Abstract
Infection with genotype 4 of the Hepatitis C virus is common in Africa and the Mediterranean area, but has also been found at increasing frequencies in injection drug users in Europe and North America. Full length viral sequences to characterize viral diversity and structure have recently become available mostly for subtype 4a, and studies in Egypt and Saudi Arabia, where high proportions of subtype 4a infected patients exist, have begun to establish optimized treatment regimens. However knowledge about other subtype variants of genotype 4 present in less developed African states is lacking. In this study the full coding region from so far poorly characterized variants of HCV genotype 4 was amplified and sequenced using a long range PCR technique. Sequences were analyzed with respect to phylogenetic relationship, possible recombination and prominent sequence characteristics compared to other known HCV strains. We present for the first time two full-length sequences from the HCV genotype 4k, in addition to five strains from HCV genotypes 4d and 4f. Reference sequences for accurate HCV genotyping are required for optimized treatment, and a better knowledge of the global viral sequence diversity is needed to guide vaccines or new drugs effective in the world wide epidemic.
Keywords: Hepatitis C Virus, subtype, long template PCR
Introduction
Hepatitis C virus (HCV) infection is a global health problem with 170 million infected people worldwide [Lauer and Walker, 2001]. Hepatitis C virus is a member of the Hepacivirus genus in the Flaviviridae family. Current classifications distinguish between 6 major genotypes and multiple subtypes [Simmonds et al., 2005], and very recently a novel seventh genotype has been described [Murphy et al., 2007a]. These HCV genotypes differ in their genetic structure, clinical features, and their geographical distribution. Genotype 1b is observed worldwide, whereas genotypes 1a and 3a are regionally concentrated in European and North American countries, genotype 2 in the Mediterranean region, far east and Western Africa, genotype 5 in South Africa, genotype 6 in South East Asia [Simmonds et al., 2005], and genotype 7 was found in patients from the Democratic Republic of Congo [Murphy et al., 2007a]. Genotype 4 is encountered throughout Africa and Mediterranean countries, but more recently has spread to Europe and North America mostly by immigrants and injection drug users [Nguyen and Keeffe, 2005]. Significant frequencies of genotype 4 infections have also been reported in India and the Caribbean [Nguyen and Keeffe, 2005].
Accurate diagnosis of HCV genotypes is clinically relevant as genotypes differ in their response rates to the current standard treatment with pegylated Interferon alpha and Ribavirin. Clinical trials indicate sustained virological response rates of up to 80% in patients infected with genotype 2 and 3 after 24 weeks of therapy, considerably higher than the 42–52% achieved with a 48 week treatment duration in genotype 1 infection [Fried et al., 2002; Hadziyannis et al., 2004; Manns et al., 2001]. While there remains a paucity of controlled studies for genotypes 5 and 6 which are therefore often treated similar to genotype 1, recent data indicate that genotype 4 infected patients may achieve viral clearance in 60–80% of cases [Alfaleh et al., 2004; El-Zayadi et al., 2005; Hasan et al., 2004]. Aside from drug response rates, knowledge regarding worldwide patterns of viral sequence diversity is also essential for research efforts to design efficient vaccines and new drugs to fight the global epidemic.
In order to provide a systematic structure to capture the global viral diversity a recent consensus conference delineated guidelines for the nomenclature of HCV variants, including a list of reference sequences to enable accurate classification of diagnostic samples and potential new strains [Simmonds et al., 2005]. Major genotypes were defined as strains that form phylogenetically distinct clusters and differ from other genotype sequences by 31% – 33% on the nucleotide level, compared to only 20% – 25% differences between subtypes. Up to 19 subtypes were assigned for each of the 6 major genotypes, however only a provisional classification was possible for many of these since only sub-genomic sequences (e.g. from NS5B or the Core/E1 region) are available for many presumed subtypes. Although in most cases the sub-genomic sequences provide sufficient information for clinical purposes, the recent discovery of inter-genotypic recombinants [Colina et al., 2004; Kalinina et al., 2004; Kalinina et al., 2002; Legrand-Abravanel et al., 2007; Noppornpanth et al., 2006] has highlighted the need for full length sequences to fully define novel viral genotypes and subtypes.
In an effort towards assisting in the definitive classification of all currently provisionally assigned subtypes this study presents five viral sequences confirming the classification of strains 4d and 4f, and for the first time characterizes two sequences covering the full open reading frame (ORF) of the HCV subtype 4k.
Methods
Genotyping
Viral RNA was extracted from 140µl of plasma using the Qiagen vRNA Extraction Kit Qiagen, Hilden, Germany). Primers HCV_Geno_F and HCV4_Geno_R (Supplementary Table 1) were used for reverse transcription and subsequent amplification of a 1kb sequence fragment in NS5B in a single RT-PCR reaction using the Qiagen One-Step RT-PCR kit with 3µl of vRNA as template. PCR conditions on an MJ PTC-200 thermal cycler (BioRad) were reverse transcription at 50°C for 60 min., denaturation at 95°C for 15 min., followed by 50 PCR amplification cycles with 94°C for 30 s, 62°C for 30 s and 72°C for 90 s, and a final extension at 68°C for 20 min. PCR products were sequenced bidirectionally on an ABI 3130x automated sequencer (Applied Biosystems, Foster City, CA, USA). Sequences were then compared by phylogenetic analysis to published reference sequences.
