow rate as previously described (Hendrickson et al, 2004). Mobile phase was 20% acetonitrile, 0.05% acetic acid and 5 mM ammonium acetate. Amphetamine signals were quantified by comparison of multiple reaction monitoring signals to the internal standard.
G Protein Measures
Five to eight days after behavioral testing, animals were killed by decapitation between 3 and 5 pm as described previously (Brodkin et al, 1998). This was performed in a separate area from animal housing, with each animal retrieved individually from the housing area, and with glove changing between animals to minimize possible stress-related alterations in mesolimbic functioning. Brains were removed rapidly and cooled in ice-cold physiological buffer (Brodkin et al, 1998). The nucleus accumbens samples were obtained from 1 mm thick coronal cross-sections by use of a 12-gauge syringe needle and were stored at −70°C until Western blotting was performed as reported previously (Brodkin et al, 1998).
DNA Extraction, Purification, Amplification and Analysis
As described previously (Potenza et al, 2004), genomic DNA was obtained from frozen liver tissue of F344 and LEW parental animals and F2 progeny via alkaline lysis and column purification strategy (Qiagen; www.qiagen.com). DNA quality was assessed by agarose gel electrophoresis. DNA amplification was performed using the polymerase chain reaction and primers obtained from Research Genetics (www.resgen.com), the National Institute of Arthritis and Musculoskeletal and Skin Diseases (Bethesda, MD) (Remmers et al, 1996), or Applied Biosystems Incorporated (ABI; www.appliedbiosystems.com). DNA analysis was performed via size fractionation, either via agarose gel electrophoresis and ethidium bromide visualization, or acrylamide gel electrophoresis and fluorescence detection using an ABI 377 semiautomated sequencer. Data from these gels were independently read by two individuals and double-entered prior to analysis.
QTL Analysis
QTL analysis was performed as described previously (Remmers et al, 1996; Potenza et al, 2004). Sample sizes were based on power analyses and prior studies that successfully identified QTLs in rats (Remmers et al, 1996). One hundred eighty-eight of 298 F2 progeny, including those with phenotypic extremes for AIL, NIL, Gαi1,2 levels, and Gαi3 levels (top and bottom 5-15%), were analyzed at 178 genetic loci distributed across the rat autosomes (average (SD) spacing of 8.65 (4.62) cM). We investigated power for the QTL analysis using the method of revolving power (Darvasi and Soller, 1997), which was calculated under the assumption of an infinite number of markers and was found to be similar to the 95% confidence interval (CI) of QTL map location using moderate marker spacing, e.g., 10 to 20 cM. Our analysis showed that with a sample size of 188, the 95% CI is 20 cM for a standardized dominant effect of d=0.63. The average spacing (8.65 cM) of the 178 genetic loci used in this study is substantially smaller than 20 cM, and with denser marker spacing, smaller effect sizes can be detected. Based on these analyses, we have a good coverage of marker variation and adequate power to detect QTL of magnitudes of d=0.63 or less.
Data were analyzed as described previously with MAPMAKER/EXP and MAPMAKER/QTL (Remmers et al, 1996) and MapManager QT (Manly and Olson, 1999). Data presented are from analyses using MapManager QT. The likelihood ratio statistic (LRS) score, a value that is 4.6 times the lod score value, is used to report magnitude of the QTLs as has been done previously when reporting results from analyses using MapManager QT (McBrearty et al, 1998; Hahn et al, 2004; Potenza et al, 2004). “Suggestive significance” and “genomewide significance” thresholds were those recommended by Lander and Kruglyak (Lander and Kruglyak, 1995), using LRS scores of 9.9 and 15.2 for “suggestive” and [genomewide] “significant” QTLs, respectively. Marker map locations from a SHRSPxBN genetic map as described in the rat genome database (www.rgd.mcw.edu/GENOMESCANNER) were used in the QTL analyses, with study data-derived distances used for six markers (D1Arb8, D1Arb11, D1Arb25, D4Arb17, D12Arb8, and D20Mgh1) not available from the map. Exploration of candidate genes in the vicinities of identified QTL and identification of corresponding regions of human and mouse genomes were performed using the NCBI Map Viewer (http://www.ncbi.nlm.nih.gov/mapview/).
Mixed data have been reported with respect to the position of one marker (D3Rat63) employed in the present study. The Rat Genome Database (http://rgd.mcw.edu/objectSearch/sslpReport.jsp?rgd_id=35218), the MIT rat genomics website (http://www.broad.mit.edu/rat/public/) and the NCBI website (http://www.ncbi.nlm.nih.gov/mapview/map_search.cgi?taxid=10116&query=d3rat63&q chr=&strain=All&advsrch=off) all list the marker as mapping to chromosome 3. The Ensembl website locates the marker on chromosome 8 but lists the map location on chromosome 3 (http://www.ensembl.org/Rattus_norvegicus/markerview?marker=oxsts6992). Analyzing with MAPMAKER/EXP and MAPMAKER/QTL (Remmers et al, 1996) and MapManager QT (Manly and Olson, 1999) the F2 genotype data that we generated, we found that by linkage analysis the D3Rat63 marker grouped with the chromosome 3 markers between D3Rat24 and D3Arb12. This mapping procedure confirms that the map location used for the D3Rat63 marker was correct. This procedure was followed for all markers used in the analyses to verify their orders and positions.
Results
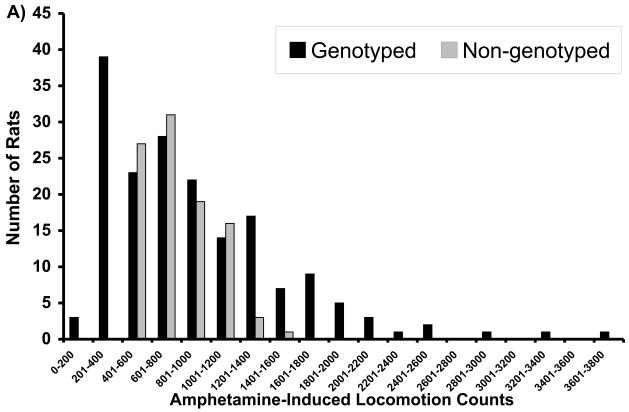
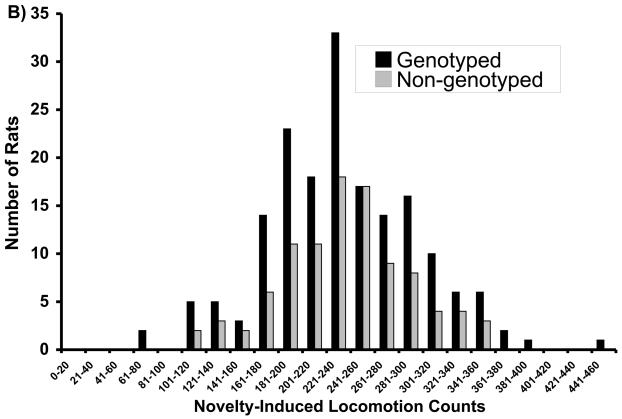
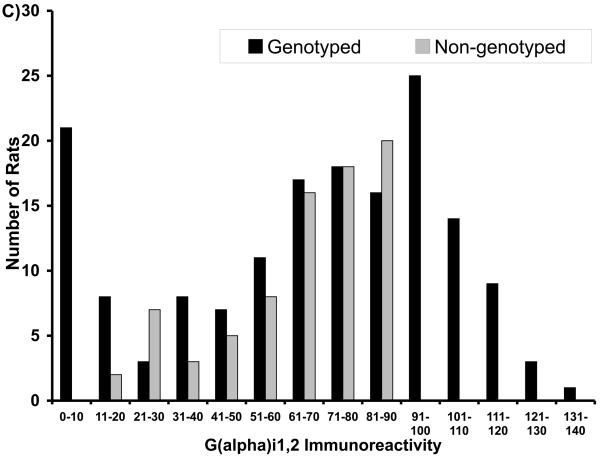
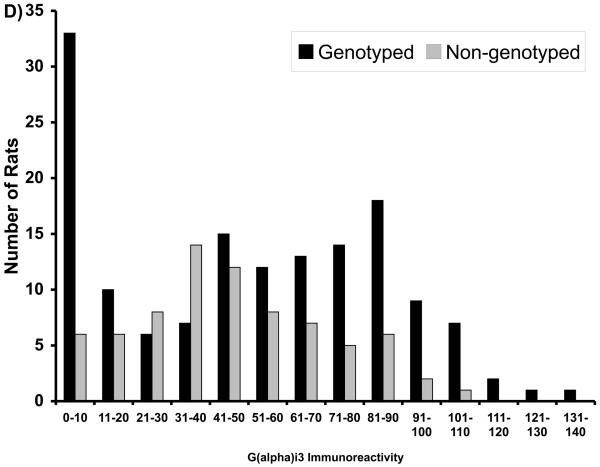
General characteristics of the F344 and LEW parental animals and F2 intercross progeny used in the QTL analysis have been described previously (Brodkin et al, 1998). F344 animals had significantly higher AIL, and higher Gαi1,2 and Gαi3 levels in the NAc, than did the LEW rats, although the ranges overlapped for the groups (Brodkin et al, 1998). Serum levels of amphetamine did not differ for F344 and LEW rats (mean ± SEM for F344 (125±10.8 ng/ml; n=10) and LEW (110±8.8 ng/ml; n=10); p=0.32) indicating that differences in AIL in F344 and LEW rats was not attributable to differences in amphetamine metabolism. No significant between-group differences in F344 and LEW rats were observed for NIL (Brodkin et al, 1998). The F2 progeny displayed AIL, Gαi1,2, and Gαi3 levels intermediate between the F344 and LEW parental strains. Mean AIL scores in F2 progeny were more similar to LEW than to F344 rats, whereas mean Gαi1,2 and Gαi3 levels were more similar to F344 rats than to LEW rats (Brodkin et al, 1998). The distributions of the F2 progeny selected for genotypic analysis are displayed (Figure 1).
