Abstract
The complexity of proteomic instrumentation for LC-MS/MS introduces many possible sources of variability. Data-dependent sampling of peptides constitutes a stochastic element at the heart of discovery proteomics. Although this variation impacts the identification of peptides, proteomic identifications are far from completely random. In this study, we analyzed interlaboratory data sets from the NCI Clinical Proteomic Technology Assessment for Cancer to examine repeatability and reproducibility in peptide and protein identifications. Included data spanned 144 LC-MS/MS experiments on four Thermo LTQ and four Orbitrap instruments. Samples included yeast lysate, the NCI-20 defined dynamic range protein mix, and the Sigma UPS 1 defined equimolar protein mix. Some of our findings reinforced conventional wisdom, such as repeatability and reproducibility being higher for proteins than for peptides. Most lessons from the data, however, were more subtle. Orbitraps proved capable of higher repeatability and reproducibility, but aberrant performance occasionally erased these gains. Even the simplest protein digestions yielded more peptide ions than LC-MS/MS could identify during a single experiment. We observed that peptide lists from pairs of technical replicates overlapped by 35–60%, giving a range for peptide-level repeatability in these experiments. Sample complexity did not appear to affect peptide identification repeatability, even as numbers of identified spectra changed by an order of magnitude. Statistical analysis of protein spectral counts revealed greater stability across technical replicates for Orbitraps, making them superior to LTQ instruments for biomarker candidate discovery. The most repeatable peptides were those corresponding to conventional tryptic cleavage sites, those that produced intense MS signals, and those that resulted from proteins generating many distinct peptides. Reproducibility among different instruments of the same type lagged behind repeatability of technical replicates on a single instrument by several percent. These findings reinforce the importance of evaluating repeatability as a fundamental characteristic of analytical technologies.
Introduction
Proteome analysis by liquid chromatography-tandem mass spectrometry (LC-MS/MS) begins with digestion of complex protein mixtures and separation of peptides by reverse phase liquid chromatography. Peptide MS/MS spectra are acquired under automated instrument control based on intensities of peptide ions. The spectra are matched to database sequences, and protein identifications are deduced from the list of identified peptides1. The complexity of these analyses leads to variation in the peptides and proteins identified. Minor differences in liquid chromatography, for example, may change the elution order of peptides2 or alter which peptides are selected for MS/MS fragmentation3. Small differences in fragmentation may cause some spectra to be misidentified by database search software4. A major source of variation is the sampling of complex mixtures for MS/MS fragmentation5.
Variation in inventories of identified peptides and proteins impacts the ability of these analyses to accurately represent biological states. If individual replicates generate different inventories, then how many replicates are needed to capture a reliable proteomic representation of the system? If inferences are to be drawn by comparing proteomic inventories, how does variation affect the ability of proteomic analyses to distinguish different biological states? This latter question is particularly relevant to the problem of biomarker discovery, in which comparative proteomic analyses of biospecimens generate biomarker candidates.
Proteomic technologies may be inherently less amenable to standardization than DNA microarrays6. Existing literature has emphasized variation in response to changes in analytical techniques. Fluctuations can occur in the autosampler, drawing peptides from sample vials7. The use of multidimensional separation can introduce more variation8, 9. Both instrument platforms10 and identification algorithms11, 12 produce variability. These contributions can be measured in a variety of ways. Several groups have evaluated the number of replicates necessary to observe a particular percentage of the proteins in a sample3, 9, 13. Others have examined how the numbers of spectra matched to proteins compared among analyses14, 15. The few comparisons across different laboratories for common samples16–18 have shown low reproducibility. On the other hand, the factors that contribute to variation in LC-MS/MS proteomics have never been systematically explored, nor have efforts been made to standardize analytical proteomics platforms. Standardization of procedures or configurations of system components would provide a means to evaluate the contributions of system variables to overall performance variation.
Analysis of variability in system performance entails two different measures that are often conflated. The first is repeatability, which represents variation in repeated measurements on the same sample and using the same system and operator19. When analyzing a particular sample the same way on the same instrumentation, the variation in results from run-to-run can be used to estimate the repeatability of the analytical technique. The second is reproducibility, which is the variation observed for an analytical technique when operator, instrumentation, time, or location is changed20. In proteomics, reproducibility could describe either the variation between two different instruments in the same laboratory or two instruments in completely different laboratories.
Here, we analyze the variation observed in a series of interlaboratory studies from the National Cancer Institute-supported Clinical Proteomic Technologies Assessment for Cancer (CPTAC) network. Overall, the six experiments for the Unbiased Discovery Working Group included seven Thermo LTQ linear ion traps (Thermo Fisher, San Jose, CA) and five Thermo LTQ-Orbitrap instruments at seven different institutions. Here we interrogate the data from the latter three of the six studies, analyzing three samples that differed in complexity and concentration. We have used these data to examine the repeatability and reproducibility for both peptide and protein levels and to examine the impact of multiple factors on these measures.
Materials and MethodsA
Study materials and design
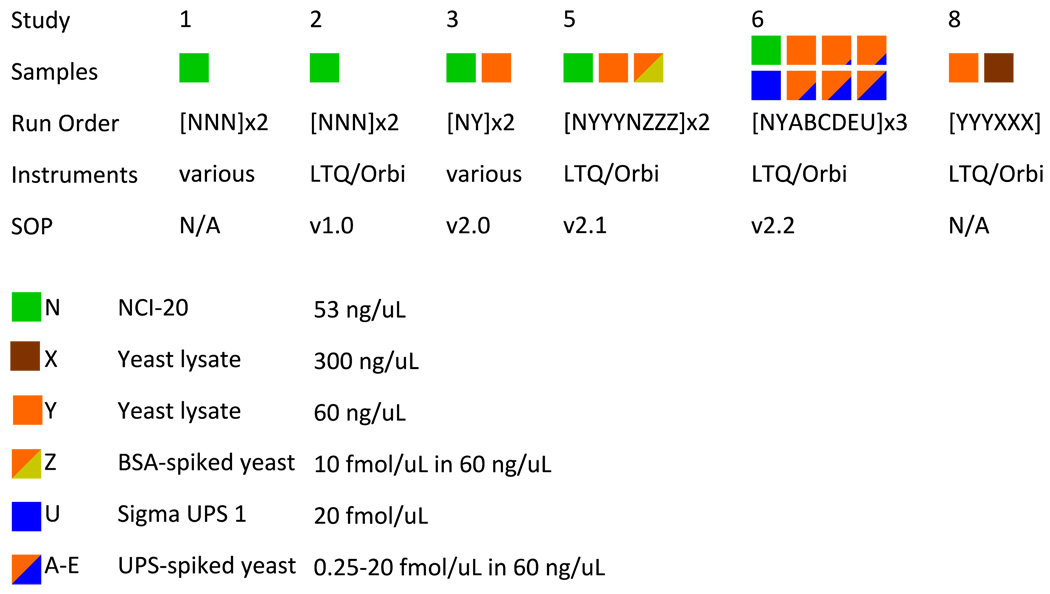
Three sample mixtures were used in these studies. The NCI-20 sample contained 20 human proteins mixed in various proportions spanning 5 g/L to 5 ng/L such that 5 proteins were routinely detected in these analyses. The Sigma UPS 1 mixture (Sigma-Aldrich, St. Louis, MO) contained 48 human proteins in equimolar concentrations. A protein extract of Saccharomyces cerevisiae was used to represent a highly complex biological proteome. In some cases, samples of the yeast extract were spiked either with bovine serum albumin (Sigma) or the Sigma UPS mixture as described below. For more detail on sample composition, see Supplementary Information. Figure 1 illustrates each of the CPTAC Studies. In Study 1, the coordinating laboratory at the National Institute of Standards and Technology (NIST) distributed three replicate vials of a tryptic digest of the NCI-20 mixture as well as undigested NCI-20 to each laboratory in two successive weeks; laboratories were to identify the contents of the vials by internal lab protocols. While Study 1 included a wide variety of instruments, all later studies focused on Thermo LTQ and LTQ-Orbitrap instruments. In Study 2, three vials of digested NCI-20 were provided to laboratories, which were instructed to use Standard Operating Procedure (SOP) version 1.0 (see Supplementary Information) to analyze the samples. Study 3 tested revision 2.0 of the SOP on this sample list: NbYb NbYb (where ‘N’ is digested NCI-20, ‘b’ is a water blank, and ‘Y’ is digested yeast reference). Study 5 extended upon Study 3 by revising the SOP to version 2.1 and adding a new sample: NbYbYbYb NbZbZbZb NbYbYbYb NbZbZbZb N, where ‘Z’ is yeast spiked with bovine serum albumin. Study 6 comprised three replicates of the following sample list: NbYbAbBbCbDbEbUbNb, where ‘A’ through ‘E’ were increasing concentrations of Sigma UPS 1 spiked into yeast and ‘U’ was the Sigma UPS 1 alone. Because some laboratories ran the sample lists back-to-back, the number of NCI-20 replicates varied from site to site. Study 8 sought to evaluate the yeast lysate at two concentrations, this time without an SOP governing RPLC or instrument configuration. All files resulting from these experiments are available at the following URL: http://cptac.tranche.proteomecommons.org/
Figure 1. Overview of CPTAC Unbiased Discovery Working Group interlaboratory studies.
All studies employed the NCI-20 reference mixture. Beginning with Study 3, a yeast lysate reference sample was introduced. Studies 2, 3, and 5 revealed ambiguities in the SOP that were corrected in subsequent versions. Studies 2–8 all prescribed blanks and washes between samples. Study 8 returned to lab-specific protocols in examining two different yeast concentrations. The full details of the SOP versions are available in Supplementary Information.
Database search pipeline
For all studies, MS/MS spectra were converted to mzML format by the ProteoWizard MSConvert tool21, set to centroid MS scans and to record monoisotopic m/z values for peptide precursor ions. Spectra from yeast proteins were identified against the Saccharomyces Genome Database orf_trans_all, downloaded March 23, 2009. These 6717 sequences were augmented by 73 contaminant protein sequences; the full database was then doubled in size by adding the reversed version of each sequence. Spectra from NCI20 and Sigma UPS 1 proteins were identified against the IPI Human database, version 3.56, with porcine trypsin added and each sequence included in both forward and reverse orientations (for a total of 153,080 protein sequences). The MyriMatch database search algorithm version 1.5.522 matched MS/MS spectra to peptide sequences. Semi-tryptic peptide candidates were included as possible matches. Potential modifications included oxidation of methionines, formation of N-terminal pyroglutamine, and carbamidomethylation of cysteines. For the LTQ, precursors were allowed to be up to 1.25 m/z from the average mass of the peptide. For the Orbitrap, precursors were required to be within 0.007 m/z of the peptide monoisotope, or of the monoisotope plus or minus one neutron. Fragment ions were uniformly required to fall within 0.5 m/z of the monoisotope. IDPicker23, 24 (April 20, 2009 build) filtered peptide matches to a 2% FDR and applied parsimony to the protein lists. Proteins were allowed to be identified by single peptides, though the number of distinct peptide sequences for each protein was recorded. Peptides were considered distinct if they differed in sequence or modifications but not if they differed only by precursor charge. IDPicker reports can be downloaded from Supplementary Information.