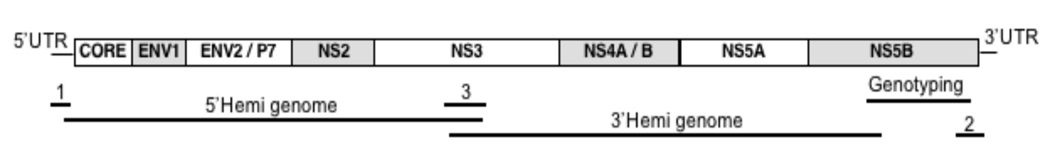
Strategy for PCR amplification of the full HCV open reading frame
First, three PCR fragments in addition to the genotyping fragment were amplified (Figure 1). Fragment 1 covered the 5’UTR, while fragment 2 covered the region between the genotyping fragment and the 3’end of the coding sequence. Fragment 3 amplified a small sequence island in NS3. Primer sequences are given in Supplementary Table1. PCR conditions were as described in the genotyping reaction above, with annealing temperatures of 58°C or 62°C. Together with the genotyping fragment sequencing of these PCR products provided small sequence islands that enabled the design of strain specific primers for the subsequent amplification of hemi-genome fragments which together covered the entire HCV ORF.
Figure 1.
Strategy for PCR amplification of the entire HCV open reading frame. A genotyping fragment in NS5B and two additional fragments 1 and 3 provide small sequence islands that enable generation of strain specific primers for the amplification of large hemi-genome fragments.
cDNA for the amplification of hemi-genome fragments was synthesized using Superscript III Reverse Transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions with 5µl of viral RNA and 1µl of 2µM strain-specific reverse primer. Hemi-genomes were amplified in a two round nested PCR using the Expand Long Range dNTPack (Roche Applied Science, Indianapolis, IN, USA) with 1µl DMSO and 0.6µl of each 25µM strain specific primer (Supplementary Table 1) in a 50µl reaction volume. PCR conditions were 94°C for 2 min, 50 cycles of denaturation at 94°C for 20 sec, annealing at 58°C for 30 sec and extension at 68°C for 3.5 min, followed by a final extension at 68°C for 7 min. After the first 10 cycles, the extension time was increased by 15 sec after each cycle. For optimal results in the first round PCR it was necessary to slow down the ramping times between the denaturation and annealing step starting with 1°C/sec from 94°C to 63°C, then 0.3°C/sec to 60°C and 0.2°C/sec to 58°C. Where necessary the results could be further improved by separating the primers from the enzyme by PCR wax during PCR setup. The size of the amplicons was checked by gel electrophoresis, and sequences were obtained by sequence walking using strain specific primers.
Generation of clonal sequences
A small region of E2 was cloned from viral RNA of patient EU392169 to evaluate a possible mixed infection suspected based on chromatograms from bulk sequences. Using primers DAV9905_F and DAV9905_R1 or DAV9905_R2 (Supplementary Table 1) two PCR fragments 393 and 695 base pairs in size were amplified in a 50 cycle one-step RT-PCR as described above. PCR fragments were cloned using the Topo TA cloning kit for sequencing (Invitrogen, Carlsbad, CA, USA) according to the manufacturers instructions. Plasmids from 16 clones were extracted using the DirectPrep 96 Miniprep Kit (Qiagen, Hilden, Germany), and were sequenced bidirectionally using M13 primers.
Sequence analysis
Sequence chromatograms were aligned and edited using Sequencher (Gene Codes Corp., Ann Arbor, MI, USA). Nucleotide and amino acid sequences from different strains were aligned using ClustalX [Thompson et al., 1997] and Se-Al v2.0a11 (http://evolve.zoo.ox.ac.uk) software, and phylogenetic trees were constructed by the neighbor-joining method with Jukes-Cantor corrected distances on PAUP4.0b10 software (Sinauer, Sunderland, MA, USA). Bootstrap values are based on 1,000 replications. Pairwise nucleotide distances between sequences were calculated using Mega 4.0 [Tamura et al., 2007]. Simplot [Lole et al., 1999] was used to evaluate possible recombinations between different viral strains.