Figure 1.
Distributions of F2 progeny with respect to (A) amphetamine-induced locomotion (AIL), (B) novelty-induced locomotion (NIL), (C) Gai1,2 levels, and (D) Gai3 levels. Progeny selected for genotyping are indicated in dark bars.
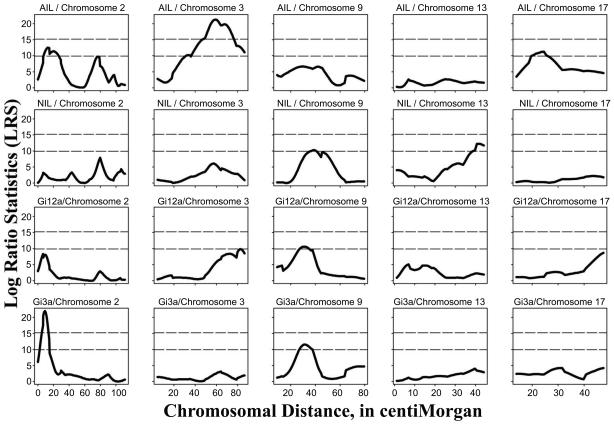
Results of an autosomal genomewide QTL linkage scan employing 178 markers are displayed in Table 1. The broadest significance peak and largest likelihood (log ratio statistics or LRS) score for AIL was observed on chromosome 3 (LRSmax=21.3) between the markers D3Rat24 and D3Rat63 (Table 1, Figure 2). This peak reached genomewide significance and was estimated to account for 12% of the phenotypic variance for AIL. QTL peaks in this region did not reach even “suggestive” significance levels for NIL (LRSmax=6.1; 4% of variance), Gαi1,2 levels (LRSmax=9.8 [slightly more distal than AIL peak]; 7% of variance), or Gαi3 levels (LRSmax=3.1; 2% of variance). Peaks reaching “suggestive” significance level thresholds for AIL were observed on the proximal region of chromosome 2 between D2Rat182 and D2Rat11 (LRSmax=12.5; 8% of variance) and on chromosome 17 between D17Rat117 and D17Rat15 (LRSmax=11.3; 7% of variance). X2 statistics for adjacent loci reaching at least p<0.05 significance for each individual locus are shown in Table 2. No other peaks reached “suggestive” significance.
Table 1.
Log Ratio Statistic Scores for Amphetamine Induced Locomotion, Novelty Induced Locomotion, and Gαi Levels in Nucleus Accumbens at Specific Genomic Locations.
| Marker | Chromosome | Map Location (cM) |
Log Ratio Statistic, AIL |
Log Ratio Statistic, NIL |
Log Ratio Statistic, Gαi1,2 |
Log Ratio Statistic, Gαi3 |
|---|---|---|---|---|---|---|
| D1Rat4 | 1 | 9.2 | 1.6 | 0.3 | 0.7 | 0.1 |
| D1Rat7 | 1 | 12.5 | 2.2 | 0.5 | 0.7 | 1.8 |
| D1Rat19 | 1 | 23.8 | 2.6 | 0.4 | 0.4 | 1 |
| D1Arb8 | 1 | 35 | 4.2 | 0 | 0.3 | 0.9 |
| D1Rat256 | 1 | 28.3 | 1.9 | 0.3 | 1.7 | 0.4 |
| D1Rat266 | 1 | 46.8 | 4.5 | 1 | 2.4 | 2.3 |
| D1Mgh7 | 1 | 53.8 | 6.8 | 0.1 | 2.2 | 3.2 |
| D1Arb11 | 1 | 57 | 4.8 | 0.8 | 4.5 | 4.2 |
| D1Rat35 | 1 | 59.4 | 3.7 | 1.2 | 5 | 5.2 |
| D1Rat215 | 1 | 74.5 | 0.1 | 0 | 0.7 | 0.6 |
| D1Rat164 | 1 | 83.5 | 0.5 | 0.9 | 0.3 | 0.4 |
| D1Rat437 | 1 | 89.1 | 0.3 | 0.7 | 1.5 | 0.9 |
| D1Rat67 | 1 | 95.9 | 0.9 | 2.1 | 1.7 | 0.8 |
| D1Rat70 | 1 | 106.1 | 1.2 | 6.4 | 2.5 | 1.4 |
| D1Rat169 | 1 | 122 | 3.2 | 7.1 | 0.4 | 0.7 |
| D1Rat76 | 1 | 125.3 | 1.9 | 8.8 | 0.6 | 1 |
| D1Mgh12 | 1 | 133.4 | 0.5 | 0.8 | 1 | 1.4 |
| D1Arb25 | 1 | 139 | 1.9 | 3.3 | 0.2 | 0.7 |
| D1Rat122 | 1 | 143.5 | 2.6 | 1.6 | 0.9 | 0.1 |
| D2Rat3 | 2 | 0.1 | 2.6 | 0.1 | 3 | 6.1 |
| D2Rat182 | 2 | 6.9 | 9.7 | 3.2 | 7.2 | 20.5 |
| D2Rat11 | 2 | 15.1 | 10.6 | 1.5 | 3.5 | 8.8 |
| D2Mit6 | 2 | 29.5 | 7.7 | 0.9 | 0.8 | 3.4 |
| D2Mgh19 | 2 | 35.5 | 3.7 | 0.9 | 0.9 | 2 |
| D2Rat217 | 2 | 43.5 | 0.9 | 3.3 | 0.7 | 2.1 |
| D2Rat34 | 2 | 57.1 | 0.1 | 0.1 | 0.3 | 1.1 |
| D2Rat170 | 2 | 68.2 | 4.9 | 1.3 | 0 | 0.7 |
| D2Rat240 | 2 | 79.7 | 6.8 | 7.9 | 2.9 | 0.7 |
| D2Rat62 | 2 | 90.8 | 1.7 | 1.4 | 0.5 | 2.1 |
| D2Rat185 | 2 | 97.7 | 4 | 1.6 | 0.2 | 0.3 |
| D2Rat69 | 2 | 106.8 | 1.2 | 4.3 | 0.4 | 0.2 |
| D2Rat168 | 2 | 111.5 | 0.9 | 2.9 | 0.3 | 0.6 |
| D3Rat53 | 3 | 4.6 | 2.8 | 1 | 0.4 | 1.4 |
| D3Rat80 | 3 | 18.9 | 4.6 | 0 | 0.9 | 0.6 |
| D3Rat75 | 3 | 36 | 9.8 | 2 | 0.7 | 0.7 |
| D3Rat24 | 3 | 49.5 | 15.6 | 3.6 | 1.6 | 0.6 |
| D3Rat63 | 3 | 65.2 | 19.5 | 4.6 | 7.5 | 2.7 |
| D3Arb12 | 3 | 79.9 | 13.1 | 2.9 | 8.9 | 1.1 |
| D3Mgh10 | 3 | 86.5 | 11.1 | 0.9 | 8.5 | 1.9 |
| D4Arb14 | 4 | 0 | 1.5 | 0.5 | 0.6 | 0.9 |
| D4Rat11 | 4 | 18.2 | 1.1 | 1.1 | 1.1 | 1.1 |
| D4Arb17 | 4 | 23 | 1.1 | 0.6 | 1.8 | 1.3 |
| D4Rat153 | 4 | 27.1 | 1.6 | 0.5 | 6 | 2.2 |
| D4Rat15 | 4 | 29.4 | 5.9 | 0.6 | 3.9 | 2.5 |
| D4Rat24 | 4 | 32.8 | 0.1 | 0.2 | 1 | 0 |
| D4Rat226 | 4 | 32.9 | 0.4 | 1.1 | 1.8 | 1.6 |
| D4Arb8 | 4 | 34.1 | 0.7 | 2.2 | 1.3 | 0.2 |
| D4Rat33 | 4 | 40.8 | 0.8 | 2 | 1.6 | 0.8 |
| D4Rat108 | 4 | 41.9 | 0.9 | 0.7 | 2.8 | 0.7 |
| D4Rat172 | 4 | 46.4 | 0.1 | 0.9 | 2 | 0.7 |
| D4Rat40 | 4 | 48.7 | 0.8 | 2.6 | 3.3 | 0.9 |
| D4Rat48 | 4 | 54.3 | 0.9 | 2.2 | 1.1 | 0.3 |
| D4Rat193 | 4 | 62.5 | 0.3 | 1.2 | 2.5 | 0 |
| D4Rat60 | 4 | 71.4 | 0 | 1.5 | 0.6 | 0.3 |
| D4Rat241 | 4 | 77 | 0.3 | 3 | 1.1 | 0.7 |
| D4Rat66 | 4 | 82.6 | 1.9 | 1.6 | 1.8 | 0.2 |
| D4Mgh30 | 4 | 86.7 | 2.6 | 1.6 | 0.3 | 0.6 |
| D4Rat112 | 4 | 98.8 | 0.9 | 3 | 0.4 | 0 |
| D5Rat121 | 5 | 3.5 | 5 | 1 | 5.1 | 3.6 |
| D5Rat126 | 5 | 16 | 8.1 | 5.3 | 4.5 | 2.6 |
| D5Rat82 | 5 | 26 | 7.8 | 4.7 | 6.1 | 8.6 |
| D5Rat10 | 5 | 37 | 8.4 | 4.5 | 5.8 | 7.6 |
| D5Rat85 | 5 | 48 | 5.9 | 3.1 | 5.8 | 9 |
| D5Rat196 | 5 | 58.1 | 5.7 | 3.4 | 3.3 | 4.3 |
| D5Rat30 | 5 | 68.3 | 3 | 2.2 | 3.2 | 0.3 |
| D5Rat171 | 5 | 78.5 | 6.4 | 2.1 | 1.6 | 0.2 |
| D5Rat93 | 5 | 85.2 | 5.4 | 0.7 | 0.4 | 0.4 |
| D5Rat49 | 5 | 105.6 | 2.4 | 0.9 | 3 | 6.4 |
| D6Rat41 | 6 | 20.6 | 1.2 | 8.4 | 1.8 | 1.6 |
| D6Rat29 | 6 | 33 | 0.8 | 1.1 | 0.5 | 0.2 |
| D6Rat133 | 6 | 37.5 | 2.8 | 0.8 | 1 | 0.9 |
| D6Rat23 | 6 | 46.6 | 1 | 1.3 | 1 | 1.2 |