To measure the effect of the database search algorithm on identification, we applied X!Tandem25 Tornado (12/01/08 build) to Orbitrap yeast data from Study 6. The software was applied to data in mzXML format, produced with otherwise identical options as above in the ProteoWizard MSConvert tool. The same FASTA database was employed as for MyriMatch. X!Tandem refinement was disabled, and the software was set for semi-tryptic cleavage. Carbamidomethylation was selected for all cysteines, with oxidation possible on methionine residues. The software performed modification refinement only on N-termini, identifying modified cysteine (+40 Da), glutamic acid (−18 Da), and glutamine (−17 Da) residues at this position. The software applied a precursor threshold of 10 ppm and a fragment mass threshold of 0.5 m/z. Identifications were converted to pepXML in Tandem2XML (www.proteomecenter.org) and processed as above in IDPicker.
Statistical analysis
IDPicker produced five reports underlying this analysis: A) the NCI20 samples of studies 1, 2, 5, and 6; B) the Sigma UPS 1 in Study 6; C) the yeast in Study 5; D) the yeast in Study 6; and E) the yeast in Study 8. The IDPicker sequences-per-protein table was used for protein analysis, with each protein required to contribute two different peptide sequences in a given LC-MS/MS replicate to be counted as detected. For peptide analyses, the IDPicker spectra-per-peptide table reported whether or not spectra were matched to each peptide sequence across the replicates. Most box plots and bar graphs were produced in Microsoft Excel 2007.
Scripts for the R statistical environment26 produced the values underlying the pair-wise reproducibility graph. This script checked each peptide for joint presence in each possible pair of LC-MS/MS runs produced on different instruments, but within the same instrument class for a particular study. The stability of spectral counts across technical replicates was evaluated using a multivariate hypergeometric model. The model computes the probability to match a number of spectra from each technical replicate to a particular protein, based on the overall numbers of spectra identified in each replicate. For each protein, the script divides the log probability for an even distribution of spectral counts by the log probability for the observed spectral counts. A larger difference between these log probabilities implies greater instability in spectral counts for that protein. These R scripts are available in Supplementary Information.
Intensity analysis
Analysis of repeatability of precursor ion intensity was performed on 5 NCI20 replicates from each instrument in Study 5. Spectrum identifiers for peptide identifications were extracted from IDPicker identification assemblies in XML format. Peptides were filtered to include only those produced by NCI-20 protein sequences that matched trypsin specificity for both peptide termini. RAW data files from each replicate were converted to MGF using ReAdW4Mascot2.exe (v. 20080803a) (NIST). This program, derived from ReAdW.exe (Institute for Systems Biology, Seattle, WA), has been modified to record additional parameters including maximum precursor abundances in the TITLE elements of the files. Briefly, MS1 peak heights for a given precursor m/z are approximated from extracted ion chromatograms using linear interpolation27. These intensities were matched to peptide identifications, reporting the maximum intensity associated with each distinct peptide sequence. When a peptide was identified at multiple charge states or at multiple retention times, the largest value was selected.
Results and Discussion
Overview of CPTAC interlaboratory studies
The studies undertaken by the CPTAC network reflect the challenges of conducting interlaboratory investigations (see Figure 1). Researchers at NIST produced a reference mixture of 20 human proteins at varying concentrations (NCI-20). Study 1 saw the distribution of sample to each participating laboratory; the teams were asked to identify the proteins using their own protocols with any available instruments. Study 2 established SOP 1.0 for both LC and MS/MS configuration and employed only Thermo LTQ and Orbitrap instruments. Substantial changes took place between Study 2 and Study 5; a new Saccharomyces cerevisiae (yeast) reference proteome was introduced28, the SOP version 2.0 was developed, and a bioinformatic infrastructure was established to collect raw data files and to identify peptides and proteins. (Study 3 tested these tools in a small-scale methodology test, whereas Studies 4 and 7 were part of a parallel CPTAC network effort directed at liquid chromatography-multiple reaction monitoring-mass spectrometry for targeted peptide quantitation29; none of these studies are considered further in this work.) Study 5 analyzed the yeast reference proteome under the new SOP. Study 6 built upon Study 5 by including spikes of the Sigma UPS 1. The inclusion of NCI-20 in all of these studies enabled the measurement of variability for this sample in a variety of SOP versions and experimental designs. Study 8 employed no SOP; individual laboratories used their own protocols in analyzing two sample loads of the yeast lysate. The same quantity of yeast was analyzed in Studies 5, 6 and 8, with an additional 5x (high load) sample analyzed in Study 8. Study 5 produced six replicates of the yeast, whereas the other studies generated triplicates. The numbers of identified spectra were consistent for individual instruments, but more variable among multiple instruments. These yeast data sets, along with the quintuplicate NCI-20 analyses from Study 5 and the triplicate Sigma UPS 1 analyses from Study 6— a total of 144 LC-MS/MS analyses from four different LTQs and four different Orbitraps— comprise the essential corpus for our analysis of repeatability and reproducibility.
Bioinformatic variability in Orbitrap data handling
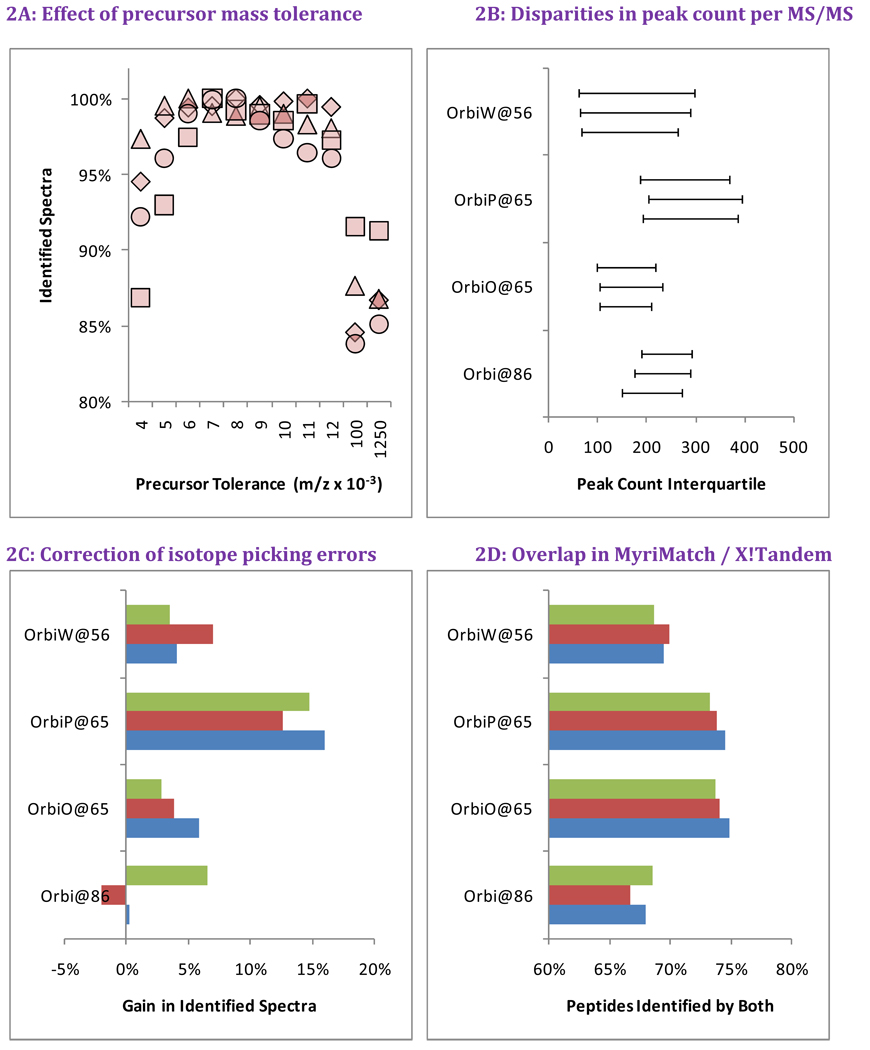
Initially, the MS/MS spectra produced in these studies were identified by database searches configured for low mass accuracy, because we expected significant differences in instrument tuning among the sites. Precursor ions were required to fall within 0.100 m/z of the values expected from peptide sequences (MyriMatch did not support configuring precursor tolerance in ppm units at the time of data analysis). However, we also recognized that higher mass accuracy of Orbitrap instruments could enable tighter mass tolerances for Orbitrap data sets. We tested MyriMatch over a range of precursor tolerances from ±0.004 to ±0.012 m/z (Figure 2A) in searches of the Orbitrap analyses of yeast from Study 6. We also included the original ±0.100 m/z setting as well as a tolerance of ±1.250 m/z, centered on the average peptide mass, which we used for searches of LTQ data. We observed an 8–16% increase in identification rate for the tight tolerances versus the original search settings and a 9–15% increase compared to the ±1.250 m/z tolerance used for the LTQ. The data indicate that the high mass accuracy of the Orbitrap produces a moderate increase in identifications, but only when precursor mass tolerance for identification is optimized. Based on these data, we used ±0.007 m/z as the precursor tolerance for all subsequent Orbitrap searches in this paper.
Figure 2. Bioinformatic variability in Orbitrap data for Study 6 Yeast.
We evaluated the impact of bioinformatic changes in evaluating Orbitrap spectra for yeast in Study 6. Panel A evaluates the best precursor tolerance for database search in m/z space. Each instrument is represented by a different shape (see legend on Figure 3), with the number of identifications normalized to the highest value produced for that instrument. Too low a tolerance prevents correct sequences from being compared to some spectra, while too high a tolerance results in excessive comparisons for each spectrum. Panel B reveals that the peaks counts from tandem mass spectra were repeatable for a given instrument but varied considerably among instruments. Panel C shows that database searches that allow for a one neutron variance from reported precursor m/z improved peptide identification in all but one of the twelve replicates. Panel D demonstrates that substituting a different search engine (X!Tandem, in this case) will dramatically change which sequences are identified, even if the total number of identified spectra is essentially the same.
Other aspects of these instruments can also impact the effectiveness of this peptide identification. We examined the number of peaks recorded per tandem mass spectrum from each of the four instruments as a measure of variability remaining after SOP refinement. Panel 2B reveals significant differences in peak count interquartile range among these instruments. These differences may reflect differences in electron multipliers, low levels of source or instrument contamination, or instrument idiosyncrasies. Many search engines make allowance for high resolution instruments mis-reporting the monoisotope for a precursor ion. We conducted the MyriMatch database search with and without the option to add or subtract one neutron from the computed precursor mass, and Panel 2C shows the extent to which this feature improved identification rates. OrbiP@65 benefited disproportionately from this feature. Of all twelve files, only one replicate from Orbi@86 failed to benefit from allowing the addition or subtraction of a neutron. Differences in peak density per spectrum and monoisotope selection can both influence peptide identification.
Many studies have shown that database search engines produce limited overlap in the peptides they identify, and the comparison shown in Panel 2D is no exception. We repeated the database search for these twelve files in X!Tandem as described in Materials and Methods. The number of identifications produced by X!Tandem typically fell within 3% of the number produced by MyriMatch, but Panel 2D shows an average overlap of only 71% between the peptide sequences from each identification algorithm for each LC-MS/MS experiment. Oddly, the degree of overlap appeared to be higher for the two Orbitraps at site 65 than for the others, though this would not appear to reflect tuning similarities, given Figure 2B. Though bioinformatics pipelines are perfectly repeatable for a given LC-MS/MS file, the choice of search engine and configuration clearly can have a tremendous impact on the identifications produced for a given data set.