Results and Discussion
Study subjects
Seven discarded plasma samples were received from clinical diagnostic laboratories in Great Britain and Cameroon. Approval to conduct these investigations for all patient samples included in the study was obtained from the institutional review board at Massachusetts General Hospital. Viral sequences from the Core and NS5B region of samples CM9905 and CM1578 originating from patients in Cameroon (new Genbank accession numbers EU392169 and EU392170) have been previously published [Ndjomou et al., 2003]. No demographic or clinical patient information was available for the other five samples that were obtained as discarded samples after HCV genotyping from diagnostic laboratories in Great Britain. Each of these samples were previously typed as either unspecified genotypes 4 other than 4a, or as the uncommon HCV genotypes 4d, 4f and 4k according to results based on restriction fragment length polymorphisms (RFLP) in the 5’UTR [Davidson et al., 1995] or sequence analysis in the Core/E1 or NS5B regions [Mellor et al., 1995; Viazov et al., 1997].
Provisional identification of HCV subtypes
The highly conserved 5’UTR allows reliable and sensitive PCR amplification for determination of viral loads, and sequences from this genomic region are therefore widely used also for HCV genotyping. However, due to the low degree of sequence diversity in the 5’UTR as compared to the more informative Core/E1 and NS5B regions, assays based on the 5’UTR are limited in their ability to distinguish some major genotypes and to differentiate many HCV subtypes [Chinchai et al., 2003; Mellor et al., 1996; Murphy et al., 2007b; Stuyver et al., 1996].
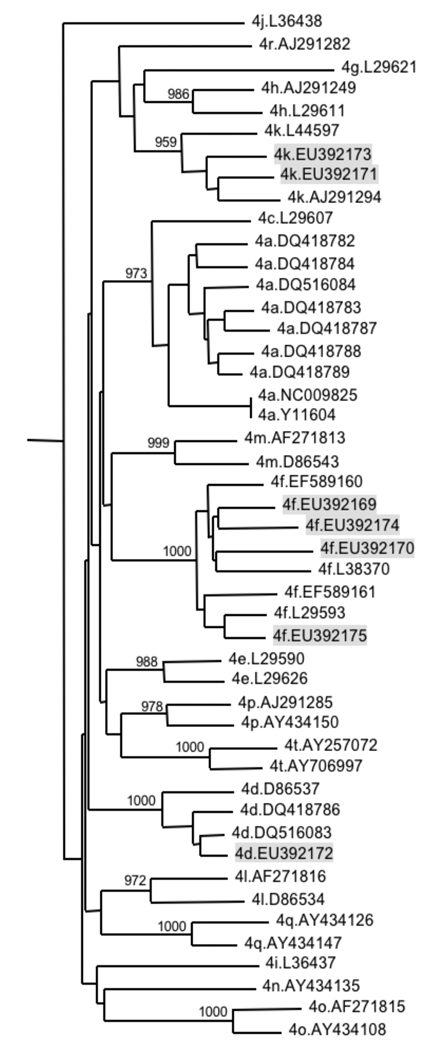
To confirm the previously determined HCV genotypes and establish provisional subtype classifications for these samples part of the NS5B region was therefore amplified and sequenced. Sequences were then compared by phylogenetic analysis to published reference sequences of all major genotypes, and of confirmed [Chamberlain et al., 1997; Franco et al., 2007; Timm et al., 2007] or provisionally assigned [Simmonds et al., 2005] HCV genotype 4 subtypes that are available from the Los Alamos National Laboratory (LANL) database (http://hcv.lanl.gov, NCBI accession numbers provided in figure 2). Clustering of the new sequences with subtype specific reference sequences suggested classification of these samples as HCV subtypes 4d, 4f and 4k (Figure 2).
Figure 2.
Neighbour joining tree based on partial NS5B sequences (nucleotide positions 7939–8269 relative to the HCV genotype 1a reference sequence H77) of HCV strains presented in this manuscript (shaded in grey), and sequences provisionally assigned as HCV subtype 4 references [Simmonds et al., 2005]. The numbers of bootstrap replicates supporting relevant nodes (total 1000 replicates) are indicated. Clusters of novel and reference nucleotide sequences suggest classification of the new strains as HCV subtypes 4d, 4f and 4k.
Definitive assignment of HCV subtypes based on sequences covering the entire HCV open reading frame
In order to characterize further the world wide sequence diversity, confirm provisionally assigned subtype classifications, and provide a set of reference sequences for each viral subtype, the entire HCV ORF and part of the 5’- and 3’UTRs from 7 samples of the putative genotypes 4d, 4f and 4k were amplified and sequenced, comprising between 9269 and 9303 nucleotides in length. Sequences are available from GenBank under accession numbers EU392169 - EU392175.