| D6Rat14 | 6 | 57.8 | 1.5 | 0.8 | 2 | 2.6 |
| D6Rat160 | 6 | 76 | 2.3 | 1.3 | 0.8 | 1.8 |
| D6Rat109 | 6 | 85.2 | 0.6 | 3.4 | 1.5 | 2.5 |
| D7Rat113 | 7 | 3.1 | 8.9 | 3.8 | 1.2 | 2 |
| D7Rat37 | 7 | 6.8 | 4.4 | 0.5 | 2.5 | 1.7 |
| D7Rat27 | 7 | 22 | 6 | 0.1 | 3.5 | 1.9 |
| D7Rat51 | 7 | 31 | 6.6 | 2.8 | 1.8 | 0.7 |
| D7Rat22 | 7 | 45.5 | 1.9 | 6.3 | 0.6 | 2 |
| D7Rat139 | 7 | 52.3 | 4.9 | 4.7 | 2 | 1.8 |
| D7Rat11 | 7 | 64.7 | 3.9 | 0.5 | 3.3 | 2 |
| D7Rat81 | 7 | 72.4 | 1 | 0.5 | 1.3 | 0.3 |
| D7Rat4 | 7 | 80.4 | 1.7 | 1.4 | 1.1 | 0.4 |
| D8Rat77 | 8 | 0 | 3 | 0.1 | 1.1 | 0.1 |
| D8Rat55 | 8 | 8.4 | 5.6 | 0.3 | 2.7 | 2.6 |
| D8Rat52 | 8 | 14.3 | 0.8 | 0.2 | 7.4 | 5 |
| D8Rat164 | 8 | 18.5 | 0.7 | 0.1 | 5.6 | 5.8 |
| D8Arb6 | 8 | 23.1 | 0.8 | 0.7 | 1.8 | 2.2 |
| D8Arb8 | 8 | 34.6 | 0.7 | 1 | 1.6 | 1.6 |
| D8Rat43 | 8 | 36.9 | 0.4 | 5.6 | 0.9 | 0.7 |
| D8Rat36 | 8 | 41.2 | 0 | 4.3 | 2.4 | 3 |
| D8Rat23 | 8 | 46 | 1.9 | 6.7 | 6.9 | 5.2 |
| D8Rat21 | 8 | 47.1 | 2.1 | 7 | 8.9 | 3.8 |
| D8Rat16 | 8 | 57.2 | 1.2 | 6 | 2.7 | 0 |
| D8Rat123 | 8 | 66.3 | 0.9 | 3.8 | 0.1 | 0.6 |
| D8Rat11 | 8 | 71.9 | 2.1 | 4.5 | 0.4 | 0.1 |
| D8Arb119 | 8 | 77.4 | 0.6 | 4.1 | 1 | 0.1 |
| D9Rat135 | 9 | 9.1 | 4 | 0.2 | 4.2 | 1.2 |
| D9Rat133 | 9 | 13.9 | 4 | 0.2 | 3.1 | 1.5 |
| D9Rat126 | 9 | 37.7 | 5.9 | 10.2 | 9.6 | 10 |
| D9Rat13 | 9 | 45.5 | 6.2 | 9.7 | 2.4 | 1.6 |
| D9Rat110 | 9 | 64.7 | 3.6 | 0.2 | 1.2 | 3.5 |
| D9Rat1 | 9 | 79.5 | 2.2 | 0.5 | 0.6 | 4.7 |
| D10Rat218 | 10 | 4.6 | 3.7 | 4 | 0.6 | 0.9 |
| D10Rat117 | 10 | 6.1 | 2.1 | 3.8 | 1.6 | 0.1 |
| D10Rat45 | 10 | 21.6 | 2.1 | 0.3 | 0.8 | 0.1 |
| D10Rat38 | 10 | 34 | 3.3 | 1.8 | 2 | 1.8 |
| D10Rat164 | 10 | 38.5 | 1.6 | 0.4 | 0.2 | 0.5 |
| D10Rat28 | 10 | 47.6 | 1.6 | 0.5 | 1.4 | 0.9 |
| D10Rat153 | 10 | 51 | 0.8 | 0.1 | 1.6 | 1.2 |
| D10Rat124 | 10 | 58.5 | 1.5 | 0.9 | 1.3 | 1.8 |
| D10Rat142 | 10 | 66.5 | 2.8 | 0.4 | 3.1 | 1.7 |
| D10Rat203 | 10 | 71.1 | 3.2 | 0.8 | 6.6 | 6.2 |
| D10Rat15 | 10 | 73.3 | 3.8 | 0.8 | 7.3 | 6.6 |
| D10Rat11 | 10 | 77.1 | 2.2 | 2.2 | 4.5 | 6 |
| D10Rat8 | 10 | 85.9 | 2.7 | 0.8 | 0.5 | 2.6 |
| D10Rat105 | 10 | 92.7 | 3.7 | 0.6 | 0.4 | 3.9 |
| D10Rat135 | 10 | 94.1 | 4.1 | 0.6 | 0.4 | 4.1 |
| D11Rat73 | 11 | 8.2 | 5.2 | 0.9 | 0.1 | 0.5 |
| D11Mit1 | 11 | 11.5 | 3.4 | 0.8 | 0.1 | 0.8 |
| D11Rat6 | 11 | 19.5 | 2.3 | 0.8 | 2.3 | 1.8 |
| D11Rat91 | 11 | 36.5 | 4.6 | 2 | 2.8 | 1.1 |
| D12Rat59 | 12 | 5.9 | 1.7 | 3.2 | 4.3 | 0.5 |
| D12Arb8 | 12 | 11 | 0.5 | 3.2 | 2.3 | 3.1 |
| D12Rat4 | 12 | 17.1 | 0.8 | 3 | 3 | 3.8 |
| D12Rat51 | 12 | 24.1 | 0.9 | 4.1 | 0.5 | 0.8 |
| D12Rat76 | 12 | 33 | 1 | 1.3 | 0.5 | 2.1 |
| D12Rat52 | 12 | 44.3 | 0.1 | 1.8 | 1.7 | 3.6 |
| D12Rat44 | 12 | 54.1 | 3.4 | 0.1 | 5 | 1.1 |
| D13Rat7 | 13 | 1.2 | 0.3 | 4 | 0.9 | 0.2 |
| D13Arb5 | 13 | 7.9 | 2.2 | 2.1 | 3.7 | 0.9 |
| D13Arb8 | 13 | 14.8 | 0.7 | 2.1 | 4.7 | 1.4 |
| D13Rat126 | 13 | 19.3 | 1.1 | 0.6 | 4 | 1.5 |
| D13Rat85 | 13 | 26.1 | 2.6 | 4.3 | 0.9 | 1.5 |
| D13Rat131 | 13 | 33.9 | 1.5 | 5.7 | 0.8 | 2.4 |
| D13Mit4 | 13 | 40.7 | 1.8 | 12.2 | 2.3 | 3.4 |
| D13Rat153 | 13 | 44.1 | 1.6 | 11.7 | 1.9 | 2.9 |
| D14Rat72 | 14 | 2.3 | 2.1 | 0.8 | 0.2 | 0.2 |
| D14Rat77 | 14 | 6.9 | 0.3 | 0.6 | 2.5 | 1 |
| D14Rat50 | 14 | 15.9 | 1 | 1.7 | 1.4 | 0.5 |
| D14Rat68 | 14 | 28.2 | 2.4 | 2.2 | 0.9 | 0.3 |
| D14Arb10 | 14 | 43 | 4.8 | 2.6 | 1.2 | 0.4 |
| D14Rat49 | 14 | 64 | 0.6 | 0.2 | 1.6 | 1 |
| D15Rat55 | 15 | 5.5 | 2.3 | 3.5 | 7.5 | 2 |
| D15Rat66 | 15 | 15.8 | 0.9 | 4.3 | 0.3 | 0.4 |
| D15Rat116 | 15 | 25 | 0 | 7 | 1.5 | 0.6 |
| D15Rat96 | 15 | 41.9 | 1.8 | 6 | 0.8 | 1.2 |
| D15Rat26 | 15 | 58.9 | 3.4 | 2 | 0.5 | 1.9 |
| D16Rat35 | 16 | 5.6 | 6.8 | 1.5 | 0.4 | 0.4 |
| D16Rat67 | 16 | 18.1 | 5 | 1.6 | 2.6 | 1.2 |
| D16Rat53 | 16 | 28.3 | 3.3 | 0.3 | 1.5 | 0.2 |
| D16Rat37 | 16 | 38.5 | 1.9 | 2.5 | 0.4 | 0.3 |
| D16Rat15 | 16 | 46.7 | 3.8 | 0.4 | 0.3 | 0.2 |
| D17Rat115 | 17 | 13.9 | 3.5 | 0.3 | 1.1 | 2.4 |
| D17Rat117 | 17 | 19.6 | 9.9 | 0.4 | 0.8 | 2.2 |
| D17Rat15 | 17 | 25.5 | 10.1 | 0.2 | 2.3 | 2.7 |
| D17Arb7 | 17 | 32.6 | 5.6 | 1.2 | 1.8 | 3 |
| D17Rat130 | 17 | 40.8 | 5.3 | 2 | 3.9 | 2.2 |
| D17Rat154 | 17 | 47.5 | 4.6 | 1.8 | 8.7 | 4.2 |
| D18Rat133 | 18 | 2.5 | 1.1 | 5 | 0.3 | 0.3 |
| D18Rat25 | 18 | 11.5 | 2.9 | 6.9 | 0.2 | 0.2 |
| D18Rat17 | 18 | 18.3 | 0.8 | 2 | 1.8 | 0.5 |
| D18Rat55 | 18 | 22.7 | 0.9 | 3.7 | 1.8 | 1.3 |
| D18Rat8 | 18 | 43.2 | 0.8 | 0.9 | 4.9 | 2.2 |
| D18Rat76 | 18 | 48.9 | 0.6 | 0.6 | 4.1 | 1.9 |
| D19Rat82 | 19 | 6.7 | 0.5 | 0.5 | 5.6 | 1.8 |
| D19Rat12 | 19 | 20.2 | 1.7 | 1.3 | 2.6 | 1.5 |
| D19Rat35 | 19 | 27.8 | 1.5 | 2 | 1 | 1 |
| D19Rat67 | 19 | 35.7 | 1.4 | 2.3 | 1 | 2.6 |
| D19Rat63 | 19 | 39.1 | 3.5 | 0.7 | 0 | 0.5 |
| D19Rat58 | 19 | 48.8 | 3.1 | 3 | 0.1 | 1 |
| D20Rat46 | 20 | 0 | 3.1 | 0.4 | 2.1 | 2 |
| D20Rat31 | 20 | 11.5 | 0.1 | 1.9 | 0.1 | 0 |
| D20Rat34 | 20 | 21.5 | 0.6 | 2.5 | 0 | 0.6 |
| D20Rat39 | 20 | 30.9 | 1.2 | 0.9 | 0.4 | 0.4 |
| D20Mgh1 | 20 | 39 | 4 | 1.2 | 0.6 | 0.7 |
| D20Arb10 | 20 | 48.2 | 3.9 | 1.3 | 0.7 | 0.8 |
Figure 2.