Variability and repeatability in identified MS/MS spectra between instruments
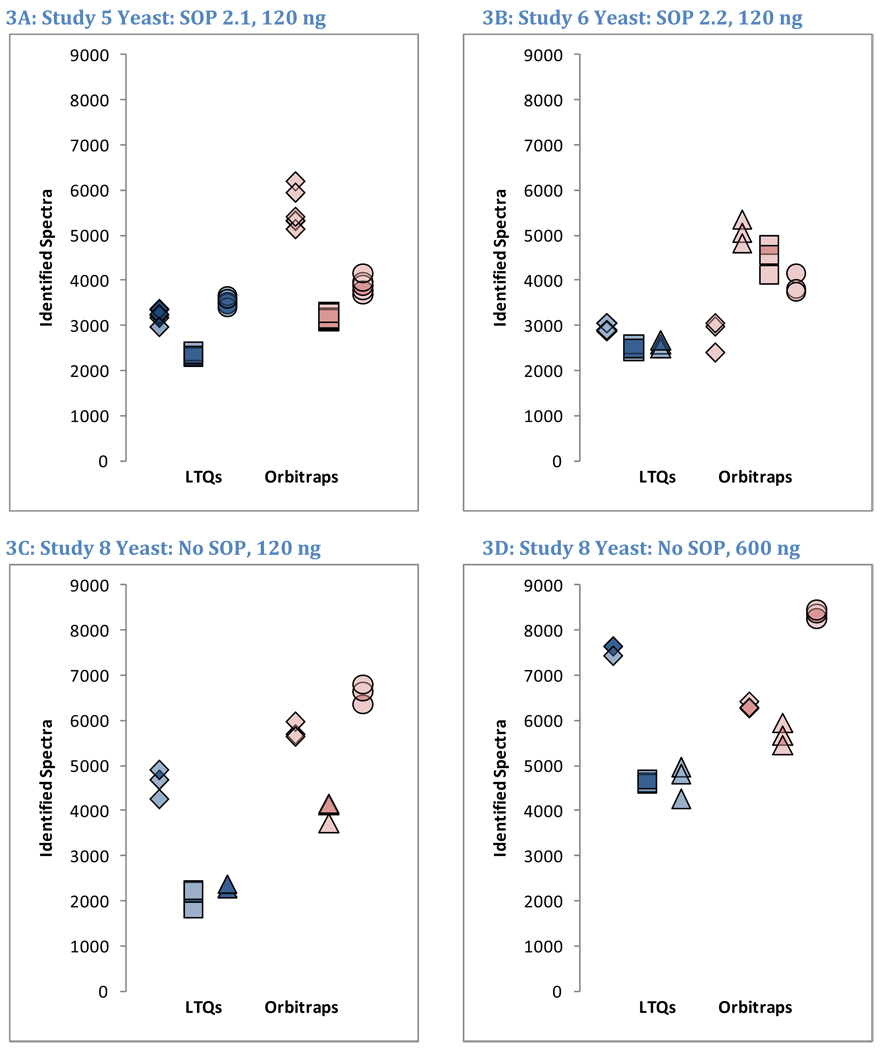
Figure 3 depicts the numbers of MS/MS spectra mapped to yeast sequences in Studies 5, 6, and 8. At a glance, Figure 3 suggests that strong instrument-dependent effects are observed in the identifications. The numbers of raw MS/MS spectra produced by LTQ instruments were approximately double the numbers of MS/MS spectra produced by Orbitraps due to the use of the charge state exclusion feature in the Orbitraps (data not shown). The numbers of MS/MS spectra that could be confidently matched to peptide sequences, however, were quite similar between instrument classes. Study 6 was most suggestive of differences in numbers of identifications between LTQs and Orbitraps (Figure 3B), and yet even for this case the p-value (0.058) was insignificant (Student’s t-test using unpaired samples, two-sided outcomes, and unequal variances). While the fraction of raw spectra that were identified successfully was much higher for Orbitraps than for LTQs, the overall numbers of identifications between instrument classes were comparable.
Figure 3. Overview of identified spectra in yeast replicates.
 LTQ@73
LTQ@73
 LTQx@65
LTQx@65
 LTQ2@95
LTQ2@95
 LTQc@65
LTQc@65
 Orbi@86
Orbi@86
 OrbiO@65
OrbiO@65
 OrbiP@65
OrbiP@65
 OrbiW@56
OrbiW@56
The number of spectra matched to peptide sequences varied considerably from instrument to instrument. These four graphs show the identification success for each yeast replicate in each instrument for Studies 5, 6, and 8 (two concentrations). LTQs are colored blue, and Orbitraps are shown in pink. A different shape represents each instrument appearing in the studies, as described in the legend. Each symbol reports identifications from an individual technical replicate. Despite SOP controls in Studies 5 and 6, instruments differed from each other by large margins. The Orbitrap at site 86 delivered the highest performance in Study 5, but performance decreased using the higher flow rate specified by the SOP in Study 6. Increasing the yeast concentration by five-fold in Study 8 increased the numbers of spectra identified.
The full set of samples extended beyond the yeast lysate. Table 1 shows the numbers of identified spectra, distinct peptides, and different proteins from each instrument from each of the studies. Spectra have been summed across all replicates for each instrument. Peptides were distinct if they differed in sequence or chemical modifications, but they were considered indistinct if they differed only by precursor charge. Proteins were only counted as identified if they matched multiple distinct peptides for a particular instrument. The data demonstrate that the NCI20 and Sigma UPS1 defined mixtures produced an order of magnitude fewer identified spectra and distinct peptides than did the more complex yeast sample. In both of these samples, contaminating proteins (e.g. keratins, trypsin) sometimes caused instruments to appear to have identified more proteins from these defined mixtures than the pure samples contained.
Table 1.
Identification counts for all included samples
| Sample and Replicates |
Instrument and Site |
Identified Spectra |
Distinct Peptides |
Multipeptide Proteins |
|---|---|---|---|---|
| Study5 NCI20 | LTQ@73 | 1085 | 358 | 8 |
| (5 replicates) | LTQ2@95 | 888 | 300 | 7 |
| LTQc@65 | 1446 | 455 | 8 | |
| Orbi@86 | 968 | 374 | 11 | |
| OrbiP@65 | 1660 | 424 | 10 | |
| OrbiW@56 | 1708 | 444 | 11 | |
| Study5 Yeast | LTQ@73 | 19341 | 6261 | 880 |
| (6 replicates) | LTQ2@95 | 14054 | 4574 | 730 |
| LTQc@65 | 21209 | 7118 | 993 | |
| Orbi@86 | 33301 | 8977 | 1280 | |
| OrbiP@65 | 19118 | 4652 | 627 | |
| OrbiW@56 | 23245 | 5708 | 813 | |
| Study6 Yeast | LTQ@73 | 8832 | 4263 | 681 |
| (3 replicates) | LTQ2@95 | 7471 | 3537 | 568 |
| LTQx@65 | 7763 | 3859 | 610 | |
| Orbi@86 | 8448 | 4354 | 795 | |
| OrbiO@65 | 15177 | 7110 | 1024 | |
| OrbiP@65 | 13465 | 6111 | 838 | |
| OrbiW@56 | 11675 | 5727 | 823 | |
| Study6 UPS 1 | LTQ@73 | 991 | 378 | 51 |
| (3 replicates) | LTQ2@95 | 1257 | 494 | 50 |
| LTQx@65 | 1367 | 562 | 56 | |
| Orbi@86 | 877 | 366 | 51 | |
| OrbiO@65 | 1619 | 692 | 58 | |
| OrbiP@65 | 2143 | 798 | 59 | |
| OrbiW@56 | 1296 | 617 | 59 | |
| Study8 Yeast | LTQ@73 | 22740 | 8322 | 1039 |
| High concentration | LTQ2@95 | 13909 | 5470 | 819 |
| (3 replicates) | LTQx@65 | 14010 | 6642 | 951 |
| Orbi@86 | 18943 | 7362 | 939 | |
| OrbiO@65 | 17037 | 6891 | 918 | |
| OrbiW@56 | 25082 | 9816 | 1111 | |
| Study8 Yeast | LTQ@73 | 13801 | 5586 | 751 |
| Low concentration | LTQ2@95 | 6274 | 2859 | 478 |
| (3 replicates) | LTQx@65 | 6899 | 3467 | 583 |
| Orbi@86 | 17304 | 7407 | 957 | |
| OrbiO@65 | 12018 | 5133 | 789 | |
| OrbiW@56 | 19782 | 8132 | 1048 |
Laboratories employing LC-MS/MS with data-dependent acquisition of MS/MS spectra expect identification variability. If peptides from a single digestion are separated on the same HPLC column twice, variations in retention times for peptides will alter the particular mix of peptides eluting from that column at a given time. These differences, in turn, impact the observed intensities for peptide ions in MS scans and influence which peptide ions will be selected for fragmentation and tandem mass spectrum collection. Spectra for a particular peptide, in turn, may differ significantly in signal-to-noise, causing some to be misidentified or to be scored so poorly as to be eliminated during protein assembly. All of these factors diminish the expected overlap in peptide identifications among replicate LC-MS/MS analyses.
Because Orbitraps targeted peptides for fragmentation on the basis of higher resolution MS scans than did the LTQs, we asked whether Orbitrap peptide identifications were more repeatable. Repeatability for each instrument was measured by determining the percentage of peptide overlap between each possible pair of technical replicates. The six replicates of yeast in Study 5 enabled fifteen such comparisons for each instrument, while the five replicates of NCI-20 yielded ten comparisons. In Studies 6 and 8, triplicates enabled only three comparisons per instrument (A vs. B, A vs. C, and B vs. C).