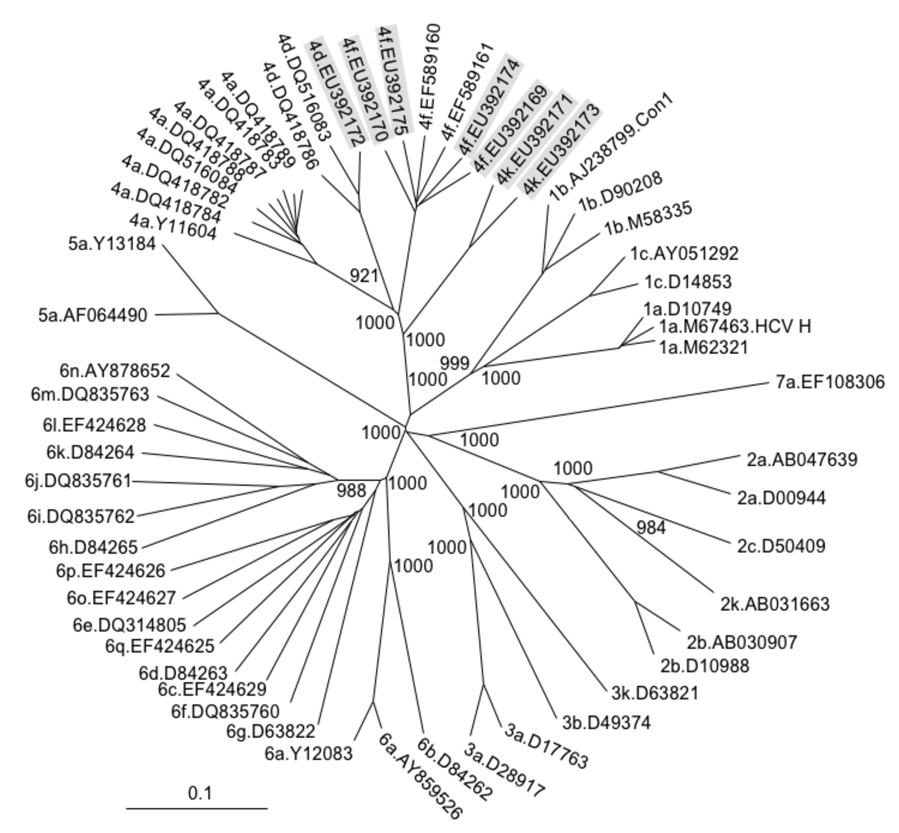
To confirm the provisional classification based on NS5B sequence, the novel strains were aligned to selected full length sequences of the major HCV genotypes and to all full length sequences of genotype 4 subtypes currently available at public databases. As expected, a phylogenetic analysis revealed that five of the novel viral strains clustered together with published sequences of subtypes 4d and 4f, whereas the sequences provisionally assigned to subtype 4k formed a separate cluster within genotype 4, but distinct from other existing genotype 4 strains. All clusters were supported by high bootstrap values (Figure 3).
Figure 3.
Neighbour joining tree based on published nucleotide sequences covering the full HCV open reading frame, with novel full-length sequences shaded in grey. The numbers of bootstrap replicates supporting relevant nodes (total 1000 replicates) are indicated. Full-length sequences provisionally assigned to subtypes 4d, 4f and 4k form distinct clusters within genotype 4 separate from other published subtypes, thus confirming their definition as separate subtypes according to consensus criteria [Simmonds et al., 2005]. HCV genotypes, subtypes and NCBI reference numbers are given for each sequence.
HCV genotypes typically differ by around 30% in their nucleotide sequence, compared to 20%–25% differences between subtypes [Simmonds et al., 2005]. To verify that the novel HCV strains, especially the genotype 4k strains, represented an independent genotype 4 subtype, pairwise genetic distances were calculated between these and the full length HCV sequences included in Figure 3. Genotype 4 sequences clustering together in Figure 3 showed between 6.8% (5.1%) and 9.0% (5.6%) pairwise nucleotide (amino acid) distances, whereas sequences from separate clusters differed by 19.5% – 21.7% (11.9% – 14.5%). In addition, all genotype 4 sequences exhibited 28.6% – 34.2% (20.8% – 28.8%) pairwise nucleotide (amino acid) distances from all other genotypes. Taken together these data confirm subtypes 4d and 4f and additionally establish the subtype 4k as an independent viral subtype within genotype 4 based on full ORF sequences according to consensus criteria [Simmonds et al., 2005].