LRS values for amphetamine-induced locomotion (AIL), novelty-induced locomotion (NIL) and G-protein (Gai1,2 and Gai3) levels on chromosomes showing suggestive or significant QTLs. Horizontal lines indicate thresholds for suggestive or significant QTL values, as described in the methods.
Table 2.
Chi-squared statistics for individual markers
| Markers | Chromosome | LRS.AIL | LRS.NIL | LRS.Gi12a | LRS.Gi3a | AIL.pvalues | NIL.pvalues | Gi12a.pvalues | Gi3a.pvalues |
|---|---|---|---|---|---|---|---|---|---|
| D2Rat3 | 2 | 2.6 | 0.1 | 3 | 6.1 | 0.27 | 0.95 | 0.22 | 0.05 |
| D2Rat182 | 2 | 9.7 | 3.2 | 7.2 | 20.5 | 0.008 | 0.2 | 0.03 | 0.00004 |
| D2Rat11 | 2 | 10.6 | 1.5 | 3.5 | 8.8 | 0.005 | 0.47 | 0.17 | 0.01 |
| D2Mit6 | 2 | 7.7 | 0.9 | 0.8 | 3.4 | 0.02 | 0.64 | 0.67 | 0.19 |
| D3Rat75 | 3 | 9.8 | 2 | 0.7 | 0.7 | 0.007 | 0.37 | 0.7 | 0.7 |
| D3Rat24 | 3 | 15.6 | 3.6 | 1.6 | 0.6 | 0.0004 | 0.17 | 0.45 | 0.74 |
| D3Rat63 | 3 | 19.5 | 4.6 | 7.5 | 2.7 | 0.00006 | 0.1 | 0.02 | 0.26 |
| D3Arb12 | 3 | 13.1 | 2.9 | 8.9 | 1.1 | 0.001 | 0.24 | 0.01 | 0.58 |
| D3Mgh10 | 3 | 11.1 | 0.9 | 8.5 | 1.9 | 0.004 | 0.64 | 0.01 | 0.39 |
| D9Rat126 | 9 | 5.9 | 10.2 | 9.6 | 10 | 0.05 | 0.006 | 0.008 | 0.007 |
| D9Rat13 | 9 | 6.2 | 9.7 | 2.4 | 1.6 | 0.05 | 0.008 | 0.3 | 0.45 |
| D13Mit4 | 13 | 1.8 | 12.2 | 2.3 | 3.4 | 0.41 | 0.002 | 0.32 | 0.18 |
| D13Rat153 | 13 | 1.6 | 11.7 | 1.9 | 2.9 | 0.45 | 0.003 | 0.39 | 0.23 |
| D17Rat117 | 17 | 9.9 | 0.4 | 0.8 | 2.2 | 0.007 | 0.82 | 0.67 | 0.33 |
| D17Rat15 | 17 | 10.1 | 0.2 | 2.3 | 2.7 | 0.006 | 0.9 | 0.32 | 0.26 |
Results presented represent findings from multipoint analyses keyed to single marker locations.
Data presented represent two or more contiguous markers at p<0.05 in a free regression model for any of the studied phenotypes.
P values significant at p<0.05 are bolded. AIL p values for D9Rat126 and D9Rat13 are 0.052 and 0.04505, respectively.
Among the other phenotypes, the most robust QTL peak was observed for Gαi3 levels in the NAc. The QTL peak was significant at a genomewide level (LRSmax=22.0; 15% of variance) and was located on the proximal end of chromosome 2 between D2Rat182 and D2Rat11. This region coincides with a “suggestive” QTL peak for AIL. Within this region, no QTL peaks for NIL (LRSmax=3.2; 2% of variance) nor for Gαi1,2 levels (LRSmax=8.1; 5% of variance) reached “suggestive” significance. No other QTL peaks for Gαi3 levels and none for NIL or Gαi1,2 reached genomewide significance. A “suggestive” QTL peak for Gαi3 levels was observed in the vicinity of D9Rat126 (LRSmax=11.5; 8% of variance), and “suggestive” peaks for NIL (LRSmax =10.3; 6% of variance) and Gαi1,2 level (LRSmax=10.6; 7% of variance) were observed in the same region. An additional “suggestive” peak for NIL was observed on chromosome 13 at 13Mit4 (LRSmax=12.2; 8% of variance).
Conclusions
Summary of Findings
The current study investigated genomic contributions to differences in AIL, NIL, and levels of inhibitory G-protein subunits (Gαi1,2 and Gαi3) in the NAc in two inbred strains of rats. Our first hypothesis, that we would identify genomic regions contributing to AIL, NIL, Gαi1,2 levels, and Gαi3 levels, was partially supported. Specifically, QTLs reaching genomewide significance were identified for AIL and Gαi3 levels, and additional QTLs reaching “suggestive” significance levels were identified for all four phenotypes. Our second hypothesis, that genomic regions contributing to AIL would be distinct from those contributing to NIL, was largely supported. No QTL for NIL reached “suggestive” significance levels in the vicinities of QTLs reaching “suggestive” or genomewide significance levels for AIL and vice versa. Our third hypothesis, that genomic regions contributing to AIL would partially overlap with those contributing to levels of Gαi1,2 and Gαi3 in the NAc, was supported. Specifically, the location of the QTL peak on chromosome 2, which reached genomewide significance for Gαi3 levels in the NAc, coincided regionally with the QTL peak reaching “suggestive” significance for AIL. Analogously, QTL peaks reaching “suggestive” significance levels were observed in a similar region of chromosome 9 for NIL, Gαi1,2 levels, and Gαi3 levels. Together, these findings suggest that distinct genetic mechanisms underlie at least some of the differences in AIL and NIL observed in F344 and LEW rats. The findings also suggest overlapping genetic influences for biochemical and behavioral measures for AIL and Gαi3 levels and for NIL and Gαi1,2 and Gαi3 levels, respectively. The implications of these findings for psychiatric disorders are described below.
Importance of Current Findings
This study represents the first to our knowledge to identify via QTL analysis in any species genomic regions linked to inhibitory G protein levels in the NAc, and to identify genomic regions associated with AIL and NIL in rats. Although QTL and other linkage analyses have been used in studies of numerous psychiatric disorders in humans and related phenotypes in animal models, few have addressed stimulant-related behaviors, and even fewer biochemical markers within the NAc, and we believe that ours is the first to do so simultaneously in any species. One prior study using recombinant inbred mice identified a region of chromosome 19 that influences dopamine transporter (DAT) binding, which correlated with cocaine- and methamphetamine-induced locomotion (Janowsky et al, 2001). Another study of recombinant inbred mice found a QTL associated with copper levels in NAc on a region of chromosomes 5 in males (Jones et al, 2006). Brain copper concentrations were associated with cocaine-related open-field behavior in these mice (Jones et al, 1999). A third study found that mice lacking the M5 muscarinic receptor gene (located on chromosome 2) showed increased dopamine D2 receptor expression in the NAc and diminished AIL (Wang et al, 2004). A fourth study used gene expression microarray analyses to identify differences in gene expression in the NAc in two mouse lines selected for differences in methamphetamine-induced locomotion (Palmer et al, 2005). Using an internet database, the authors identified a region on chromosome 15 that co-mapped with the behavioral QTL for methamphetamine-induced locomotion. Similar regions of murine chromosome 15 have been associated with stimulant-, phencyclidine- and ethanol-induced locomotion (Alexander et al, 1996; Grisel, Belknap et al, 1997; Phillips et al, 1998; Jones et al, 1999; Boyle and Gill, 2001; Downing et al, 2003). Together, these studies indicate that stimulant-induced locomotion is a poly-genetically determined behavior. With respect to the present findings, only the chromosome 2 falls within a homologous region implicated in the current analysis, corresponding to the region of rat chromosome 3 with the most robust QTL for AIL.