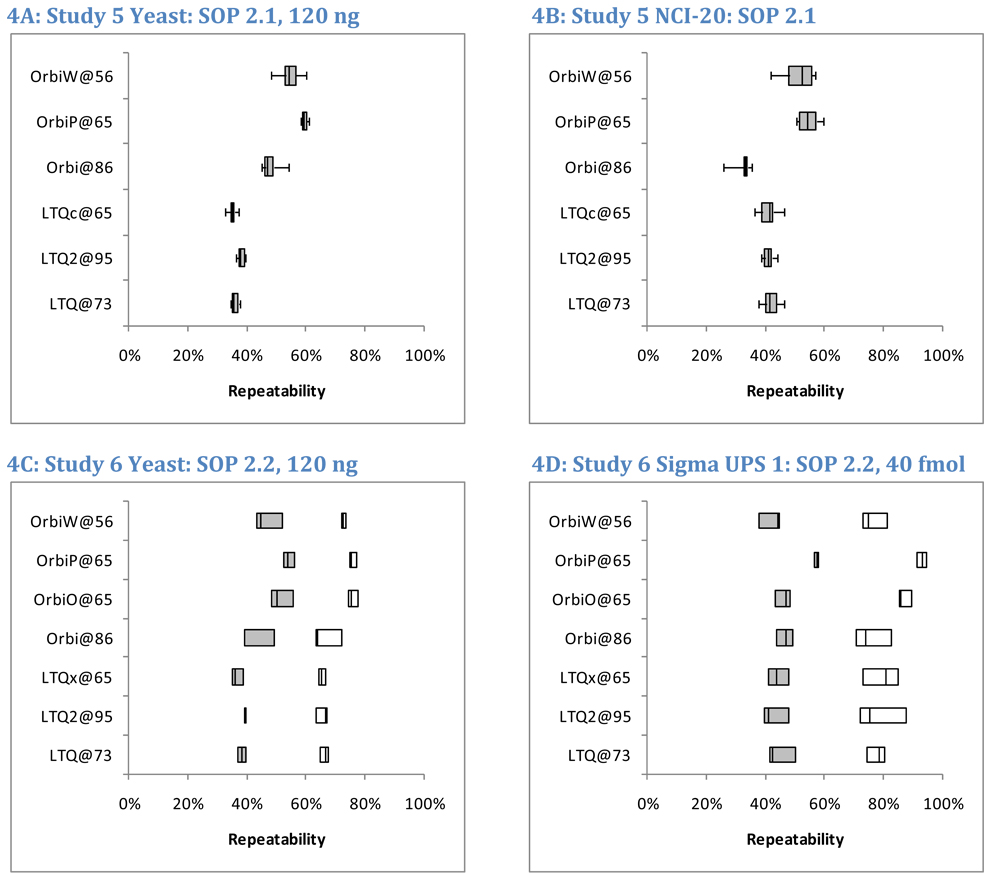
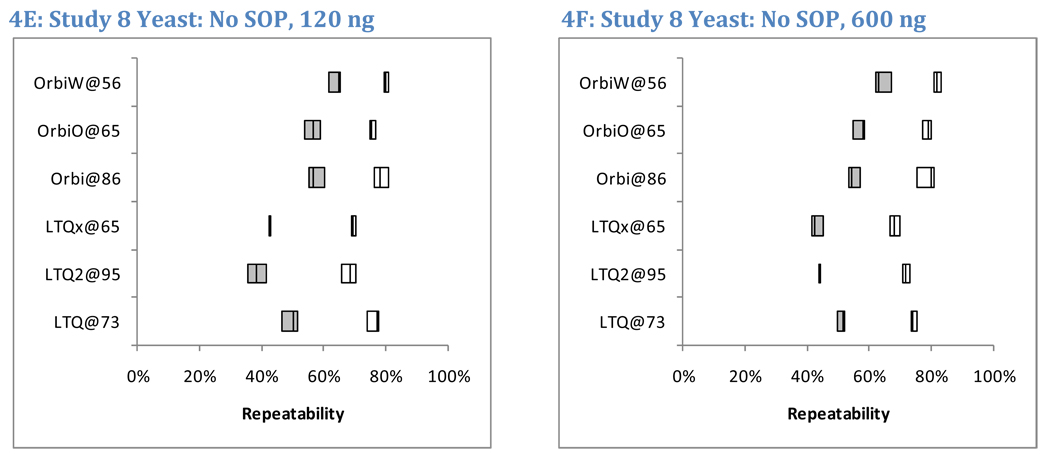
Figure 4 reports the pair-wise repeatability for peptide identifications in Studies 5, 6, and 8. By including six replicates of yeast and five replicates of NCI-20, Study 5 yielded the most information for comparison among technical replicates (panels 4A and 4B); Student’s t-test produced a p-value of 0.035 in comparing LTQ to Orbitraps in Study 5 yeast peptide repeatability (using unpaired samples, two-sided outcomes, and unequal variances). Significant differences were not observed in Study 6 analyses of yeast peptides (p = 0.057, panel 4C); this anomaly is traceable to the low repeatability (39%) observed for the Orbitrap at site 86, a set of runs that also suffered from low sensitivity (Figure 3B). Yeast data from Study 8 (panels 4E and 4F) differentiated instrument classes with a p-value of 0.027. The averages of medians for LTQ yeast peptide repeatability were 36%, 38%, and 44% in Studies 5, 6, and 8, respectively. The corresponding values for Orbitraps were 54%, 47%, and 59%. Orbitrap instruments achieved 9–18% greater peptide repeatability than LTQs for the yeast samples.
Figure 4. Peptide and protein repeatability.
To assess repeatability, we evaluated the overlapping fraction of identified peptides in pairs of technical replicates. For example, if 2523 and 2663 peptides were identified in two different replicates and 1362 of those sequences were common to both lists, the overlap between these replicates was 35.6%. Shaded boxes represent peptide repeatability, and open boxes represent protein repeatability (where two distinct peptide sequences were required for protein to be detected). For Study 5, only peptide repeatability was characterized; the boxes represent the inter-quartile range, while the whiskers represent the full range of observed values (Panels A and B). The mid-line in each box is the median. The six replicates of yeast in Study 5 enabled fifteen pair-wise comparisons per instrument, while the five replicates of NCI-20 enabled ten comparisons for that sample. Studies 6 (Panels C and D) and 8 (Panels E and F) produced triplicates, enabling only three pair-wise comparisons for repeatability. These images show all three values.
Identification repeatability and sample complexity
We hypothesized that increased sample complexity would decrease peptide repeatability. A complex mixture such as yeast should yield a more diverse peptide mixture than the 48 proteins of Sigma UPS 1, which in turn should yield a more diverse peptide mixture than the simple NCI-20 sample.
This hypothesis can be tested by returning to Figure 4. On average, the simple NCI-20 yielded a median 44% overlap in the peptides identified between pairs of replicates (Figure 4B). The Sigma UPS 1 produced almost the same average— 46% overlap for peptides (Figure 4D). Yeast repeatability fell between the two, giving a 45% median overlap (Figure 4A). The interquartile ranges across all instruments for these three mixtures were 39–50% for NCI-20, 42–48% for Sigma UPS 1, and 36–54% for yeast. These similar overlaps are all the more striking when one considers the numbers of peptides observed for each sample. In total, 977 different peptide sequences (in 7755 spectra) were identified from the NCI-20 (this includes a number from contaminant proteins), and the Sigma UPS 1 generated 1292 peptides from 9550 identified spectra. The yeast, on the other hand, produced 14,969 peptides from 130,268 identified spectra. These data demonstrate that repeatability in peptide identification is independent of the complexity of the sample mixture and is robust against significant changes in the numbers of identifications.
Observing these consistent but low repeatability values conveys a key message: all digestions of protein mixtures are complex at the peptide level. Clearly NCI-20 and yeast are widely separated on a scale of protein complexity. The peptides resulting from both digestions, however, are still diverse enough to preclude comprehensive identification in a single LC-MS/MS. The peptides identified from NCI-20 included numerous semi-tryptic peptides in addition to the canonical tryptic peptides. Likewise, the concentration of a sample may change the number of identified spectra dramatically without changing the fractional overlap between successive replicates. Peptide identification repeatability may be a better metric for judging the particular instruments or analytical separations than for particular samples.
While peptide repeatability was essentially unchanged in response to sample complexity, protein repeatability appeared different for the equimolar Sigma UPS 1 and complex yeast samples (open boxes, Figures 4C and 4D). Student’s t-test, comparing the protein repeatabilities for yeast and Sigma UPS 1 in each instrument, produced p-values below 0.05 for the LTQ@73, LTQx@65, OrbiO@65, and OrbiP@65, but the p-values for LTQ2@95, Orbi@86, and OrbiW@56 were all in excess of 0.10 (unpaired samples, two-sided outcomes, and unequal variances). The wide spread of protein repeatability observed for Sigma UPS 1 in Figure 4D prevented a strong claim that protein repeatability differed between these two samples.
Identification repeatability and sample load
A similar result appears when high concentrations of yeast are compared to low concentrations. Study 8 differs from the others in that each laboratory employed lab-specific protocols rather than a common SOP. Figure 4, panels E–F display the peptide and protein repeatability for both high and low sample loads of yeast. A five-fold increase in sample on-column increased identified peptide counts an average of 48% per instrument, whereas protein counts increased by an average of 35%. The median peptide repeatability, however, was essentially identical between the low and high loads, both with median values of 53% (with an interquartile range of 43–58% for low load and 44–58% for high load). The repeatability for proteins was always higher than for peptides in Studies 6 and 8, but it, too, was unaffected by protein concentration. The median value for protein repeatability at the low load was 76%, while the median for the high load was 75%. The stability of repeatability between low and high concentrations helps reinforce the findings for Studies 5 and 6, which used the same sample load as the low concentration in Study 8. The 120 ng load was intended to be high enough for good identification rates but low enough to forestall significant carryover between replicates. Though larger numbers of identifications resulted from a higher sample load, repeatability for peptides and proteins was unchanged by the difference in concentration.
Study 8 also provided an opportunity to examine the problem of peptide oversampling in response to sample load. All instruments in this study were configured to employ the “Dynamic Exclusion” feature to reduce the extent to which multiple tandem mass spectra were collected from each ion, though the specific configurations differed. For each replicate in each instrument, we computed the fraction of peptides for which multiple spectra were collected. Several possible reasons can account for the collection of multiple spectra: each precursor ion may appear at multiple charge states, dominant peptides may produce wide chromatographic peaks, or a different isotope of a peptide may be selected for fragmentation. In LTQs, an average of 15.3% of peptides matched to multiple scans for each replicate of the low concentration samples, while this statistic was 18.5% for the high concentration samples. Orbitraps were more robust against collecting redundant identifications, with 8.8% of peptides matching to multiple spectra in the low concentration samples and 11.5% matching to multiples in the high concentration samples. Across all instruments, the five-fold increase in sample concentration corresponded to 2.9% more peptides matching to multiple spectra. This difference produced significant p-values (below 0.05) for all but one of the instruments by t-test (using unpaired samples, allowing for two sided outcomes, and assuming unequal variances). Although more spectra were identifiable when sample concentration increased, the redundancy of these identifications was also higher.
Properties that influence repeatability in peptide identification
Given that peptide-level repeatability rarely approached 60%, it may seem that the appearance of peptides across replicates is largely hit-or-miss. In fact, some peptides are far more repeatable than others. We examined three factors for their correlation with peptide repeatability: trypsin specificity, peptide ion intensity, and protein of origin.
