Summary
Monoubiquitination of proliferating cell nuclear antigen (PCNA) is a critical post-translational modification essential for DNA repair by translesion DNA synthesis (TLS). The Rad18 E3 ubiquitin ligase cooperates with the E2 Rad6 to monoubiquitinate PCNA in response to DNA damage. How PCNA is monoubiquitinated in unperturbed cells and whether this plays a role in the repair of DNA associated with replication is not known. We show that the CRL4Cdt2 E3 ubiquitin ligase complex promotes PCNA monoubiqutination in proliferating cells in the absence of external DNA damage independent of Rad18. PCNA monoubiquitination via CRL4Cdt2 is constitutively antagonized by the action of the ubiquitin-specific protease 1 (USP1). In vitro, CRL4Cdt2 monoubiquitinates PCNA at Lys164, the same residue that is monoubiquitinated by Rad18. Significantly, CRL4Cdt2 is required for TLS in non-damaged cells via a mechanism that is dependent on PCNA monoubiquitination. We propose that CRL4Cdt2 regulates PCNA-dependent TLS associated with stresses accompanying DNA replication.
Introduction
Protein ubiquitination is an essential post-translational modification involved in a variety of physiological processes including cell cycle control, DNA replication and repair. Whereas the covalent attachment of polyubiquitin chains to proteins frequently results in the targeted proteolysis of the ubiquitinated protein, the attachment of a single ubiquitin moiety often results in the modification of its activity by regulating its interactions with other proteins, cellular localization or enzymatic activity. The monoubiquitination of PCNA at a conserved lysine residue (Lys164 in human PCNA) in response to DNA damage increases its affinity for members of the Y-family of DNA bypass polymerases and allows DNA synthesis across DNA lesions (Andersen et al., 2008). DNA damage-dependent PCNA monoubiquitination requires the activities of the Rad6 E2 ubiquitin-conjugating enzyme and the Rad18 E3 ubiquitin ligase (Hoege et al., 2002; Kannouche et al., 2004) and cells deficient of either of these enzymes are extremely sensitive to a variety of DNA damaging agents (Andersen et al., 2008). Recently, RNF8 has been suggested to function as an E3 ligase for PCNA monoubiquitination in cells irradiated with UV (Zhang et al., 2008). Huang et al., however, showed that UV-induced PCNA monoubiquitination is not affected in HeLa cells depleted of RNF8 or in MEFs deficient of RNF8 (Huang et al., 2009). The basis for these conflicting results are currently unclear, but the available evidence suggest that at least under some conditions, other ubiquitin ligases are capable of promoting PCNA monoubiquitination. Indeed, several reports demonstrated the presence of residual monoubiqutinated PCNA in Rad18-deficient cells (Brun et al., 2008; Huang et al., 2006; Simpson et al., 2006).
The CRL4Cdt2 E3 ubiquitin ligase complex is a member of the cullin-RING family that promotes the polyubiquitination and degradation of the replication licensing factor Cdt1 (Arias and Walter, 2006; Higa et al., 2006; Jin et al., 2006; Senga et al., 2006). CRL4Cdt2 consists of Cul4A or Cul4B, DDB1 (damage-specific DNA binding protein 1), the RING-finger protein ROC1 and the DCAF (DDB1 and Cul4-associated factor) and substrate recognition factor/WD40 protein Cdt2. Additional substrates for CRL4Cdt2 have been recently described including the CDK inhibitor p21 (Abbas et al., 2008; Kim et al., 2008; Nishitani et al., 2008), the C.elegans polymerase eta (Kim and Michael, 2008), and the D. melanogaster E2f1 transcription factor (Shibutani et al., 2008). Notably, most identified CRL4Cdt2 substrates require their interaction with PCNA for their polyubiquitination.
Because of the physical association of CRL4Cdt2 with PCNA in the context of substrate ubiquitination we examined whether CRL4Cdt2 monoubiquitinates PCNA. We report here that in normally cycling mammalian cells, CRL4Cdt2 monoubiquitinates PCNA and that this activity is constitutively antagonized by the deubiquitinating enzyme USP1 (Huang et al., 2006). In response to DNA damage however, Rad6/Rad18 is most critical for PCNA monoubiquitination with Cdt2 depletion affecting this monoubiquitination in some cell lines but not all. We finally show that PCNA monoubiquitination via CRL4Cdt2 synergizes with Rad6/18 in promoting TLS in undamaged cells.