Genomic Regions, Candidate Genes and Implications
Our study, unlike any prior report, directly searched for QTL associated with AIL using F2 progeny generated from an intercross of two rat strains showing parental differences in AIL. Consistent with prior studies, multiple genomic regions were identified in association with AIL. QTL analyses define relatively large intervals that are highly likely to contain multiple candidate genes that might contribute to the phenotype under investigation. Consequently, candidate genes within the region, while important to consider, should be viewed cautiously.
The most robust QTL for AIL was found on chromosome 3 in a location corresponding to regions of chromosome 11p, 15q and 20q in humans and chromosome 2 in mice. Regions of human chromosome 11p have been identified in autosome/genomewide scans as contributing to tobacco smoking, alcoholism and opioid dependence (Long et al, 1998; Gelernter, Liu et al, 2004; Gelernter et al, 2006). The region of human chromosome 15 homologous to the identified region of rat chromosome 3 has been implicated in multiple studies of alcoholism and tobacco smoking and contains a GABA receptor gene cluster and the gene encoding the nicotinic acetylcholine α7 subunit (Leonard et al, 2000; Song et al, 2003). The region of human chromosome 20q homologous to the identified region of rat chromosome 3 has been implicated in studies of cigarette traits (Saccone et al, 2003). Other regions of the human genome map to the region of rat chromosome 3 containing the QTL. For example, the gene encoding brain-derived neurotrophic factor (BDNF), located on human chromosome 11p, is within the identified region of rat chromosome 3. BDNF is induced by chronic cocaine (Kumar et al, 2005), regulated by CREB (Choi et al, 2006), and implicated in cocaine withdrawal (Grimm et al, 2003; Pu et al, 2006) and cocaine-induced locomotion and reward (Horger et al, 1999; Hall et al, 2003), and possibly associated with substance dependence (e.g., (Zhang et al, 2006)). Other candidate genes including ones included within the identified region of rat chromosome 3 include those coding for the M5 muscarinic cholinergic receptor and ΔFosB/FosB. The muscarinic M5 receptor has been previously implicated in NAc function and AIL in mice (Wang et al, 2004). ΔFosB has been widely implicated in addictive processes, including the rewarding and locomotor effects of stimulants (Nestler et al, 2001; Zhu et al, 2007).
Among the genomic regions reaching suggestive significance for AIL, the one on chromosome 2 overlapped in proximity with the most robust QTL identified in the study, that for Gαi3 in the NAc. This region is homologous to regions of chromosome 5q in humans and chromosome 13 in mice. Although this region appears distinct from a GABA(A) gene cluster implicated in alcoholism (Radel et al, 2005), regions of human chromosome 5q closer to the homologous region of rat chromosome have been implicated in alcohol craving (Ehlers and Wilhelmsen, 2005) and event-related brain potentials in families with a history of alcoholism (Almasy et al, 2001). One of the genes in this region of rat chromosome 2 is that encoding the serotonin 1A receptor is expressed in the NAc (Luna-Munguia et al, 2005), couples through Gαi with strongest affinity for Gαi3 (Pucadyil et al, 2005), influences cocaine-induced dopamine levels in the NAc (Andrews et al, 2005) and cocaine-induced locomotion (Carey et al, 2005), and has been implicated in aggressive behaviors, including those induced by cocaine (Knyshevski et al, 2005). A nearby gene is that for tyrosine hydroxylase, the rate-limiting enzyme in dopamine synthesis, which has been implicated in cocaine self-administration (Self et al, 2004). Also in this genomic region is the gene for the peptide CART (cocaine and amphetamine related transcript), which has been implicated in addictive processes including cocaine dependence (Jaworski and Jones, 2006). CART is expressed in mesolimbic regions including the NAc (Philpot and Smith, 2006), is regulated by stimulant exposure, dopaminergic transmission, and the cAMP pathway in the NAc (Hunter et al, 2006; Jones and Kuhar, 2006), influences cocaine-induced locomotion (Jaworski et al, 2003), and is elevated the NAc of people who have abused cocaine (Albertson et al, 2004).
A region of chromosome 17 corresponding to areas of chromosome 6p in humans and 13 in mice reached suggestive significance for AIL. In humans, chromosome 6p has been implicated in tobacco smoking (Fust et al, 2004) and intelligence in individuals with alcoholism and their families (Dick et al, 2006). Of several genes in this region of rat chromosome 17, prolactin has been implicated in multiple studies of people with cocaine dependence. Prolactin levels have been associated with severity of cocaine use (Patkar et al, 2006), cocaine administration increases prolactin levels (Elman and Lukas, 2005), serotonergically induced prolactin release is blunted in cocaine dependent subjects (Patkar et al, 2006), and this effect is associated with high behavioral disinhibition and aggression (Patkar et al, 2006). Another gene in this vicinity is that encoding protein phosphatase 1, which has been implicated in the function of NAc neurons and in their response to cocaine (Hu et al, 2005; Svenningsson et al, 2005; Zachariou et al, 2006). Also present is the gene for Cdk5, a protein kinase, which has been implicated in cocaine-mediated dopamine signaling (Chergui et al, 2004; Takahashi et al, 2005), is increased following chronic cocaine or methamphetamine exposure (Bibb et al, 2001; Benavides and Bibb, 2004; Chen and Chen, 2005), interacts with tyrosine hydroxylase (Kansy et al, 2004), is regulated by ΔFosB (Kumar et al, 2005), and has been implicated in NAc-mediated methamphetamine-induced locomotion (Chen and Chen, 2005).
Dopamine systems contribute to exploratory behaviors in rodents (Kliethermes and Crabbe, 2006) and differences in the genetic influences that contribute to exploratory and drug-induced locomotion have been reported (Hiroi and Agatsuma, 2005; Agatsuma et al, 2006). A region of chromosome 9 contained suggestive QTL for NIL and levels of Gαi1,2 and Gαi3 in the NAc. This region corresponds to areas of chromosome 2q in humans and 1 in mice. In humans, a similar genomic region of chromosome 2q has been implicated in co-occurring alcoholism and depression (Nurnberger et al, 2001). Among genes in this region of rat chromosome 9 is that for CREB, which is widely implicated in G-protein-related signal transduction in the NAc and animal models of stress and addiction (Barrot et al, 2005; Carlezon et al, 2005; Green et al, 2006; Nestler and Carlezon, 2006).
Study Limitations and Future Directions
There exist multiple limitations in the present study. First, we only examined males, and future research in female populations is needed. Second, other inbred rat strains may also show differences in AIL, NIL, and levels of Gα subunits in NAc, and in their relationship to other specific behavioral or biochemical measures of psychiatric relevance; we cannot apply any direct conclusions to these others strains at this point. However, although certain QTLs may be strain-specific, we speculate that common QTLs will influence phenotypes across strains and outbred rats. Also, it is likely that other strains may be used to identify additional QTLs relevant to stimulant responsiveness and biochemical measures in the NAc. A third limitation is that the findings were obtained in rats and the extent to which they are applicable to humans warrants further investigation. A fourth limitation is inherent to QTL analyses like the one employed here. Specifically, large genomic regions are identified that contain many candidate genes. Additional research is necessary to determine the extent to which specific genes are implicated and to identify the nature of the genetic differences generating the biochemical and behavioral differences observed in the parental rat strains. Such work could involve gene expression investigations or the identification of strain-related differences in coding regions of candidate genes within the regions defined by the QTLs. A fifth limitation involves the use of an F2 design with respect to the investigation of dominant versus recessive loci or on loci with effects dependent on interactions with strain specific alleles at the X-chromosome or maternal environment. A sixth limitation involves the complex nature of behavioral responses like AIL and NIL, and future investigations could examine the extent to which specific genetic factors contribute to specific aspects (e.g., the temporal magnitudes) of the responses.
Despite the limitations of the present study, the findings identify rat QTLs underlying differences in stimulant responsiveness and NAc functioning. As such, the investigation is important in several ways. Few prior studies have attempted to map neurochemical QTLs in inbred strains of rats. Since much work has been devoted to defining the neurochemistry in rat models of psychiatric disorders, this line of research is important in that it circumvents the need to translate phenotypes well-described in rats to genetic models in mice. As rat genomics become increasingly well-defined and more frequently used, this line of research will become increasingly important. Likewise, few prior studies have simultaneously mapped behavioral and neurochemical QTLs concurrently. The simultaneous assessment of QTLs provides insight into the molecular mechanisms underlying behaviors with psychiatric relevance. Finally, and most importantly, the findings of specific genomic regions influencing stimulant responsiveness provide a basis for future investigations into the genetic basis for multiple psychiatric conditions in humans.
Grant Support and Other Acknowledgments
This work was supported by a Young Investigator Award from the National Alliance for Research in Schizophrenia and Depression (NARSAD), a Drug Abuse Research Scholar Program in Psychiatry Award from the American Psychiatric Association and the National Institute on Drug Abuse (K12-DA00366), the Clinician Scientist Training Program (K12-DA00167), the U.S. Department of Veterans Affairs (the VA Connecticut-Massachusetts Mental Illness Research, Education and Clinical Center (MIRECC), VA Research Enhancement Award program (REAP) and the Veterans Affairs Neuroscience and Traumatic Brain Injuries Post-doctoral Fellowship), NIDA R01 DA12849, NIAAA R01 AA11330, NIDA P01 DA08227, NIMH P50 MH66172, Burroughs Wellcome Fund Career Award in the Biomedical Sciences, and NIMH KO8-MH068586. The authors thank Xingguang Luo, Eric Londin, Michael Bernabeo, Yong Huang and Anne Marie Lacobelle for technical assistance and Elaine F. Remmers and Ronald L. Wilder for advice on QTL methodologies.