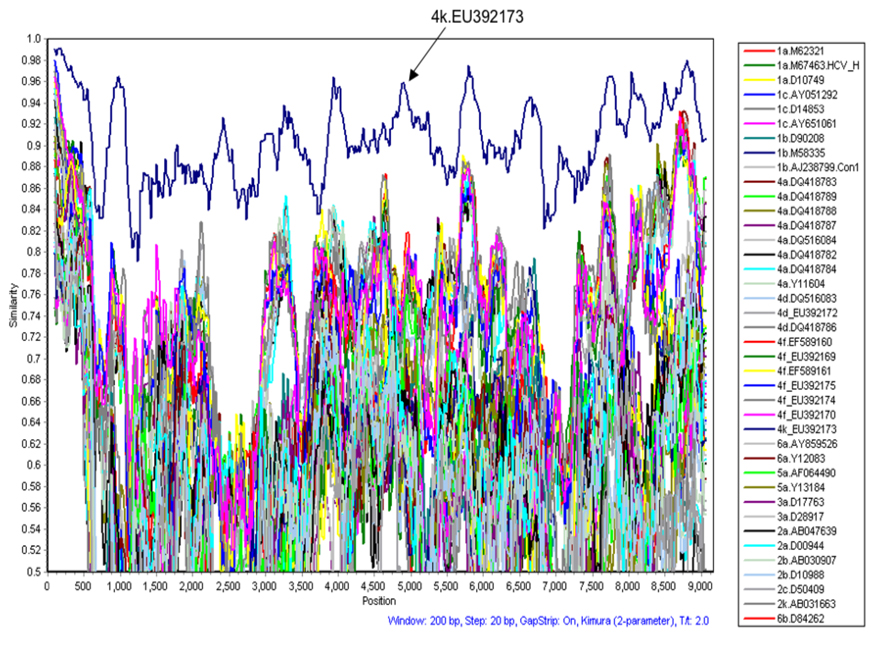
Recombinations between viral strains of different genotypes and subtypes have been described in patients from Russia, Peru, France and Vietnam [Colina et al., 2004; Kalinina et al., 2004; Kalinina et al., 2002; Legrand-Abravanel et al., 2007; Noppornpanth et al., 2006]. In order to examine the new sequences for inter-genotypic sequence recombination each sequence was compared to all other nucleotide sequences included in Figure 3. Percent similarities between sequences were calculated in a sliding window of 200 residues shifted in 20 nucleotide steps across the genome using Simplot software. No evidence for recombination was found (Figure 4).
Figure 4.
Simplot graph displaying pairwise nucleotide similarities between the test sequence 4k.EU392171 and full length sequences of all sequences included in Figure 3. Pairwise similarities are plotted based on calculations from a 200 nucleotide window sliding in 20 nucleotide steps across the genome. As illustrated, only the sequence of the second genotype 4k patient EU392173 shows consistently higher nucleotide similarities with 4k.EU392171 compared to all other sequences, with no evidence for recombination between this genotype 4k sequence and any other HCV genotype.
Prominent sequence characteristics in comparison with other HCV genotypes and subtypes
Prominent characteristics were examined that distinguish these new sequences from other HCV genotypes and subtypes. Amino acid positions are reported relative to the genotype 1a strain H77 (NCBI accession number A009606) with lower case letters indicating insertions relative to this reference strain according to published guidelines [Kuiken et al., 2006].
Compared to other genotype 4 sequences several alterations in the envelope genes were observed in some of these new sequences, namely a single amino acid deletion in position 479a in all genotype 4d sequences, and a two amino acid insertion in position 575 and 576 in all genotype 4f sequences. Notably, in the latter position in patient EU392169 unclear patterns in bulk sequence chromatograms indicated a possible mixed infection of viral quasispecies with and without this insertion. Indeed the presence of two quasispecies populations differing by a two amino acid insertion and an adjacent single amino acid substitution was confirmed in clonal sequences (Figure 5). Otherwise the two populations were highly similar to each other within the cloned area, but differed from sequences of other patients infected with genotype 4f by several amino acids (data not shown). Thus, the two amino acid insertion in patient EU392169 appeared to represent a polymorphism between viral quasispecies, but not a superinfection with two different viral strains.
Figure 5.
Amino acid alignment of clonal sequences from patient EU392169 documenting a mixed infection with two different viral quasispecies with and without a two amino acid insertion in position 575 and 576.
Moreover, in NS5A deletion of one (genotype 4d & 4k) or two (genotype 4f) amino acids was found in position 2263a and 2264 (Figure 6). Of note, genotype 4k sequences differed from all other genotype 4 sequences by a four amino acid insertion in position 2235–2238 that was identical to genotype 1 and 3 sequences (Figure 6). Last, there was a single amino acid insertion in all genotype 4f sequences at position 2331a in NS5A. Together with the >23% pairwise amino acid distances between genotypes these data illustrate considerable structural and potential functional differences between viral genotypes and subtypes.
Figure 6.
Amino acid alignment of reference sequences and novel strains in the NS5A region, including the Interferon Sensitivity Determining Region (ISDR, shaded in grey). Novel strains and noticeable parts of the sequences are marked in grey. Dashes indicate gaps or deletions. Amino acid positions are indicated relative to the HCV genotype 1a reference sequence H77.