Results
Cdt2 associates with CRL4 to promote PCNA monoubiquitination in normally cycling cells
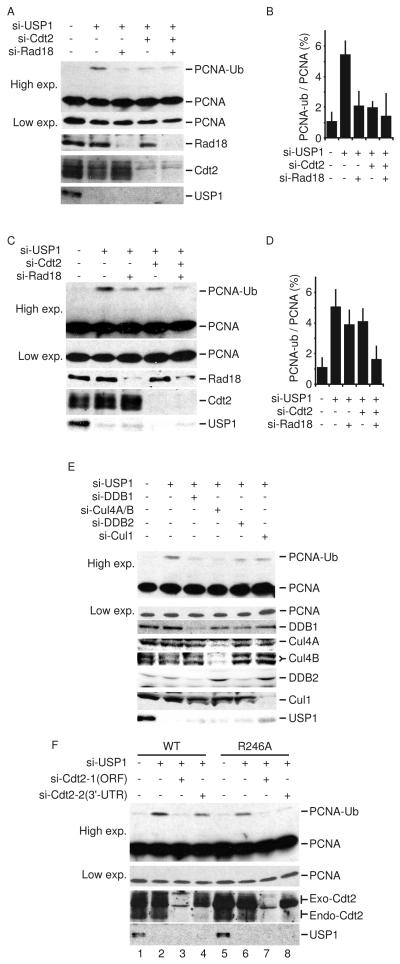
We and others have recently shown that Cdt1 and p21 ubiquitination via the CRL4Cdt2 requires association with PCNA (Abbas et al., 2008; Arias and Walter, 2006; Kim et al., 2008; Nishitani et al., 2008; Senga et al., 2006), raising the possibility that CRL4Cdt2 may regulate PCNA ubiquitination.. It has been reported that USP1 depletion from Rad18-deficient cells induces PCNA monoubiqutinatiton, indicating that other E3 ubiquitin ligase(s) carry out this function independent of Rad18 (Simpson et al., 2006). Thus, we tested whether CRL4Cdt2 inactivation may affect basal PCNA monoubiqutination under USP1-depleted condition. As expected, transfection of HeLa cells with si-USP1 enhanced PCNA monoubiquitination, which was reduced by the codepletion of Rad18 (Figure 1A, B). Surprisingly, depletion of Cdt2 also reduced PCNA monoubiquitination to levels observed with Rad18 depletion without affecting the steady state level of Rad18 (Figure 1A, B). However, we did not detect an additive effect when both Cdt2 and Rad18 are depleted in this cell line, most likely because each of the depletions repressed PCNA monoubiquitination to near base-line levels (see below). Similar results were observed in U2OS cells although the best decrease in PCNA monoubiquitination in this cell line was seen when Rad18 and Cdt2 were codepleted (Figure 1C, D). Thus in these cells Rad18 and Cdt2 appear to have equal and independent roles in basal monoubiquitination of PCNA.
Figure 1. CRL4Cdt2 depletion reduces the steady-state level of PCNA monoubiquitination.
(A-D) HeLa cells (A, B) or U2OS cells (C, D) were transfected with the indicated siRNA and analyzed by immunoblotting with anti-PCNA, anti-Rad18, anti-Cdt2, or anti-USP1 antibody. A low exposure of the non-ubiquitinated PCNA is also shown. The ratio of monoubiquitinated PCNA to total PCNA is blotted in B and D respectively. Mean ± S.D. of 3 experiments. The quantitative analysis of PCNA monoubiquitination is shown in Supplementary Figure 1. (E) HeLa cells transfected with the indicated si-RNA and the level of PCNA monoubiquitination analyzed as in A and C. (F) U2OS cells stably expressing flag-tagged WT Cdt2 or Cdt2-R246A were transfected with si-Cdt2-1 (targeting the ORF) or with si-Cdt2-2 (targeting the 3′-UTR). Where indicated, cells were also transfected with si-USP1. Protein lysates were separated by SDS-PAGE and analyzed by immunoblotting with anti-PCNA, anti-Cdt2, or anti-USP1 antibody.