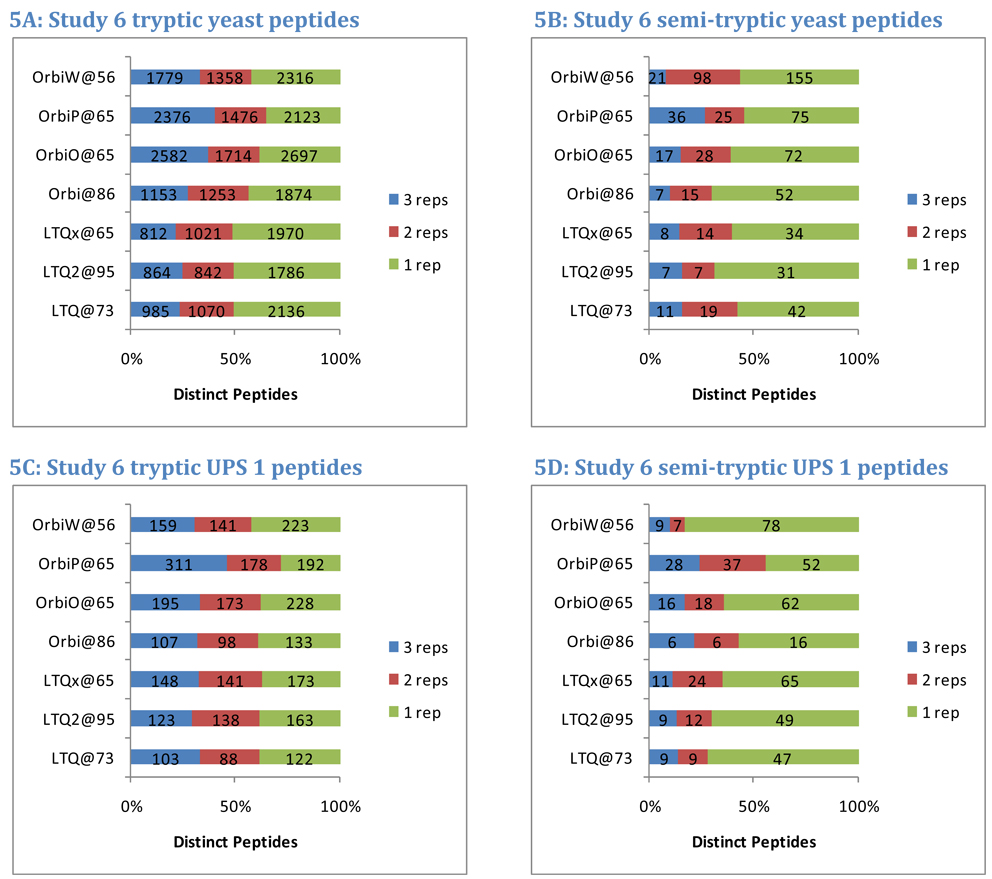
Many laboratories routinely consider only fully-tryptic peptide identifications in database searches, expecting only a minimal amount of nonspecific enzymatic cleavage and in-source fragmentation in their samples. Allowance for semi-tryptic matches (peptides that match trypsin specificity only at one terminus) has been shown to improve identification yield23, 30. In Study 6, semi-tryptic peptides constituted 2.2% of the identifications from yeast per instrument and 16.9% of the peptides identified from the Sigma UPS 1 sample. In both samples, semi-tryptic peptides were less likely to appear in multiple replicates than fully tryptic peptides (Figure 5). In yeast, an average of 45% of the fully tryptic peptides appeared in only one replicate, but 62% of the semi-tryptic peptides appeared in only one replicate. Comparing percentages by instrument produced a p-value of 0.000763 by paired, two-sided t-test. A similar trend appeared in Sigma UPS 1, with 38% of fully tryptic peptides appearing in only one replicate and 65% of semi-tryptic peptides appearing in only one replicate (p=0.00014). Although these two samples produced different percentages of semi-tryptic peptide identifications, both results are evidence that semi-tryptics are less repeatable in discovery experiments.
Figure 5. Peptide tryptic specificity impacts repeatability.
Enzymatic digestion by trypsin favors the production of peptides that conform to standard cleavages after Lys or Arg on both termini. Fully tryptic peptides feature such cleavages on both termini, while semi-tryptic peptides match this cleavage motif on only one terminus. As shown in panel A, an average of 29% of fully tryptic yeast peptides appeared in all three replicates from Study 6. Semi-tryptic peptides were detected with lower probability. On average, only 15% of these peptides appeared in all three replicates. Though a higher percentage of semi-tryptic peptides were observed in Sigma UPS 1 (panels C and D), the repeatability for semi-tryptic peptides was lower than for tryptic sequences.
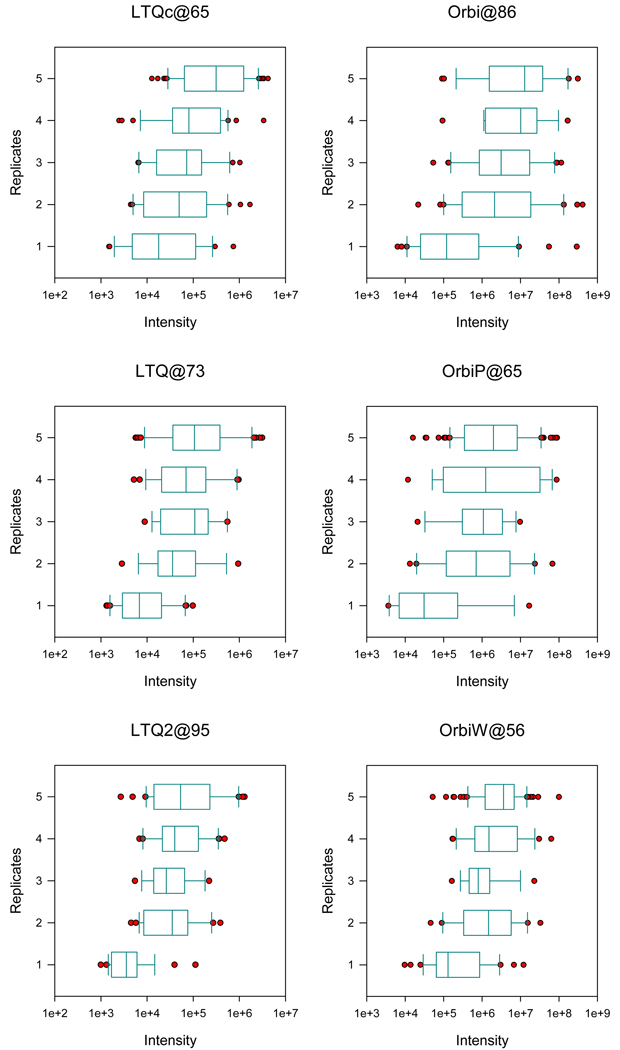
Precursor ion intensity drives selection for MS/MS and would be expected to correlate with repeatability of peptide identifications. We analyzed the five replicates of NCI-20 from Study 5 to measure this relationship. Peptides from each instrument were separated into classes by the number of replicates in which they were identified, and the maximum intensity of each tryptic precursor ion was computed from the MS scans (see Methods). Box plots were used to summarize the results (see Figure 6). For both the LTQs and Orbitraps, repeatability positively correlated with precursor intensity, with peptides identified in only one replicate yielding much lower intensities. We also observed that low intensity peptides were less reproducible across instruments (data not shown). As expected, intense ions are more consistently identified from MS/MS scans.
Figure 6. Precursor ion intensity affects peptide repeatability.
We examined the MS ion intensity of peptides from the Study 5 NCI-20 quintuplicates in three LTQ and three Orbitrap instruments. When a peptide was observed in multiple replicates, we recorded the median intensity observed. These graphs depict the distribution of intensities for peptides by the number of replicates in which they were identified. Peptides that were observed in only one replicate were considerably less intense than those appearing in multiple replicates. Orbitrap and LTQ instruments report intensities on different scales as reflected by the x-axes of the graphs.
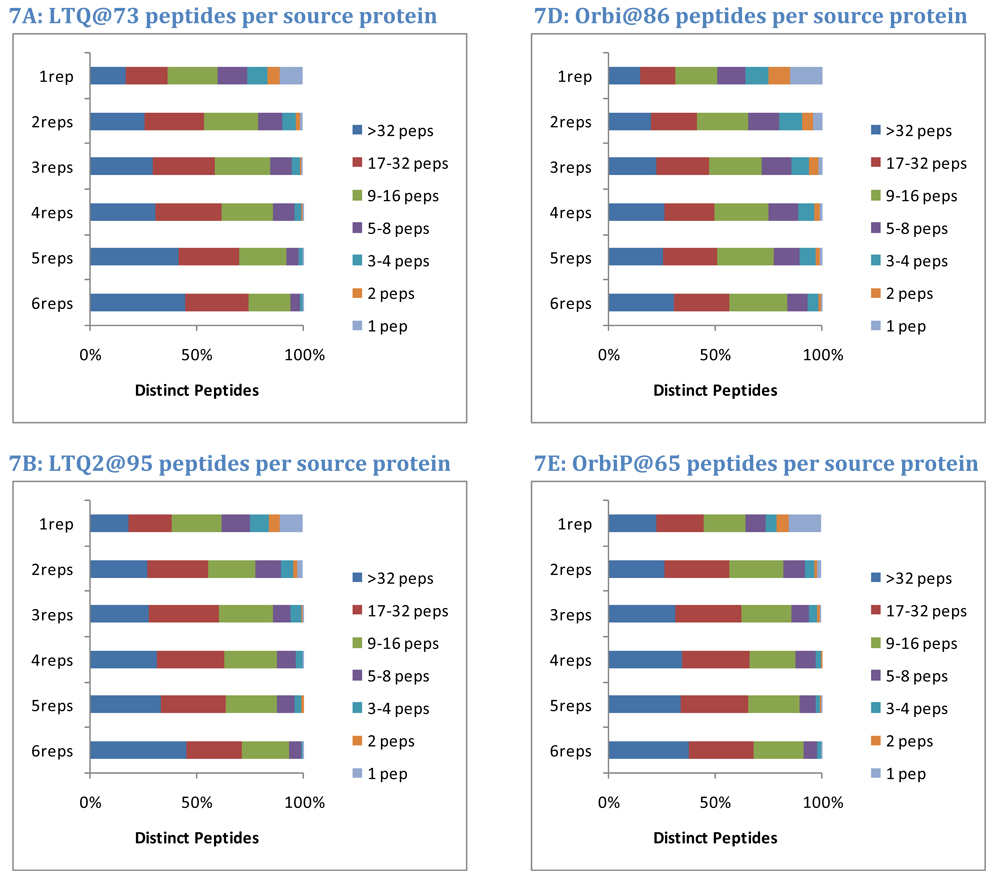
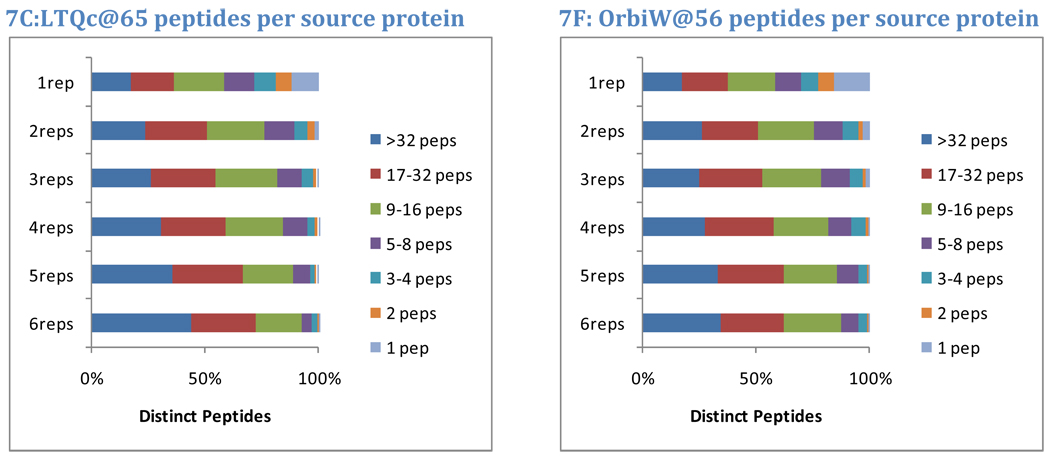
The protein of origin may also impact the repeatability of a peptide. Intuitively, a peptide that results from the digestion of a major protein is more likely to repeat across replicates than a peptide from a minor protein. In this analysis, the overall number of distinct peptide sequences observed for each protein was used to segregate them into seven classes. Proteins with only one peptide observed comprised the most minor class. The number of peptides required for each successive higher class was doubled, yielding classes of proteins identified by 2 peptides, 3–4 peptides, 5–8 peptides, 9–16 peptides, and 17–32 peptides. Proteins with more than 32 peptides were classed as major proteins. For each instrument in Study 5, observed yeast peptides were split into classes by the number of replicates (out of six) in which they appeared. The graphs in Figure 7 show how protein class corresponds to peptide repeatability.