HCV sequence variation is not only relevant for epidemiological or evolutionary studies, but also has important clinical implications as HCV genotypes differ considerably in their susceptibility to pegylated Interferon-α and Ribavirin [Fried et al., 2002; Hadziyannis et al., 2004; Manns et al., 2001]. Sequence differences in a 40 amino acid region in NS5A (termed the “Interferon Sensitivity Determining Region”, ISDR) in genotype 1 infected patients have been associated with treatment response rates in several Asian studies [Enomoto et al., 1996; Hung et al., 2003; Watanabe et al., 2001]. However many studies have failed to confirm this observation in European or North American populations [Airoldi et al., 2004; Chung et al., 1999; Halfon et al., 2000; Pascu et al., 2004; Squadrito et al., 1997; Squadrito et al., 1999; Squadrito et al., 2002; Veillon et al., 2004]. Therefore, although the clinical significance of ISDR mutations remains unclear, it is striking that considerable differences between genotypes with accumulation of insertions and deletions are seen within or in close proximity to the ISDR. While these differences thus far appeared to be genotype-specific, a novel aspect highlighted by this study is that considerable changes may also be observed between subtypes, as shown here in Figure 6 within genotype 4 for subtype 4k sequences. Only a handful of controlled studies to explore optimal treatment conditions are currently available for genotype 4 [Alfaleh et al., 2004; El-Zayadi et al., 2005; Hasan et al., 2004], however none of these have included a subtype analysis. Therefore it will be important to evaluate whether treatment results are generally applicable to the entire spectrum of HCV subtypes especially in areas with larger sequence diversity such as Africa or South East Asia.
Summary
Viral sequence diversity of HCV strains present in Europe and the United States is well characterized and established treatment regimens for the locally common variants of genotypes 1, 2 and 3 are intensively studied and optimized. In contrast, viral strains that predominate in less developed regions such as Africa or South East Asia remain poorly defined. This study presents a set of full length reference sequences of genotypes 4d and 4f, and provides the first two full length sequences for genotype 4k. These data will enable more accurate genotyping and subtyping approaches and provide a basis for in depth studies of treatment options suitable for these genotypes. More extensive studies are needed to characterize the global sequence diversity as a basis for drug design and worldwide vaccination efforts.
Supplementary Material
Acknowledgement
This project has been funded in whole or in part with funds from the Deutsche Forschungsgemeinschaft grant DFG KU2250/1-1 (TK), and with Federal funds from the National Institutes of Health grant RO1-AI067926-01 (TMA) and the National Institute of Allergy and Infectious Disease, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN266200400001C (BWB).
Footnotes
The authors have no conflicting financial interests.
References
- Airoldi A, Zavaglia C, Silini E, Tinelli C, Martinetti M, Asti M, Rossini A, Vangeli M, Salvaneschi L, Pinzello G. Lack of a strong association between HLA class II, tumour necrosis factor and transporter associated with antigen processing gene polymorphisms and virological response to alpha-interferon treatment in patients with chronic hepatitis C. Eur J Immunogenet. 2004;31(6):259–265. doi: 10.1111/j.1365-2370.2004.00478.x. [DOI] [PubMed] [Google Scholar]
- Alfaleh FZ, Hadad Q, Khuroo MS, Aljumah A, Algamedi A, Alashgar H, Al-Ahdal MN, Mayet I, Khan MQ, Kessie G. Peginterferon alpha-2b plus ribavirin compared with interferon alpha-2b plus ribavirin for initial treatment of chronic hepatitis C in Saudi patients commonly infected with genotype 4. Liver Int. 2004;24(6):568–574. doi: 10.1111/j.1478-3231.2004.0976.x. [DOI] [PubMed] [Google Scholar]
- Chamberlain RW, Adams N, Saeed AA, Simmonds P, Elliott RM. Complete nucleotide sequence of a type 4 hepatitis C virus variant, the predominant genotype in the Middle East. J Gen Virol. 1997;78(Pt 6):1341–1347. doi: 10.1099/0022-1317-78-6-1341. [DOI] [PubMed] [Google Scholar]
- Chinchai T, Labout J, Noppornpanth S, Theamboonlers A, Haagmans BL, Osterhaus AD, Poovorawan Y. Comparative study of different methods to genotype hepatitis C virus type 6 variants. J Virol Methods. 2003;109(2):195–201. doi: 10.1016/s0166-0934(03)00071-5. [DOI] [PubMed] [Google Scholar]
- Chung RT, Monto A, Dienstag JL, Kaplan LM. Mutations in the NS5A region do not predict interferon-responsiveness in american patients infected with genotype 1b hepatitis C virus. J Med Virol. 1999;58(4):353–358. [PubMed] [Google Scholar]