Cdt2 is the substrate recognition subunit of the CRL4Cdt2 complex that includes Cul4A or B and DDB1. Depletion of DDB1, or Cul4A and Cul4B also inhibited PCNA monoubiquitination in the absence of USP1 (Figure 1E), demonstrating that the effect of Cdt2 on PCNA monoubiquitination is mediated via its assembly in CRL4 ubiquitin ligase complexes. The requirement of CRL4Cdt2 in promoting PCNA monoubiquitination was specific since there was significant PCNA monoubiquitination in cells depleted of Cul1, a Cul4 related E3 ligase, or DDB2, another DCAF that assembles with the CRL4 ligase (Figure 1E). Furthermore, we tested whether exogenous siRNA-resistant Cdt2 can restore PCNA monoubiquitination in the absence of endogenous Cdt2. We developed U2OS cell which express either wild type Cdt2 or Cdt2 protein deficient in binding DDB1 (Cdt2-R246A) (Jin et al., 2006). Both versions of the protein are expressed from a cDNA without the 3′-UTR. si-Cdt2-1, targeting the ORF of Cdt2 and so targeting both endogenous and exogenous Cdt2, reduced PCNA monoubiquitination in the absence of USP1 (Figure 1F). si-Cdt2-2 was designed to target the 3′-UTR of Cdt2 and so exclusively targets the endogenous gene while sparing exogenous Cdt2. Indeed, si-Cdt2-2 did not decrease PCNA monoubiquitination in cells overexpressing wild type Cdt2 (Figure 1F, lane 4). On the other hand, Cdt2-R246A could not restore PCNA monoubiquitination in cells transfected with si-Cdt2-2 (Figure 1F, lane 8). Collectively, these results demonstrate that CRL4Cdt2 specifically promotes basal PCNA monoubuiquitylation and that this activity is counteracted by USP1.
CRL4Cdt2 contributes to DNA damage-induced PCNA monoubiquitination
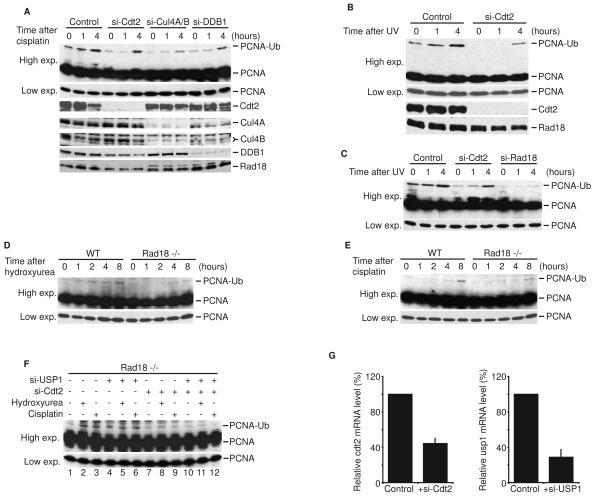
ES2 ovarian cancer cells, 293T kidney cancer cells, and MCF7 breast cancer cells showed higher levels of basal monoubiquitinated PCNA that could be detected without USP1 depletion (0 hr lanes in Figure 2A-C). Interestingly, in these cell lines, Cdt2 depletion reduced both basal and damage-dependent PCNA monoubiquitination. Furthermore, depletion of Cul4 and DDB1 in ES2 cells also inhibited cisplatin-induced PCNA monoubiquitination (Figure 2A) without affecting Rad18 expression levels. The depletion of Cul4A or DDB1 did not alter cell cycle profile, suggesting that the reduction of PCNA monoubiquitination is independent from cell cycle effects (Supplementary Figure 2). Although the kinetics of PCNA monoubiquitination after DNA damage was delayed, all these cells eventually monubiquitinated PCNA in the absence of Cdt2, consistent with the well-accepted role of Rad18 in this process. Thus, in several human cell-lines with a high level of basal PCNA monoubiquitination, CRL4Cdt2 seems to be important for both basal monoubiquitination and the kinetics of the DNA damage-induced monoubiquitination of PCNA.
Figure 2. CRL4Cdt2 contributes to DNA damage-induced PCNA monoubiquitination.