Figure 7. Protein of origin impacts repeatability.
Peptide identifications from major proteins are more repeatable than those from minor proteins. Yeast data from Study 5, however, reveal that peptides from major proteins (here defined as those producing more than 32 peptides in the data accumulated for all instruments) constitute 40% of the peptides observed in all six replicates and 18% of the peptides observed in only one replicate. Peptides that are the sole evidence for a protein constitute 0% of the peptides observed in all six replicates but 13% of the peptides observed only once. These trends illustrate that major proteins contribute peptides across the entire range of repeatability. Achieving optimal sensitivity requires the acceptance of less-repeated peptides; in this data set, single-observation peptides were more than twice as numerous as any other set.
On average, 40% of peptides appearing in all six replicates matched to the major proteins (more than 32 distinct peptide sequences). At the other extreme, peptides appearing in only one replicate matched to major proteins only 18% of the time. Almost none of the peptides that were sole evidence for a protein were observed in all six replicates, but 13% of the peptides observed in only one replicate were of this class, and only 3% of the peptides observed in two of the six replicates were sole evidence for a protein. These data are consistent with a model in which digestion of any protein produces peptides with both high and low probabilities of detection; peptides for a major protein may be observed in the first analysis of the sample, but additional peptides from the same protein will be identified in subsequent analyses. The highest probability peptides from minor proteins must compete for detection with less detectable peptides from major proteins.
Factors governing peptide and protein identification reproducibility
The analyses of repeatability described above establish the level of variation among technical replicates, but they do not address the reproducibility of shotgun proteomics across multiple laboratories and instruments. What degree of variability should be expected for analyses of the same samples across multiple instruments? Studies 6 and 8 provide data to address this question. Study 6 was conducted under SOP version 2.2, with a comprehensive set of specified components, operating procedures, and instrument settings shared across laboratories and instruments. Study 8, on the other hand, was performed with substantial variations in chromatography (e.g. different column diameters, self-packed or monolithic columns and various gradients) and instrument configuration (e.g. varied dynamic exclusion, injection times, and MS/MS acquisitions). See Supplementary Information for additional detail.
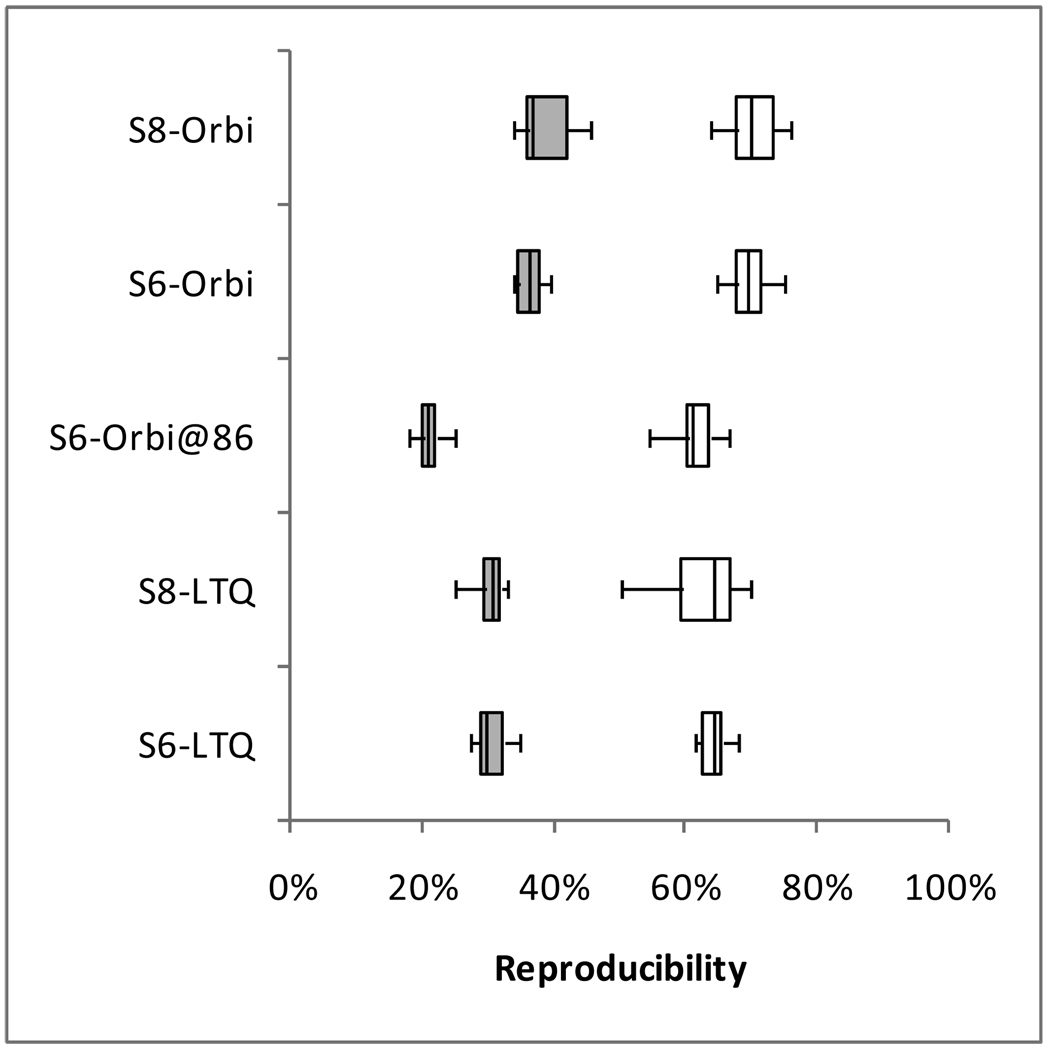
Figure 8 compares the yeast peptides and proteins identified from each replicate LC-MS/MS analysis on one instrument to all other analyses on other instruments of the same type in the same study. The peptide median of 30% for Study 6 LTQs, for example, indicates that typically 30% of the peptides identified from a single analysis on one LTQ were also identified in individual analyses on the other LTQs. As observed previously in repeatability, protein reproducibility was always higher than the corresponding peptide reproducibility. Figure 8 separates the Orbitrap at site 86 from the others in Study 6 because it was a clear outlier; all comparisons including this instrument yielded lower reproducibility than comparisons that excluded it. The remaining three Orbitraps in Study 6 were cross-compared to produce the Study 6 Orbitrap evaluation.
Figure 8. Reproducibility of yeast identifications among instruments.
In this analysis, the identifications from each replicate are compared to the identifications of replicates from other instruments of the same type in the same study to determine the overlap in identifications. For example “S6-LTQ” shows the overlaps between pairs of RAW files from LTQs in Study 6, where each pair was required to come from two different LTQs. Shaded boxes represent peptides, while white boxes represent proteins. Because the Orbitrap at site 86 yielded abnormally low reproducibility in Study 6, the comparisons including this instrument were separated from the other three Orbitraps in this Study.
Surprisingly, the reproducibility observed with and without an SOP was unchanged. The median reproducibility insignificantly increased by 0.8% for LTQs from Study 6 to Study 8 and by 0.4% for Orbitraps. The inter-quartile range of reproducibility increased slightly for proteins (both LTQ and Orbitrap) and for peptides observed by Orbitraps. While the median reproducibility was unaffected by the SOP, the range of reproducibility broadened when no SOP was employed. This test of reproducibility is limited in scope to include only the identifications resulting from these data; an examination of retention time reproducibility or elution order might reveal considerably more detail about the reproducibility gains achieved through this SOP. Because Study 8 followed the experiments incorporating the SOP, lab-specific protocols may have been altered to incorporate some elements of the SOP, thus diminishing any observable effect.
The comparison between LTQ and Orbitrap platforms shows two contrary phenomena. First, the Orbitrap at site 86 shows a potential disadvantage of these instruments; an Orbitrap that is not in top form can produce peptide identifications that do not reproduce well on other instruments. When this instrument is excluded, however, the remaining Orbitrap analyses were more reproducible at both the peptide and protein levels than were analyses on LTQ instruments. The difference in median reproducibility by instrument class ranged between 5.2% and 6.6%, depending on which study was analyzed and whether peptides or proteins were examined. Although Orbitraps can produce more reproducible identifications than LTQs, the difference is not large, and Orbitraps that are not operating in peak form lack this advantage.
Impact of repeatability on discrimination of proteomic differences
One of the most important applications of LC-MS/MS-based shotgun proteomics is to identify protein differences between different biological states. This approach is exemplified by the analysis of proteomes from tissue or biofluid samples that represent disease states, such as tumor tissues and normal controls. These studies typically compare multiple technical replicates for one sample type to technical replicates for a second sample type. Identified proteins that differ in spectral counts or intensities between samples may be considered biomarker candidates, which may be further evaluated in a biomarker development pipeline31. Repeatability of the analyses is a critical determinant of the ability to distinguish true differences between sample types.
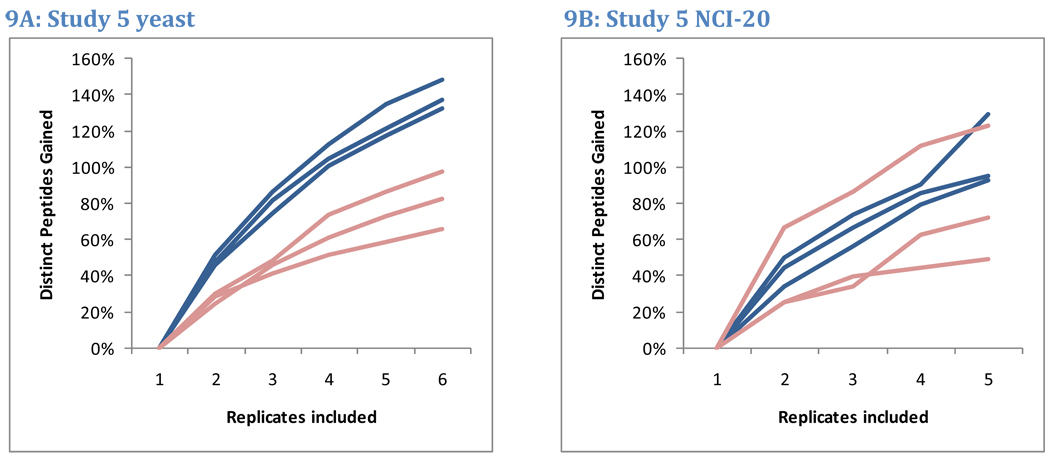
The repeatability analyses described above revealed that approximately half the peptides observed in LC-MS/MS analysis will be identified in a second replicate. This low degree of overlap implies that the total number of peptides identified grows rapidly for the first few replicates and then slows. A plot of this growth for the six yeast replicates and five NCI-20 replicates of Study 5 is presented in Figure 9. The rates at which newly-identified peptides accumulated for NCI-20 and yeast replicates were indistinguishable. The first two replicates for NCI-20 identified an average of 41% more peptides per instrument than did the first replicate alone. Similarly, two replicates for yeast contributed an average of 38% more peptides than a single analysis. Three replicates identified 59% more peptides for NCI-20 than did the first replicate alone, whereas three replicates for yeast identified 63% more peptides than one replicate. The similarity of these trends implies that one cannot choose an appropriate number of replicates based on sample protein complexity alone. By collecting triplicates, researchers will, at a minimum, be able to estimate the extent to which new replicates improve the sampling of these complex mixtures.