- Colina R, Casane D, Vasquez S, Garcia-Aguirre L, Chunga A, Romero H, Khan B, Cristina J. Evidence of intratypic recombination in natural populations of hepatitis C virus. J Gen Virol. 2004;85(Pt 1):31–37. doi: 10.1099/vir.0.19472-0. [DOI] [PubMed] [Google Scholar]
- Davidson F, Simmonds P, Ferguson JC, Jarvis LM, Dow BC, Follett EA, Seed CR, Krusius T, Lin C, Medgyesi GA, et al. Survey of major genotypes and subtypes of hepatitis C virus using RFLP of sequences amplified from the 5' non-coding region. J Gen Virol. 1995;76(Pt 5):1197–1204. doi: 10.1099/0022-1317-76-5-1197. [DOI] [PubMed] [Google Scholar]
- El-Zayadi AR, Attia M, Barakat EM, Badran HM, Hamdy H, El-Tawil A, El-Nakeeb A, Selim O, Saied A. Response of hepatitis C genotype-4 naive patients to 24 weeks of Peg-interferon-alpha2b/ribavirin or induction-dose interferonalpha2b/ribavirin/amantadine: a non-randomized controlled study. Am J Gastroenterol. 2005;100(11):2447–2452. doi: 10.1111/j.1572-0241.2005.00253.x. [DOI] [PubMed] [Google Scholar]
- Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334(2):77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- Franco S, Tural C, Clotet B, Martinez MA. Complete nucleotide sequence of genotype 4 hepatitis C viruses isolated from patients co-infected with human immunodeficiency virus type 1. Virus Res. 2007;123(2):161–169. doi: 10.1016/j.virusres.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Goncales FL, Jr, Haussinger D, Diago M, Carosi G, Dhumeaux D, Craxi A, Lin A, Hoffman J, Yu J. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347(13):975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- Hadziyannis SJ, Sette H, Jr, Morgan TR, Balan V, Diago M, Marcellin P, Ramadori G, Bodenheimer H, Jr, Bernstein D, Rizzetto M, Zeuzem S, Pockros PJ, Lin A, Ackrill AM. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann Intern Med. 2004;140(5):346–355. doi: 10.7326/0003-4819-140-5-200403020-00010. [DOI] [PubMed] [Google Scholar]
- Halfon P, Halimi G, Bourliere M, Ouzan D, Durant J, Khiri H, Mercier L, Gerolami V, Cartouzou G. Integrity of the NS5A (amino acid 2209 to 2248) region in hepatitis C virus 1b patients non-responsive to interferon therapy. Liver. 2000;20(5):381–386. doi: 10.1034/j.1600-0676.2000.020005381.x. [DOI] [PubMed] [Google Scholar]
- Hasan F, Asker H, Al-Khaldi J, Siddique I, Al-Ajmi M, Owaid S, Varghese R, Al-Nakib B. Peginterferon alfa-2b plus ribavirin for the treatment of chronic hepatitis C genotype 4. Am J Gastroenterol. 2004;99(9):1733–1737. doi: 10.1111/j.1572-0241.2004.40077.x. [DOI] [PubMed] [Google Scholar]
- Hung CH, Lee CM, Lu SN, Lee JF, Wang JH, Tung HD, Chen TM, Hu TH, Chen WJ, Changchien CS. Mutations in the NS5A and E2-PePHD region of hepatitis C virus type 1b and correlation with the response to combination therapy with interferon and ribavirin. J Viral Hepat. 2003;10(2):87–94. doi: 10.1046/j.1365-2893.2003.00414.x. [DOI] [PubMed] [Google Scholar]
- Kalinina O, Norder H, Magnius LO. Full-length open reading frame of a recombinant hepatitis C virus strain from St Petersburg: proposed mechanism for its formation. J Gen Virol. 2004;85(Pt 7):1853–1857. doi: 10.1099/vir.0.79984-0. [DOI] [PubMed] [Google Scholar]
- Kalinina O, Norder H, Mukomolov S, Magnius LO. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J Virol. 2002;76(8):4034–4043. doi: 10.1128/JVI.76.8.4034-4043.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken C, Combet C, Bukh J, Shin IT, Deleage G, Mizokami M, Richardson R, Sablon E, Yusim K, Pawlotsky JM, Simmonds P. A comprehensive system for consistent numbering of HCV sequences, proteins and epitopes. Hepatology. 2006;44(5):1355–1361. doi: 10.1002/hep.21377. [DOI] [PubMed] [Google Scholar]
- Lauer GM, Walker BD. Hepatitis C virus infection. N Engl J Med. 2001;345(1):41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- Legrand-Abravanel F, Claudinon J, Nicot F, Dubois M, Chapuy-Regaud S, Sandres-Saune K, Pasquier C, Izopet J. New natural intergenotypic (2/5) recombinant of hepatitis C virus. J Virol. 2007;81(8):4357–4362. doi: 10.1128/JVI.02639-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73(1):152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358(9286):958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- Mellor J, Holmes EC, Jarvis LM, Yap PL, Simmonds P. Investigation of the pattern of hepatitis C virus sequence diversity in different geographical regions: implications for virus classification. The International HCV Collaborative Study Group. J Gen Virol. 1995;76(Pt 10):2493–2507. doi: 10.1099/0022-1317-76-10-2493. [DOI] [PubMed] [Google Scholar]