(A) ES2 cells were transfected with the indicated siRNA. After 48 hours, cells were treated with 500 uM cisplatin for the indicated time. Cell lysates were analyzed by immunoblotting with anti-PCNA, anti-Cdt2, anti-Cul4, anti-DDB1, and anti-Rad18 antibodies. (B and C) 293T cells (B) and MCF7 cells (C) were transfected with si-Cdt2. After 48 hours, cells were irradiated with UV (50 J/m2) and harvested at the indicated time post-radiation. Cell lysates were analyzed as above. (D and E) DT40 cells or DT40 deficient of Rad18 (Rad18−/−) were treated with 50 uM cisplatin or 1 mM hydroxyurea for the indicated time and analyzed as above. (F) DT40Rad18−/− cells were transfected with the indicated siRNA. Three days after transfection, cells were either left untreated or treated with 50 uM cisplatin or 1 mM hydroxyurea for 8 hours before harvesting. Lysates were immunoblotted with anti-PCNA antibody. (G) Total RNA was prepared from DT40Rad18−/− cells with or without siRNA treatment. Using β-actin as a control, cdt2 and usp1 mRNA were analyzed by quantitative real-time PCR and expressed relative to the control si-GL2 sample. Mean ± S.D. of 3 experiments.
CRL4Cdt2 promotes PCNA monoubiquitination independent of Rad18
It is conceivable that CRL4Cdt2 may affect PCNA indirectly via the modulation of Rad18 activity. To rule out this possibility, we tested the effect of CRL4Cdt2 on PCNA monoubiquitination in DT40 chicken cells which do not express Rad18 (DT40Rad18−/−). Wild type and Rad18-deficient DT40 cells were treated with the DNA damaging agents, cisplatin or hydroxyurea. Consistent with previous reports, DT40Rad18−/− cells exhibited PCNA monoubiquitination that was significantly less than that observed in Rad18-proficient cells (Figure 2D and 2E). Importantly, Cdt2 depletion in DT40Rad18−/− cells decreased DNA-damage-induced PCNA monoubiquitination (Figure 2F, lanes 8, 9 compared with lanes 2, 3) as well as basal monoubiquitination seen after depleting cells of USP1 (lane 10 compared with lane 4). Thus, CRL4Cdt2 promotes PCNA monoubiquitination independent of Rad18 and can substitute for its activity in response to DNA-damage in Rad18-deficient cells.
CRL4Cdt2 promotes PCNA monoubiquitination in vitro
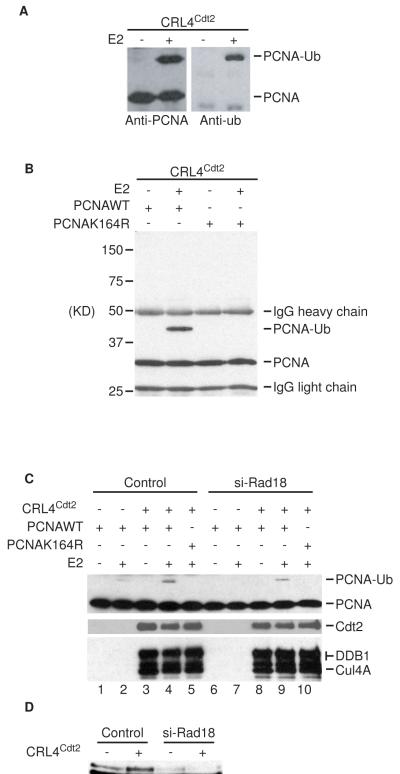
To address whether the CRL4Cdt2 can directly monoubiqutinate PCNA, we tested whether PCNA can serve as a direct substrate for CRL4Cdt2 in vitro. In the presence of E1, E2, and ATP, PCNA was efficiently monoubiquitinated by the CRL4Cdt2 complex immunopurified from 293T cells (Figure 3A, B). Furthermore, CRL4Cdt2 monoubiquitinated PCNA specifically at the conserved lys164, as a mutant PCNA in which this residue was mutated to arginine (PCNA-K164R) was not monoubiquitinated (Figure 3B, lane 4). Thus, CRL4Cdt2 monoubiquitinates PCNA at the same site that is monoubiquitinated via Rad18 (Hoege et al., 2002). Importantly, depletion of 293T cells of Rad18 did not prevent CRL4Cdt2-dependent PCNA monoubiquitination (Figure 3C, lane 9), ruling out that the in vitro enzymatic activity of CRL4Cdt2 is indirectly mediated by Rad18.
Figure 3. CRL4Cdt2 complex promotes PCNA monoubiquitination at Lys164 in vitro.
(A and B) Immunopurified CRL4Cdt2 complexes were mixed with recombinant WT or K164R mutant PCNA, E1 and UbcH5. Ubiqutiniated PCNA were then detected by Western blotting after separating the reaction products by SDS-PAGE (C) Same as in A except that the CRL4Cdt2 immunocomplexes were either purified from 293T cells or from 293T cells that were pre-depleted of Rad18 by siRNA. Immunoblots for Cdt2 (anti-Cdt2), Cul4A and DDB1 (anti-myc) are also shown. (D) Western blot of Rad18 to show effectiveness of si-Rad18 in (C).