Figure 9. Peptide accumulation with additional replicates.
Because peptides are partially sampled in each LC-MS/MS analysis, repeated replicates can build peptide inventories. Data from Study 5 reveal that this growth is not limited to the complex yeast samples but is also observed in the simple NCI-20 mixture. Blue lines represent growth in LTQ peptide lists, while pink lines represent Orbitrap peptide lists. The second NCI-20 replicate for the Orbitrap at site 86 identified more peptides than any other, producing a substantial increase in peptides from the first to the second replicate.
In this analysis, independent technical replicates were evaluated for cumulative identifications. If maximizing distinct peptide identifications were the priority, however, one might instead minimize repeated identifications among replicates to as great an extent as possible. Some instrument vendors have implemented features to force subsequent replicates to avoid the ions sampled for tandem mass spectrometry in an initial run (such as RepeatIDA from Applied Biosystems). Researchers have also used accurate mass exclusion lists32 or customized instrument method generation33 to reduce the re-sampling of peptides in repeated LC-MS/MS analysis. The use of such strategies would lead to reduced repeatability for peptide ions and steeper gains in identifications from replicate to replicate.
Discrimination of proteomic differences between samples based on spectral count data requires relatively low variability in counts observed across replicates of the same samples. To characterize spectral count variation, we examined the counts observed for each protein across six replicates of yeast for each instrument in Study 5. Coefficients of variation (CVs), which compare the standard deviation to the mean for these counts, have two significant drawbacks for this purpose. First, they do not take into account the variation in overall sensitivity for some replicates (as illustrated by the spread of data points for each instrument in Figure 3). Second, CVs are generally highest for proteins that produce low spectral counts. We developed a statistic based on the multivariate hypergeometric (MVH) distribution instead (see Methods) that addressed both of these problems. In this approach, we compute the ratio between two probabilities. The first is the probability that spectral counts for a given protein would be distributed across replicates as actually observed, given the number of all identified spectra in each replicate. The second is the probability associated with the most likely distribution of these spectral counts across replicates. The ratio, expressed in natural log scale, asks how much less likely a particular distribution is than the most common distribution of spectral counts.
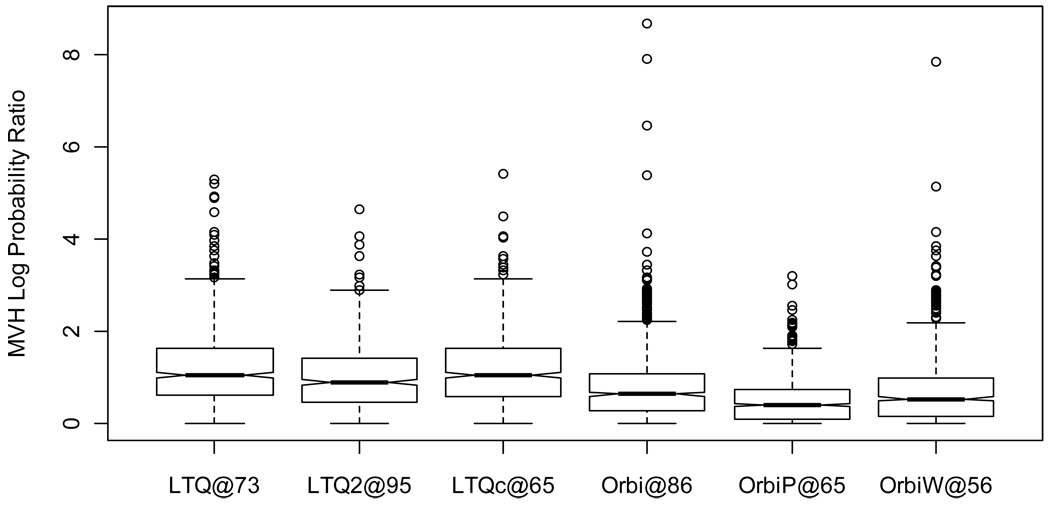
The results, illustrated in Figure 10 and Table 2 and Table 3, show considerable spread in the probability ratios for each instrument. The highest value (i.e. least stable spectral counts) for a protein in LTQ@73, for example, is 5.29 on a natural log scale, indicating that the observed spectral count distribution for this protein was approximately 200 times less likely than the most equitable distribution of spectral counts (mostly due to the low spectral counts observed in replicates 5 and 6). The proteins with extreme MVH scores for each instrument reflect that examining spectral counts for large numbers of proteins invariably reveals a set of proteins with uneven spectral counts, even when the sample is unchanged. This phenomenon reflects the need for multiple-testing correction in the statistical techniques employed for spectral count differentiation.
Figure 10. Study 5 yeast protein spectral count stability.
Spectral count differentiation attempts to detect differences between samples by recognizing changes in the numbers of spectra identified to those proteins. This image depicts the stability of spectral counts across six replicates when the sample is unchanged. A value of zero represents spectral counts that are spread across the replicates as evenly as possible. A value of 5 indicates that the ratio of probabilities for the observed spread of spectral counts versus the even distribution is e5=148. LTQs showed greater instability of spectral counts than did Orbitraps.
Table 2.
Study 5 yeast protein extreme spectral count variation
| Instrument | Accession | MVH1 | Rep1 | Rep2 | Rep3 | Rep4 | Rep5 | Rep6 |
|---|---|---|---|---|---|---|---|---|
| LTQ@73 | YKL060C | 5.29 | 35 | 32 | 31 | 28 | 16 | 18 |
| LTQ2@95 | YHR025W | 4.65 | 2 | 6 | 5 | 1 | 4 | 0 |
| LTQc@65 | YMR045C | 5.41 | 1 | 10 | 3 | 2 | 2 | 2 |
| Orbi@86 | YKL060C | 8.70 | 35 | 43 | 43 | 82 | 55 | 68 |
| OrbiP@65 | YDR158W | 3.20 | 3 | 5 | 6 | 0 | 4 | 4 |
| OrbiW@56 | rev_YLR057W | 7.85 | 10 | 0 | 2 | 1 | 5 | 6 |
Natural log of multivariate hypergeometric probability ratio: observed distribution versus expected distribution
Table 3.
Study 5 yeast protein median spectral count variation
| Instrument | Accession | MVH1 | Rep1 | Rep2 | Rep3 | Rep4 | Rep5 | Rep6 |
|---|---|---|---|---|---|---|---|---|
| LTQ@73 | YDR012W | 1.05 | 28 | 27 | 29 | 23 | 25 | 18 |
| LTQ2@95 | YGL062W | 0.90 | 2 | 2 | 2 | 2 | 5 | 3 |
| LTQc@65 | YLR441C | 1.06 | 12 | 14 | 10 | 8 | 10 | 9 |
| Orbi@86 | YDL124W | 0.64 | 5 | 3 | 3 | 4 | 2 | 3 |
| OrbiP@65 | YLR150W | 0.41 | 4 | 4 | 4 | 4 | 3 | 2 |
| OrbiW@56 | YBR109C | 0.54 | 3 | 4 | 3 | 4 | 2 | 5 |
Natural log of multivariate hypergeometric probability ratio: observed distribution versus expected distribution
The median cases for these scores are examples of routine disparities in spectral counts. The medians for Orbitrap instruments were approximately half of the LTQ medians (Student’s t-Test p-value=0.00617, using unpaired samples, two-sided outcomes, and unequal variances); spectral counts vary more in LTQ instruments. As a result, biomarker studies employing LTQ spectrum counts need to include more replicates than those employing Orbitraps to achieve the same degree of differentiation.
Conclusion
The CPTAC network undertook a series of interlaboratory studies to systematically evaluate LC-MS/MS proteomics platforms. We employed defined protein mixtures and SOPs to enable comparisons under conditions where key system variables were held constant, in order to discern inherent variability in peptide and protein detection for Thermo LTQ and Orbitrap instruments. The data documented repeatability and reproducibility of LC-MS/MS analyses and quantified contributions of instrument class and performance, protein and peptide characteristics and sample load and complexity. Several of our observations are consistent with broad experience in the field. For example, peptide detection is more variable than protein detection. High resolution Orbitraps outperform lower resolution LTQs in repeatability and reproducibility and in stability of spectral count data, provided that the Orbitrap is optimally tuned and that database searches are optimized. On the other hand, our data revealed some surprising insights. The first is that peptide and protein identification repeatability is independent of both sample complexity and sample load, at least in the range of sample types we studied—defined protein mixtures to complex proteomes. This indicates that essentially all protein digestions produce sufficiently complex peptide mixtures to challenge comprehensive LC-MS/MS identification.
Nevertheless, a standardized analysis platform yields a high degree of repeatability and reproducibility (~70–80%) in protein identifications, even from complex mixtures. Taken together with the accumulation of identifications with replicate analyses, this indicates that LC-MS/MS systems can generate consistent inventories of proteins even in complex biological samples. How this translates into consistent detection of proteomic differences between different phenotypes remains to be evaluated and will be the subject of future CPTAC studies.
Supplementary Material
Acknowledgment
This work was supported by grants from the National Cancer Institute (U24 CA126476, U24 126477, U24 126480, U24 CA126485, and U24 126479), part of NCI Clinical Proteomic Technologies for Cancer (http://proteomics.cancer.gov) initiative. A component of this initiative is the Clinical Proteomic Technology Assessment for Cancer (CPTAC) teams that include the Broad Institute of MIT and Harvard (with the Fred Hutchinson Cancer Research Center, Massachusetts General Hospital, the University of North Carolina at Chapel Hill, the University of Victoria and the Plasma Proteome Institute); Memorial Sloan-Kettering Cancer Center (with the Skirball Institute at New York University); Purdue University (with Monarch Life Sciences, Indiana University, Indiana University-Purdue University Indianapolis and the Hoosier Oncology Group); University of California, San Francisco (with the Buck Institute for Age Research, Lawrence Berkeley National Laboratory, the University of British Columbia and the University of Texas M.D. Anderson Cancer Center); and Vanderbilt University School of Medicine (with the University of Texas M.D. Anderson Cancer Center, the University of Washington and the University of Arizona). An interagency agreement between the National Cancer Institute and the National Institute of Standards and Technology also provided important support. Contracts from National Cancer Institute through SAIC to Texas A&M University funded statistical support for this work.
Footnotes
Certain commercial equipment, instruments, or materials are identified in this document. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the products identified are necessarily the best available for the purpose.