Footnotes
Conflicts of Interest/Disclosure
All of the authors report that they have no conflicts of interest over the past three years to report as related to the subject of the report. Dr. Potenza has received financial support or compensation for the following: Dr. Potenza consults for and is an advisor to Boehringer Ingelheim; has consulted for and has financial interests in Somaxon; has received research support from the National Institutes of Health, Veteran's Administration, Mohegan Sun, and Forest Laboratories, Ortho-McNeil and Oy-Control/Biotie pharmaceuticals; has participated in surveys, mailings or telephone consultations related to drug addiction, impulse control disorders or other health topics; has consulted for law offices and the federal public defender's office in issues related to impulse control disorders; has performed grant reviews for the National Institutes of Health and other agencies; has given academic lectures in grand rounds, CME events and other clinical or scientific venues; has generated books or book chapters for publishers of mental health texts; and provides clinical care in the Connecticut Department of Mental Health and Addiction Services Problem Gambling Services Program. Dr. Brodkin has received financial support or compensation for the following: Dr. Brodkin has performed grant reviews for the U.S. Civilian Research and Development Foundation, the New Jersey Governor,s Council on Autism, and the U.S. Department of Defense Autism Spectrum Disorder Research Program (grant review organized by Constella Group, Inc); is a member of the Gerson Lehrman Group Healthcare Council (although have not performed any services or received any compensation from Gerson Lehrman Group to date); has received research support from the National Institutes of Health, the Burroughs Wellcome Fund, the Cure Autism Now Foundation, NARSAD, the Philadelphia Foundation, and the Department of Veteran Affairs; and has given academic lectures in grand rounds and other clinical or scientific venues. Dr. Gelernter has received financial support or compensation from the following: related to consultation for Columbia University, the Thailand Center for Excellence for Life Sciences (TCELS), the University of CT Health Center, NIH, and Faegre & Benson; related to grant reviews for the National Institutes of Health; and related to academic lectures and editorial functions in various scientific venues (including for the ACNP). Drs. Yang, Birnbaum and Nestler do not have any additional financial support, compensation or personal financial holdings to disclose according to journal policy.
References
- Agatsuma S, Lee M, Zhu H, Chen K, Shih JC, Seif I, Hiroi N. Monoamine oxidase A knockout mice exhibit impaired nicotine preference but normal responses to novel stimuli. Human Mol Genet. 2006;15:2721–2731. doi: 10.1093/hmg/ddl206. [DOI] [PubMed] [Google Scholar]
- Albertson DN, Pruetz B, Schmidt CJ, Kuhn DM, Kapatos G, Bannon MJ. Gene expression profile of the nucleus accumbens of human cocaine abusers: evidence for dysregulation of myelin. J Neurochem. 2004;88:1211–1219. doi: 10.1046/j.1471-4159.2003.02247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander RC, Wright R, Freed W. Quantitative trait loci contributing to phencyclidine-induced and amphetamine-induced locomotor behavior in inbred rats. Neuropsychopharmacol. 1996;15:484–490. doi: 10.1016/S0893-133X(96)00058-9. [DOI] [PubMed] [Google Scholar]
- Almasy L, Porjesz B, Blangero J, Goate A, Edenberg HJ, Chorlian DB, et al. Genetics of event-related brain potentials in response to a semantic priming paradigm in families with a history of alcoholism. Am J Human Genet. 2001;68:128–135. doi: 10.1086/316936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews CM, Kung HF, Lucki I. The 5-HT1A receptor modulates the effects of cocaine on extracellular serotonin and dopamine levels in the nucleus accumbens. Eur J Pharmacol. 2005;508:123–130. doi: 10.1016/j.ejphar.2004.12.033. [DOI] [PubMed] [Google Scholar]
- Barr AM, Panenka WJ, MacEwan GW, Thornton AE, Lang DJ, Honer WG, et al. The need for speed: an update on methamphetamine addiction. J Psychiatry Neurosci. 2006;331:301–313. [PMC free article] [PubMed] [Google Scholar]
- Barrot M, Wallace DL, Bolanos CA, Graham DL, Perrotti LI, Neve RL, et al. Regulation of anxiety and initiation of sexual behavior by CREB in the nucleus accumbens. Proc Nat Acad Sci USA. 2005;102:8357–8362. doi: 10.1073/pnas.0500587102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benavides DR, Bibb JA. Role of Cdk5 in drug abuse and plasticity. Ann NY Acad Sci. 2004;1025:335–344. doi: 10.1196/annals.1316.041. [DOI] [PubMed] [Google Scholar]
- Bibb JA, Chen J, Taylor JR, Svenningsson P, Nishi A, Snyder GL, et al. Effects of chronic exposure to cocaine are regulated by the neuronal protein Cdk5. Nature. 2001;410:376–380. doi: 10.1038/35066591. [DOI] [PubMed] [Google Scholar]
- Bice P, Foroud T, Bo R, Castelluccio P, Lumeng L, Li T-K, et al. Genomic screen for QTLs underlying alcohol consumption in the P and NP rat lines. Mammalian Genome. 1998;9:949–955. doi: 10.1007/s003359900905. [DOI] [PubMed] [Google Scholar]
- Biederman J, Spencer T, Wilens T. Evidence-based pharmacotherapy for attention-deficit hyperactivity disorder. Int J Neuropsychopharmacol. 2004;7:293–300. doi: 10.1017/S1461145703003973. [DOI] [PubMed] [Google Scholar]
- Blackburn JR, Szumlinski KK. Ibogaine effects on sweet preference and amphetamine induced locomotion: implications for drug addiction. Behav Brain Res. 1997;89:99–106. doi: 10.1016/s0166-4328(97)00050-8. [DOI] [PubMed] [Google Scholar]
- Boyle AEL, Gill K. Sensitivity of AXB/BXA recombinant inbred lines of mice to the locomotor activating effects of cocaine: a quantitative trait loci analysis. Pharmacogenetics. 2001;11:254–264. doi: 10.1097/00008571-200104000-00009. [DOI] [PubMed] [Google Scholar]
- Brodkin ES, Carlezon WA, Jr, Haile CN, Kosten TA, Heninger GR, Nestler EJ. Genetic analysis of behavioral, neuroendocrine, and biochemical parameters in inbred rodents: Initial studies in Lewis and Fischer 344 rats and in A/J and C57BL/6J mice. Brain Res. 1998;805:55–68. doi: 10.1016/s0006-8993(98)00663-5. [DOI] [PubMed] [Google Scholar]
- Brodkin ES, Goforth SA, Keene AH, Fossella JA, Silver LM. Identification of quantitative trait loci that affect aggressive behavior in mice. J Neurosci. 2002;22:1165–1170. doi: 10.1523/JNEUROSCI.22-03-01165.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey RJ, DePalma G, Damianopoulos E, Shanahan A, Muller CP, Huston JP. Evidence that the 5-HT1A autoreceptor is an important pharmacological target for the modulation of cocaine behavioral stimulant effects. Brain Res. 2005;1034:162–171. doi: 10.1016/j.brainres.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Carlezon WA, Jr, Duman RS, Nestler EJ. The many faces of CREB. Trends Neurosci. 2005;28:436–445. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Chausmer A, Ettenberg A. Intraaccumbens raclopride attenuates amphetamine-induced locomotion, but fails to prevent the response-reinstating properties of food reinforcement. Pharmacology, Biochemistry & Behavior. 1999;62:299–305. doi: 10.1016/s0091-3057(98)00165-8. [DOI] [PubMed] [Google Scholar]
- Chen PC, Chen JC. Enhanced Cdk5 activity and p35 translocation in the ventral striatum of acute and chronic methamphetamine-treated rat. Neuropsychopharmacol. 2005;30:538–549. doi: 10.1038/sj.npp.1300604. [DOI] [PubMed] [Google Scholar]
- Chergui K, Svenningsson P, Greengard P. Cyclin-dependent kinase 5 regulates dopaminergic and glutamatergic transmission in the striatum. Proc Nat Acad Sci USA. 2004;101:2191–2196. doi: 10.1073/pnas.0308652100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KH, Whisler K, Graham DL, Self DW. Antisense-induced reduction in nucleus accumbens cyclic AMP response element binding protein attenuates cocaine reinforcement. Neurosci. 2006;137:373–383. doi: 10.1016/j.neuroscience.2005.10.049. [DOI] [PubMed] [Google Scholar]
- Conversi D, Bonito-Oliva A, Orsini C, Cabib S. Habituation to the test cage influences amphetamine-induced locomotion and Fos expression and increases FosB/DeltaFosB-like immunoreactivity in mice. Neurosci. 2006;141:597–605. doi: 10.1016/j.neuroscience.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Corda MG, Piras G, Lecca D, Fernandez-Teruel A, Driscoll P, Giorgi O. The psychogenetically selected Roman rat lines differ in the susceptibility to develop amphetamine sensitization. Behav Brain Res. 2005;157:147–156. doi: 10.1016/j.bbr.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Darvasi A, Soller M. A simple method to calculate resolving power and confidence interval of QTL map location. Behav Genet. 1997;27:125–132. doi: 10.1023/a:1025685324830. [DOI] [PubMed] [Google Scholar]