- Mellor J, Walsh EA, Prescott LE, Jarvis LM, Davidson F, Yap PL, Simmonds P. Survey of type 6 group variants of hepatitis C virus in Southeast Asia by using a core-based genotyping assay. J Clin Microbiol. 1996;34(2):417–423. doi: 10.1128/jcm.34.2.417-423.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DG, Chamberland J, Dandavino R, Sablon E. A new genotype of Hepatitis C Virus originating from Central Africa. Hepatology. 2007a;46(4) Suppl.1:623A. doi: 10.1128/JCM.02831-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DG, Willems B, Deschenes M, Hilzenrat N, Mousseau R, Sabbah S. Use of sequence analysis of the NS5B region for routine genotyping of hepatitis C virus with reference to C/E1 and 5' untranslated region sequences. J Clin Microbiol. 2007b;45(4):1102–1112. doi: 10.1128/JCM.02366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndjomou J, Pybus OG, Matz B. Phylogenetic analysis of hepatitis C virus isolates indicates a unique pattern of endemic infection in Cameroon. J Gen Virol. 2003;84(Pt 9):2333–2341. doi: 10.1099/vir.0.19240-0. [DOI] [PubMed] [Google Scholar]
- Nguyen MH, Keeffe EB. Prevalence and treatment of hepatitis C virus genotypes 4, 5, and 6. Clin Gastroenterol Hepatol. 2005;3(10 Suppl 2):S97–S101. doi: 10.1016/s1542-3565(05)00711-1. [DOI] [PubMed] [Google Scholar]
- Noppornpanth S, Lien TX, Poovorawan Y, Smits SL, Osterhaus AD, Haagmans BL. Identification of a naturally occurring recombinant genotype 2/6 hepatitis C virus. J Virol. 2006;80(15):7569–7577. doi: 10.1128/JVI.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascu M, Martus P, Hohne M, Wiedenmann B, Hopf U, Schreier E, Berg T. Sustained virological response in hepatitis C virus type 1b infected patients is predicted by the number of mutations within the NS5A-ISDR: a meta-analysis focused on geographical differences. Gut. 2004;53(9):1345–1351. doi: 10.1136/gut.2003.031336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P, Bukh J, Combet C, Deleage G, Enomoto N, Feinstone S, Halfon P, Inchauspe G, Kuiken C, Maertens G, Mizokami M, Murphy DG, Okamoto H, Pawlotsky JM, Penin F, Sablon E, Shin IT, Stuyver LJ, Thiel HJ, Viazov S, Weiner AJ, Widell A. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42(4):962–973. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- Squadrito G, Leone F, Sartori M, Nalpas B, Berthelot P, Raimondo G, Pol S, Brechot C. Mutations in the nonstructural 5A region of hepatitis C virus and response of chronic hepatitis C to interferon alfa. Gastroenterology. 1997;113(2):567–572. doi: 10.1053/gast.1997.v113.pm9247477. [DOI] [PubMed] [Google Scholar]
- Squadrito G, Orlando ME, Cacciola I, Rumi MG, Artini M, Picciotto A, Loiacono O, Siciliano R, Levrero M, Raimondo G. Long-term response to interferon alpha is unrelated to "interferon sensitivity determining region" variability in patients with chronic hepatitis C virus-1b infection. J Hepatol. 1999;30(6):1023–1027. doi: 10.1016/s0168-8278(99)80255-4. [DOI] [PubMed] [Google Scholar]
- Squadrito G, Raffa G, Restuccia T, Pollicino T, Brancatelli S, Raimondo G. Is investigation of hepatitis C virus NS5A gene heterogeneity a tool for predicting long-lasting response to interferon therapy in patients with HCV-1b chronic hepatitis? J Viral Hepat. 2002;9(5):360–369. doi: 10.1046/j.1365-2893.2002.00379.x. [DOI] [PubMed] [Google Scholar]
- Stuyver L, Wyseur A, van Arnhem W, Hernandez F, Maertens G. Second-generation line probe assay for hepatitis C virus genotyping. J Clin Microbiol. 1996;34(9):2259–2266. doi: 10.1128/jcm.34.9.2259-2266.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm J, Neukamm M, Kuntzen T, Kim AY, Chung RT, Brander C, Lauer GM, Walker BD, Allen TM. Characterization of full-length hepatitis C virus genotype 4 sequences. J Viral Hepat. 2007;14(5):330–337. doi: 10.1111/j.1365-2893.2006.00792.x. [DOI] [PubMed] [Google Scholar]
- Veillon P, Payan C, Gaudy C, Goudeau A, Lunel F. Mutation analysis of ISDR and V3 domains of hepatitis C virus NS5A region before interferon therapy with or without ribavirin. Pathol Biol (Paris) 2004;52(9):505–510. doi: 10.1016/j.patbio.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Viazov S, Kuzin S, Paladi N, Tchernovetsky M, Isaeva E, Mazhul L, Vasychova F, Widell A, Roggendorf M. Hepatitis C virus genotypes in different regions of the former Soviet Union (Russia, Belarus, Moldova, and Uzbekistan) J Med Virol. 1997;53(1):36–40. [PubMed] [Google Scholar]
- Watanabe H, Enomoto N, Nagayama K, Izumi N, Marumo F, Sato C, Watanabe M. Number and position of mutations in the interferon (IFN) sensitivity-determining region of the gene for nonstructural protein 5A correlate with IFN efficacy in hepatitis C virus genotype 1b infection. J Infect Dis. 2001;183(8):1195–1203. doi: 10.1086/319674. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.