References
- 1.Steen H, Mann M. The ABC's (and XYZ's) of peptide sequencing. Nat Rev Mol Cell Biol. 2004;5(9):699–711. doi: 10.1038/nrm1468. [DOI] [PubMed] [Google Scholar]
- 2.Prakash A, Mallick P, Whiteaker J, Zhang H, Paulovich A, Flory M, Lee H, Aebersold R, Schwikowski B. Signal maps for mass spectrometry-based comparative proteomics. Mol Cell Proteomics. 2006;5(3):423–432. doi: 10.1074/mcp.M500133-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76(14):4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 4.Tabb DL, MacCoss MJ, Wu CC, Anderson SD, Yates JR., 3rd Similarity among tandem mass spectra from proteomic experiments: detection, significance, and utility. Anal Chem. 2003;75(10):2470–2477. doi: 10.1021/ac026424o. [DOI] [PubMed] [Google Scholar]
- 5.de Godoy LM, Olsen JV, de Souza GA, Li G, Mortensen P, Mann M. Status of complete proteome analysis by mass spectrometry: SILAC labeled yeast as a model system. Genome Biol. 2006;7(6):R50. doi: 10.1186/gb-2006-7-6-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bammler T, Beyer RP, Bhattacharya S, Boorman GA, Boyles A, Bradford BU, Bumgarner RE, Bushel PR, Chaturvedi K, Choi D, Cunningham ML, Deng S, Dressman HK, Fannin RD, Farin FM, Freedman JH, Fry RC, Harper A, Humble MC, Hurban P, Kavanagh TJ, Kaufmann WK, Kerr KF, Jing L, Lapidus JA, Lasarev MR, Li J, Li YJ, Lobenhofer EK, Lu X, Malek RL, Milton S, Nagalla SR, O'Malley JP, Palmer VS, Pattee P, Paules RS, Perou CM, Phillips K, Qin LX, Qiu Y, Quigley SD, Rodland M, Rusyn I, Samson LD, Schwartz DA, Shi Y, Shin JL, Sieber SO, Slifer S, Speer MC, Spencer PS, Sproles DI, Swenberg JA, Suk WA, Sullivan RC, Tian R, Tennant RW, Todd SA, Tucker CJ, Van Houten B, Weis BK, Xuan S, Zarbl H. Standardizing global gene expression analysis between laboratories and across platforms. Nat Methods. 2005;2(5):351–356. doi: 10.1038/nmeth754. [DOI] [PubMed] [Google Scholar]
- 7.van Midwoud PM, Rieux L, Bischoff R, Verpoorte E, Niederlander HA. Improvement of recovery and repeatability in liquid chromatography-mass spectrometry analysis of peptides. J Proteome Res. 2007;6(2):781–791. doi: 10.1021/pr0604099. [DOI] [PubMed] [Google Scholar]
- 8.Delmotte N, Lasaosa M, Tholey A, Heinzle E, van Dorsselaer A, Huber CG. Repeatability of peptide identifications in shotgun proteome analysis employing off-line two-dimensional chromatographic separations and ion-trap MS. J Sep Sci. 2009;32(8):1156–1164. doi: 10.1002/jssc.200800615. [DOI] [PubMed] [Google Scholar]
- 9.Slebos RJ, Brock JW, Winters NF, Stuart SR, Martinez MA, Li M, Chambers MC, Zimmerman LJ, Ham AJ, Tabb DL, Liebler DC. Evaluation of Strong Cation Exchange versus Isoelectric Focusing of Peptides for Multidimensional Liquid Chromatography-Tandem Mass Spectrometry. J Proteome Res. 2008;7(12):5286–5294. doi: 10.1021/pr8004666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods. 2005;2(9):667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 11.Resing KA, Meyer-Arendt K, Mendoza AM, Aveline-Wolf LD, Jonscher KR, Pierce KG, Old WM, Cheung HT, Russell S, Wattawa JL, Goehle GR, Knight RD, Ahn NG. Improving reproducibility and sensitivity in identifying human proteins by shotgun proteomics. Anal Chem. 2004;76(13):3556–3568. doi: 10.1021/ac035229m. [DOI] [PubMed] [Google Scholar]
- 12.Kapp EA, Schutz F, Connolly LM, Chakel JA, Meza JE, Miller CA, Fenyo D, Eng JK, Adkins JN, Omenn GS, Simpson RJ. An evaluation, comparison, and accurate benchmarking of several publicly available MS/MS search algorithms: sensitivity and specificity analysis. Proteomics. 2005;5(13):3475–3490. doi: 10.1002/pmic.200500126. [DOI] [PubMed] [Google Scholar]
- 13.Kislinger T, Gramolini AO, MacLennan DH, Emili A. Multidimensional protein identification technology (MudPIT): technical overview of a profiling method optimized for the comprehensive proteomic investigation of normal and diseased heart tissue. J Am Soc Mass Spectrom. 2005;16(8):1207–1220. doi: 10.1016/j.jasms.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 14.Balgley BM, Wang W, Song T, Fang X, Yang L, Lee CS. Evaluation of confidence and reproducibility in quantitative proteomics performed by a capillary isoelectric focusing-based proteomic platform coupled with a spectral counting approach. Electrophoresis. 2008;29(14):3047–3054. doi: 10.1002/elps.200800050. [DOI] [PubMed] [Google Scholar]
- 15.Washburn MP, Ulaszek RR, Yates JR., 3rd Reproducibility of quantitative proteomic analyses of complex biological mixtures by multidimensional protein identification technology. Anal Chem. 2003;75(19):5054–5061. doi: 10.1021/ac034120b. [DOI] [PubMed] [Google Scholar]
- 16.Bell AW, Deutsch EW, Au CE, Kearney RE, Beavis R, Sechi S, Nilsson T, Bergeron JJ, Beardslee TA, Chappell T, Meredith G, Sheffield P, Gray P, Hajivandi M, Pope M, Predki P, Kullolli M, Hincapie M, Hancock WS, Jia W, Song L, Li L, Wei J, Yang B, Wang J, Ying W, Zhang Y, Cai Y, Qian X, He F, Meyer HE, Stephan C, Eisenacher M, Marcus K, Langenfeld E, May C, Carr SA, Ahmad R, Zhu W, Smith JW, Hanash SM, Struthers JJ, Wang H, Zhang Q, An Y, Goldman R, Carlsohn E, van der Post S, Hung KE, Sarracino DA, Parker K, Krastins B, Kucherlapati R, Bourassa S, Poirier GG, Kapp E, Patsiouras H, Moritz R, Simpson R, Houle B, Laboissiere S, Metalnikov P, Nguyen V, Pawson T, Wong CC, Cociorva D, Yates Iii JR, Ellison MJ, Lopez-Campistrous A, Semchuk P, Wang Y, Ping P, Elia G, Dunn MJ, Wynne K, Walker AK, Strahler JR, Andrews PC, Hood BL, Bigbee WL, Conrads TP, Smith D, Borchers CH, Lajoie GA, Bendall SC, Speicher KD, Speicher DW, Fujimoto M, Nakamura K, Paik YK, Cho SY, Kwon MS, Lee HJ, Jeong SK, Chung AS, Miller CA, Grimm R, Williams K, Dorschel C, Falkner JA, Martens L, Vizcaino JA. A HUPO test sample study reveals common problems in mass spectrometry-based proteomics. Nat Methods. 2009;6:423–430. doi: 10.1038/nmeth.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chamrad D, Meyer HE. Valid data from large-scale proteomics studies. Nat Methods. 2005;2(9):647–648. doi: 10.1038/nmeth0905-647. [DOI] [PubMed] [Google Scholar]
- 18.Omenn GS, States DJ, Adamski M, Blackwell TW, Menon R, Hermjakob H, Apweiler R, Haab BB, Simpson RJ, Eddes JS, Kapp EA, Moritz RL, Chan DW, Rai AJ, Admon A, Aebersold R, Eng J, Hancock WS, Hefta SA, Meyer H, Paik YK, Yoo JS, Ping P, Pounds J, Adkins J, Qian X, Wang R, Wasinger V, Wu CY, Zhao X, Zeng R, Archakov A, Tsugita A, Beer I, Pandey A, Pisano M, Andrews P, Tammen H, Speicher DW, Hanash SM. Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics. 2005;5(13):3226–3245. doi: 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- 19.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1(8476):307–310. [PubMed] [Google Scholar]
- 20.McNaught AD, Wilkinson A. Compendium of Chemical Terminology (the "Gold Book") 2.02 ed. Oxford: Blackwell Scientific Publications; 1997. [Google Scholar]
- 21.Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24(21):2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tabb DL, Fernando CG, Chambers MC. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J Proteome Res. 2007;6(2):654–661. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma ZQ, Dasari S, Chambers MC, Litton MD, Sobecki SM, Zimmerman LJ, Halvey PJ, Schilling B, Drake PM, Gibson BW, Tabb DL. IDPicker 2.0: Improved Protein Assembly with High Discrimination Peptide Identification Filtering. J Proteome Res. 2009 doi: 10.1021/pr900360j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang B, Chambers MC, Tabb DL. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J Proteome Res. 2007;6(9):3549–3557. doi: 10.1021/pr070230d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Craig R, Beavis RC. A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun Mass Spectrom. 2003;17(20):2310–2316. doi: 10.1002/rcm.1198. [DOI] [PubMed] [Google Scholar]
- 26.Team RDC. R: A language and environment for statistical computing. 2.9.0 ed. Vienna, Austria: R Foundation for Statistical Computing; 2009. [Google Scholar]
- 27.Rudnick PA, Clauser KR, Kilpatrick LE, Tchekhovskoi DV, Neta P, Blounder N, Billheimer DD, Blackman RK, Bunk DM, Cardasis HL, Ham AJ, Jaffe JD, Kinsinger CR, Mesri M, Neubert TA, Schilling B, Tabb DL, Tegeler TJ, Vega-Montoto L, Variyath AM, Wang M, Wang P, Whiteaker JR, Zimmerman LJ, Carr SA, Fisher SJ, Gibson BW, Paulovich AG, Regnier FE, Rodriguez H, Spiegelman C, Tempst P, Liebler DC, Stein SE. Performance metrics for liquid chromatography- tandem mass spectrometry systems in proteomic analyses and evaluation by the CPTAC Network. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M900223-MCP200. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paulovich AG, Billheimer DD, Ham AJ, Vega-Montoto L, Rudnick PA, Tabb DL, Wang P, Blackman RK, Bunk DM, Cardasis HL, Clauser KR, Kinsinger CR, Schilling B, Tegeler TJ, Variyath AM, Wang M, Whiteaker JR, Zimmerman LJ, Fenyo D, Carr SA, Fisher SJ, Gibson BW, Mesri M, Neubert TA, Regnier FE, Rodriguez H, Spiegelman C, Stein SE, Tempst P, Liebler DC. A yeast reference proteome for benchmarking LC-MS platform performance. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M900222-MCP200. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, Spiegelman CH, Zimmerman LJ, Ham AJ, Keshishian H, Hall SC, Allen S, Blackman RK, Borchers CH, Buck C, Cardasis HL, Cusack MP, Dodder NG, Gibson BW, Held JM, Hiltke T, Jackson A, Johansen EB, Kinsinger CR, Li J, Mesri M, Neubert TA, Niles RK, Pulsipher TC, Ransohoff D, Rodriguez H, Rudnick PA, Smith D, Tabb DL, Tegeler TJ, Variyath AM, Vega-Montoto LJ, Wahlander A, Waldemarson S, Wang M, Whiteaker JR, Zhao L, Anderson NL, Fisher SJ, Liebler DC, Paulovich AG, Regnier FE, Tempst P, Carr SA. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009;27(7):633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 31.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24(8):971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 32.Rudomin EL, Carr SA, Jaffe JD. Directed sample interrogation utilizing an accurate mass exclusion-based data-dependent acquisition strategy (AMEx) J Proteome Res. 2009;8(6):3154–3160. doi: 10.1021/pr801017a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoopmann MR, Merrihew GE, von Haller PD, MacCoss MJ. Post analysis data acquisition for the iterative MS/MS sampling of proteomics mixtures. J Proteome Res. 2009;8(4):1870–1875. doi: 10.1021/pr800828p. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.