- Dellu-Hagedorn F. Spontaneous individual differences in cognitive performances of young adult rats predict locomotor response to amphetamine. Neurobio Learning Memory. 2005;83:43–47. doi: 10.1016/j.nlm.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Dick DM, Aliev F, Bierut L, Goate A, Rice J, Hinrichs A, et al. Linkage analyses of IQ in the collaborative study on the genetics of alcoholism (COGA) sample. Behav Genet. 2006;36:77–86. doi: 10.1007/s10519-005-9009-8. [DOI] [PubMed] [Google Scholar]
- Downing C, Rodd-Henricks KK, Flaherty L, Dudek BC. Genetic analysis of the psychomotor stimulant effect of ethanol. Genes Brain Behav. 2003;2:140–151. doi: 10.1034/j.1601-183x.2003.00022.x. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Wilhelmsen KC. Genomic scan for alcohol craving in Mission Indians. Psychiatric Genet. 2005;15:71–75. doi: 10.1097/00041444-200503000-00012. [DOI] [PubMed] [Google Scholar]
- Elman I, Lukas SE. Effects of cortisol and cocaine on plasma prolactin and growth hormone levels in cocaine-dependent volunteers. Addict Behav. 2005;30:859–864. doi: 10.1016/j.addbeh.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Ferraro TN, Golden GT, Smith GG, Martin JF, Schwebel CL, Doyle GA, et al. Confirmation of a major QTL influencing oral morphine intake in C57 and DBA mice using reciprocal congenic strains. Neuropsychopharmacol. 2005;30:742–746. doi: 10.1038/sj.npp.1300592. [DOI] [PubMed] [Google Scholar]
- Fust G, Arason GJ, Kramer J, Szalai C, Duba J, Yang Y, et al. Genetic basis of tobacco smoking: strong association of a specific major histocompatibility complex haplotype on chromosome 6 with smoking behavior. Int Immunology. 2004;16:1507–1514. doi: 10.1093/intimm/dxh152. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Liu X, Hesselbrock V, Page GP, Goddard A, Zhang H. Results of a genomewide linkage scan: Support for chromosomes 9 and 11 loci increasing risk for cigarette smoking. Am J Med Genet B Neuropsychiatr Genet. 2004;129:94–101. doi: 10.1002/ajmg.b.30019. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Panhuysen C, Weiss R, Brady KT, Hesselbrock V, Rounsaville B, et al. Genomewide linkage scan for cocaine dependence and related traits: significant linkages for a cocaine-related trait and cocaine-induced paranoia. Am J Med Genet. 2005;136:45–52. doi: 10.1002/ajmg.b.30189. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Panhuysen C, Wilcox MM, Hesselbrock V, Rounsaville B, Poling J, et al. Genomewide linkage scan for opioid dependence and related traits. Am J Human Genet. 2006;78:759–769. doi: 10.1086/503631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green TA, Alibhai IN, Hommel JD, DiLeone RJ, Kumar A, Theobold DE, et al. Induction of inducible cAMP early repressor expression in nucleus accumbens by stress or amphetamine increases behavioral responses to emotional stimuli. J Neurosci. 2006;26:8235–8242. doi: 10.1523/JNEUROSCI.0880-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm JW, Lu L, Hayashi T, Hope BT, Su TP, Shaham Y. Time-dependent increases in brain-derived neurotrophic factor protein levels within the mesolimbic dopamine system after withdrawal from cocaine: implications for incubation of cocaine craving. J Neurosci. 2003;23:742–747. doi: 10.1523/JNEUROSCI.23-03-00742.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisel JE, Belknap JK, O'Toole LA, Helms ML, Wenger CD, Crabbe JC. Quantitative trait loci affecting methamphetamine responses in BXD recombinant inbred mouse strains. J Neurosci. 1997;17:745–754. doi: 10.1523/JNEUROSCI.17-02-00745.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn H, Nitzki F, Schorban T, Hemmerlein B, Threadgill D, Rosemann M. Genetic mapping of a Ptch1-associated rhabdomyosarcoma susceptibility locus on mouse chromosome 2. Genomics. 2004;84:853–858. doi: 10.1016/j.ygeno.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Hall FS, Drgonova J, Goeb M, Uhl GR. Reduced behavioral effects of cocaine in heterozygous brain-derived neurotrophic factor (BDNF) knockout mice. Neuropsychopharmacol. 2003;28:1485–1490. doi: 10.1038/sj.npp.1300192. [DOI] [PubMed] [Google Scholar]
- Hendrickson HP, Milesi-Halle A, Laurenzana EM, Owens SM. Development of a liquid chromatography–tandem mass spectrometric method for the determination of methamphetamine and amphetamine using small volumes of rat serum. J Chromatography B. 2004;806:81–87. doi: 10.1016/j.jchromb.2004.03.038. [DOI] [PubMed] [Google Scholar]
- Hiroi N, Agatsuma S. Genetic susceptibility to substance dependence. Mol Psychiatry. 2005;10:336–344. doi: 10.1038/sj.mp.4001622. [DOI] [PubMed] [Google Scholar]
- Horger BA, Iyasere CA, Berhow MT, Messer CJ, Nestler EJ, Taylor JR. Enhancement of locomotor activity and conditioned reward to cocaine by brain-derived neurotrophic factor. J Neurosci. 1999;19:4110–4122. doi: 10.1523/JNEUROSCI.19-10-04110.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu XT, Ford K, White F. Repeated cocaine administration decreases calcineurin (PP2B) but enhances DARPP-32 modulation of sodium currents in rat nucleus accumbens neurons. Neuropsychopharmacol. 2005;30:916–926. doi: 10.1038/sj.npp.1300654. [DOI] [PubMed] [Google Scholar]
- Hunter R, Jones D, Vicentic A, Hue G, Rye D, Kuhar MJ. Regulation of CART mRNA in the rat nucleus accumbens via D3 dopamine receptors. Neuropharmacol. 2006;50:858–864. doi: 10.1016/j.neuropharm.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Ikemoto S, Witkin BM. Locomotor inhibition induced by procaine injections into the nucleus accumbens core, but not the medial ventral striatum: implication for cocaine-induced locomotion. Synapse. 2003;47:117–122. doi: 10.1002/syn.10151. [DOI] [PubMed] [Google Scholar]
- Janowsky A, Mah C, Johnson RA, Cunningham CL, Phillips TJ, Crabbe JC, et al. Mapping genes that regulate density of dopamine transporters and correlated behaviors in recombinant inbred mice. J Pharmacol Expt Ther. 2001;298:634–643. [PubMed] [Google Scholar]
- Jaworski JN, Jones D. The role of CART in the reward/reinforcing properties of psychostimulants. Peptides. 2006;27:1993–2004. doi: 10.1016/j.peptides.2006.03.034. [DOI] [PubMed] [Google Scholar]
- Jaworski JN, Kozel MA, Philpot KB, Kuhar MJ. Intra-accumbal injection of CART (cocaine-amphetamine regulated transcript) peptide reduces cocaine-induced locomotor activity. J Pharmacol Expt Ther. 2003;307:1038–1044. doi: 10.1124/jpet.103.052332. [DOI] [PubMed] [Google Scholar]
- Jones BC, Tarantino LM, Rodriquez LA, Reed CL, McClearn GE, Plomin R, et al. Quantitative-trait loci analysis of cocaine-related behaviours and neurochemistry. Pharmacogenetics. 1999;9:607–617. [PubMed] [Google Scholar]
- Jones DC, Kuhar MJ. Cocaine-amphetamine-regulated transcript expression in the rat nucleus accumbens is regulated by adenylyl cyclase and the cyclic adenosine 5′-monophosphate/protein kinase a second messenger system. J Pharmacol Expt Ther. 2006;317:454–461. doi: 10.1124/jpet.105.096123. [DOI] [PubMed] [Google Scholar]
- Jones LC, McCarthy KA, Beard JL, Keen CL, Jones BC. Quantitative genetic analysis of brain copper and zinc in BXD recombinant inbred mice. Nutrition Neurosci. 2006;9:81–92. doi: 10.1080/00268970600691365. [DOI] [PubMed] [Google Scholar]
- Kansy JW, Daubner SC, Nishi A, Sotogaku N, Lloyd MD, Ngyuen C, et al. Identification of tyrosine hydroxylase as a physiological substrate for Cdk5. J Neurochem. 2004;91:374–384. doi: 10.1111/j.1471-4159.2004.02723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Vezina P. Metabotropic glutamate receptors in the rat nucleus accumbens contribute to amphetamine-induced locomotion. J Pharmacol Exper Ther. 1998;284:317–322. [PubMed] [Google Scholar]
- Kliethermes CL, Crabbe JC. Pharmacological and genetic influences on hole-board behaviors in mice. Pharmacol Biochem Behav. 2006;85:57–65. doi: 10.1016/j.pbb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Knyshevski I, Ricci LA, McCann TE, Melloni RH. Serotonin type-1A receptors modulate adolescent, cocaine-induced offensive aggression in hamsters. Physiol Behav. 2005;85:167–176. doi: 10.1016/j.physbeh.2005.03.020. [DOI] [PubMed] [Google Scholar]
- Kosten T, Ambrosio E. HPA axis function and drug addictive behaviors: Insights from studies with Lewis and Fischer 344 inbred rats. Psychoneuroendocrinology. 2002;27:35–69. doi: 10.1016/s0306-4530(01)00035-x. [DOI] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, Tsankova NM, Theobold DE, Truong HT, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Lahmame A, Grigoriadas DE, De Souza EB, Armario A. Brain corticotropin-releasing factor immunoreactivity and receptors in five inbred rat strains: Relationship to forced swimming behavior. Brain Res. 1997;750:285–292. doi: 10.1016/s0006-8993(96)01368-6. [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nature Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Leonard S, Breese C, Adams C, Benhammou K, Gault J, Stevens K, et al. Smoking and schizophrenia: abnormal nicotinic receptor expression. Eur J Pharmacol. 2000;393:237–242. doi: 10.1016/s0014-2999(00)00035-2. [DOI] [PubMed] [Google Scholar]
- Lipska B, Weinberger DR. Genetic variation in the vulnerability to the behavioral effects of neonatal hippocampal damage in rats. Proc Natl Acad Sci USA. 1996;92:8906–8910. doi: 10.1073/pnas.92.19.8906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E, et al. Evidence for genetic linkage to alcohol depencence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J Med Genet. 1998;81:216–221. doi: 10.1002/(sici)1096-8628(19980508)81:3<216::aid-ajmg2>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Luna-Munguia H, Manuel-Apolinar L, Rocha L, Meneses A. 5-HT1A receptor expression during memory formation. Psychopharmacol. 2005;181:309–318. doi: 10.1007/s00213-005-2240-4. [DOI] [PubMed] [Google Scholar]
- Manly K, Olson JM. Overview of QTL mapping software and introduction to Map Manager QT. Mammalian Genome. 1999;10:327–334. doi: 10.1007/s003359900997. [DOI] [PubMed] [Google Scholar]
- Marley RJ, Arros DM, Henricks KK, Marley ME, Miner LL. Sensitivity to cocaine and amphetamine among mice selectively bred for differential cocaine sensitivity. Psychopharmacol. 1998;140:42–51. doi: 10.1007/s002130050737. [DOI] [PubMed] [Google Scholar]
- McBrearty BA, Clark LD, Zhang X-M, Blankenhorn EP, Heber-Katz E. Genetic analysis of a mammalian wound-healing trait. Proc Natl Acad Sci USA. 1998;95:11792–11797. doi: 10.1073/pnas.95.20.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millan MJ, Brocco M, Gobert A, Joly F, Bervoets K, Rivet J, et al. Contrasting mechanisms of action and sensitivity to antipsychotics of phencyclidine versus amphetamine: importance of nucleus accumbens 5-HT2A sites for PCP-induced locomotion in the rat. Eur J Neurosci. 1999;11:4419–4432. doi: 10.1046/j.1460-9568.1999.00858.x. [DOI] [PubMed] [Google Scholar]
- Moisan MP, Courvoisier H, Bihoreau MT, Gauguier D, Hendley ED, Lathrop M, et al. A major quantitative trait locus influences hyperactivity in the WKHA rat. Nature Genet. 1996;14:471–473. doi: 10.1038/ng1296-471. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Berhow MT, Brodkin ES. Molecular mechanisms of drug addiction: Adaptations in signal transduction pathways. Mol Psychiatry. 1996;1:190–199. [PubMed] [Google Scholar]
- Nestler EJ, Barrot M, Self DW. Delta FosB: A sustained molecular switch for addiction. Proc Natl Acad Sci USA. 2001;98:11042–11046. doi: 10.1073/pnas.191352698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ, Carlezon WA., Jr The mesolimbic dopamine reward circuit in depression. Biol Psychiatry. 2006;59:1151–1159. doi: 10.1016/j.biopsych.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Nurnberger JI, Jr, Foroud T, Flury L, Su J, Meyer ET, Hu K, et al. Evidence for a locus on chromosome 1 that influences vulnerability to alcoholism and affective disorder. Am J Psychiatry. 2001;158:718–724. doi: 10.1176/appi.ajp.158.5.718. [DOI] [PubMed] [Google Scholar]
- Palmer AA, Verbitsky M, Suresh R, Kamens HM, Reed CL, Li N, et al. Gene expression differences in mice divergently selected for methamphetamine sensitivity. Mammal Genome. 2005;16:291–305. doi: 10.1007/s00335-004-2451-8. [DOI] [PubMed] [Google Scholar]
- Patkar AA, Mannelli P, Hill KP, Peindl K, Pae CU, Lee TH. Relationship of prolactin response to meta-chlorophenylpiperazine with severity of drug use in cocaine dependence. Human Psychopharmacol. 2006a;21:367–375. doi: 10.1002/hup.780. [DOI] [PubMed] [Google Scholar]
- Patkar AA, Mannelli P, Peindl K, Hill KP, Gopalakrishnan R, Beretini WH. Relationship of disinhibition and aggression to blunted prolactin response to meta-chlorophenylpiperazine in cocaine-dependent patients. Psychopharmacol. 2006b;185:123–132. doi: 10.1007/s00213-005-0261-7. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Huson MG, McKinnon CS. Localization of genes mediating acute and sensitized locomotor responses to cocaine in BXD/Ty recombinant inbred mice. J Neurosci. 1998;18:3023–3034. doi: 10.1523/JNEUROSCI.18-08-03023.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpot K, Smith Y. CART peptide and the mesolimbic dopamine system. Peptides. 2006;27:1987–1992. doi: 10.1016/j.peptides.2005.11.028. [DOI] [PubMed] [Google Scholar]
- Potenza MN, Brodkin ES, Joe B, Luo X, Remmers EF, Wilder RL, et al. Genomic regions controlling corticosterone levels in rats. Biol Psychiatry. 2004;55:634–641. doi: 10.1016/j.biopsych.2003.11.005. [DOI] [PubMed] [Google Scholar]
- Pu L, Liu QS, Poo MM. BDNF-dependent synaptic sensitization in midbrain dopamine neurons after cocaine withdrawal. Nature Neurosci. 2006;9:605–607. doi: 10.1038/nn1687. [DOI] [PubMed] [Google Scholar]
- Pucadyil TJ, Kalipatnapu S, Chattopadhyay A. The serotonin1A receptor: a representative member of the serotonin receptor family. Cell Mol Neurobio. 2005;25:1721–1753. doi: 10.1007/s10571-005-3969-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radel M, Vallejo RL, Iwata N, Aragon R, Long JC, Virkunen M, et al. Haplotype-based localization of an alcohol dependence gene to the 5q34 {gamma}-aminobutyric acid type A gene cluster. Arch Gen Psychiatry. 2005;62:47–55. doi: 10.1001/archpsyc.62.1.47. [DOI] [PubMed] [Google Scholar]
- Rajakumar N, Leung LS, Ma J, Rajakumar B, Rushlow W. Altered neurotrophin receptor function in the developing prefrontal cortex leads to adult-onset dopaminergic hyperresponsivity and impaired prepulse inhibition of acoustic startle. Biol Psychiatry. 2005;55:797–803. doi: 10.1016/j.biopsych.2003.12.015. [DOI] [PubMed] [Google Scholar]
- Ramos A, Moisan MP, Chaouloff F, Mormede C, Mormede P. Identification of female-specific QTLs affecting an emotionality-related behavior in rats. Mol Psychiatry. 1999;4:453–462. doi: 10.1038/sj.mp.4000546. [DOI] [PubMed] [Google Scholar]
- Remmers EF, Longman RE, Du Y, O'Hare A, Cannon GW, Griffiths MM, et al. A genome scan localizes five non-MHC loci controlling collagen-induced arthritis in rats. Nature Genetics. 1996;14:82–85. doi: 10.1038/ng0996-82. [DOI] [PubMed] [Google Scholar]
- Saccone NL, Goode EL, Bergen AW. Genetic analysis workshop 13: Summary of analyses of alcohol and cigarette use phenotypes in the Framingham Heart Study. Genet Epi. 2003;25:s90–s97. doi: 10.1002/gepi.10289. [DOI] [PubMed] [Google Scholar]
- Self DW, Choi KH, Simmons D, Walker JR, Smagula CS. Extinction training regulates neuroadaptive responses to withdrawal from chronic cocaine self-administration. Learning Memory. 2004;11:648–657. doi: 10.1101/lm.81404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofuoglu M, Kosten TR. Emerging pharmacological strategies in the fight against cocaine addiction. Expert Op Emerg Drugs. 2006;11:91–98. doi: 10.1517/14728214.11.1.91. [DOI] [PubMed] [Google Scholar]
- Song J, Koller DL, Foroud T, Carr K, Zhau J, Rice J, et al. Association of GABA(A) receptors and alcohol dependence and the effects of genetic imprinting. Am J Med Genet. 2003;117:39–45. doi: 10.1002/ajmg.b.10022. [DOI] [PubMed] [Google Scholar]
- Stohr T, Schulte Wermeling D, Weiner I, Feldon J. Rat strain differences in open-field behavior and the locomotor stimulating and rewarding effects of amphetamine. Pharmacology, Biochemistry & Behavior. 1998;59:813–818. doi: 10.1016/s0091-3057(97)00542-x. [DOI] [PubMed] [Google Scholar]
- Svenningsson P, Nairn AC, Greengard P. DARPP-32 mediates the actions of multiple drugs of abuse. AAPS J. 2005;7:E353–360. doi: 10.1208/aapsj070235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Ohshima T, Cho A, Sreenath T, Iadarola MJ, Pant HC, et al. Increased activity of cyclin-dependent kinase 5 leads to attenuation of cocaine-mediated dopamine signaling. Proc Nat Acad Sci USA. 2005;102:1737–1742. doi: 10.1073/pnas.0409456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuang M, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J, et al. Co-occurrence of abuse of different drugs in men. Arch Gen Psychiatry. 1998;55:967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- Wang H, Ng K, Hayes D, Gao X, Forster G, Blaha C, et al. Decreased amphetamine-induced locomotion and improved latent inhibition in mice mutant for the M5 muscarinic receptor gene found in the human 15q schizophrenia region. Neuropsychopharmacol. 2004;29:2126–2139. doi: 10.1038/sj.npp.1300502. [DOI] [PubMed] [Google Scholar]
- Zachariou V, Sgambato-Faure V, Sasaki T, Svenningsson P, Fienberg AA, Nairn AC, et al. Phosphorylation of DARPP-32 at threonine-34 is required for cocaine action. Neuropsychopharmacol. 2006;31:555–562. doi: 10.1038/sj.npp.1300832. [DOI] [PubMed] [Google Scholar]
- Zhang H, Ozbay F, Lappalainen J, Kranzler HR, van Dyck CH, Charney DS, et al. Brain Derived Neurotrophic Factor (BDNF) Genetic Variants and Alzheimer's Disease, Affective Disorders, Posttraumatic Stress Disorder, Schizophrenia and Substance Dependence. Am J Med Genet B. 2006;141B:387–393. doi: 10.1002/ajmg.b.30332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Lee MS, Agatsuma S, Hiroi N. Pleiotropic impact of constitutive fosB inactivation on nicotine-induced behavioral alterations and stress-related traits in mice. Hum Mol Genet. 2007;16:820–836. doi: 10.1093/hmg/ddm027. [DOI] [PubMed] [Google Scholar]