CRL4Cdt2 promotes PCNA-dependent translesion DNA synthesis
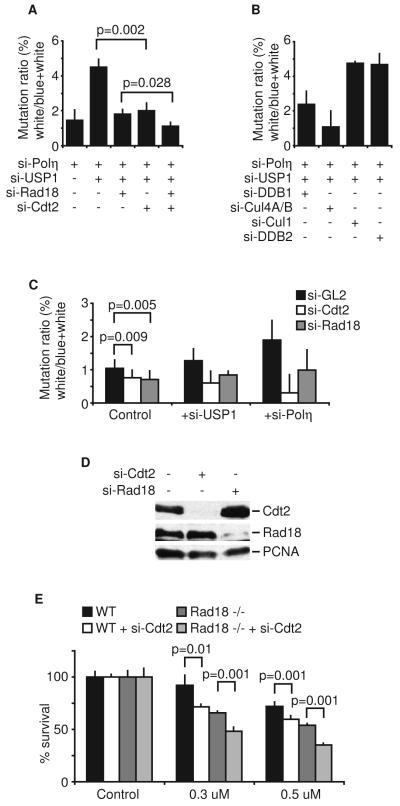
PCNA monoubiquitination plays a significant role in TLS by recruiting the error-prone translesion polymerases to sites of DNA damage (Andersen et al., 2008). Because CRL4Cdt2 promotes PCNA monoubiquitination under basal conditions, we tested whether such an activity impacts TLS in the absence of extrinsic DNA damage. We measured TLS activity by measuring the mutation frequency in a supF gene (in a shuttle vector) (Parris and Seidman, 1992). Error-prone TLS activity in 293T cells will be dependent on basal level of PCNA monoubiquitination and will mutate the damaged supF gene. The mutation rate in replicated plasmids is scored by a blue-white colony screen after recovering the supF shuttle vector from the mammalian cells and transfection into bacteria. To increase the sensitivity of the assay, we performed these experiments first after depleting 293T cells of the high fidelity TLS enzyme, DNA polymerase eta. We measured mutation frequency in cells depleted of USP1 and polymerase eta using non-UV treated supF plasmid. However, no white colony was observed among more than 3000 colonies. Thus, we used plasmids subjected to UV-induced DNA damage before its introduction into undamaged cells. Consistent with previous reports, cells depleted of USP1 showed a five-fold increase in mutation frequency compared to cells with USP1 (Figure 4A). Sequencing of the mutant supF genes revealed a mutation spectrum consistent with error-prone TLS (Supplemental Table 1). Interestingly, Cdt2 depletion significantly reduced the mutation frequency, indicating that the reduction in basal PCNA monoubiquitination seen after Cdt2 depletion in 293T cells (Figure 2B, the 0 hr time points) inhibits TLS activity in vivo. Similar results were obtained when 293T cells were depleted of DDB1 or Cul4A/B, but not upon depletion of Cul1 or DDB2 (Figure 4B). These results demonstrate that the inhibition of TLS activity was a consequence of the specific inactivation of CRL4Cdt2 complex. As shown in Figure 1C-D, codepletion of Cdt2 and Rad18 decreased basal PCNA monoubiquitination in a synergistic manner in some cells. Indeed, in 293T cells codepletion of Cdt2 and Rad18 reduced the mutagenic frequency further than the individual depletions to almost background levels (Figure 4A). Thus, CRL4Cdt2 and Rad18 independently promote TLS in normal cycling 293T cells.
Figure 4. CRL4Cdt2 E3 ubiquitin ligase complex promotes translesion DNA synthesis in unperturbed proliferating cells.
(A, B, and C) 293T cells were transfected with the indicated siRNA for twenty-four hours. UV-irradiated reporter plasmid was transfected into 293T cells. Replicated plasmids were recovered, Dpn1-digested and used to transform bacteria. Mutation frequency (see materials and methods) is expressed as the ratio of white to total (blue and white) colonies. Mean ± S.D. of 3 experiments (A and B) or 10 experiments (C). (D) Western blot of Cdt2 and Rad18 to show effectiveness of si-Cdt2 and si-Rad18 in 293T cells. (E) DT40 cells were transfected with si-Cdt2 for twenty-four hours and treated with cisplatin for seventy-two hours. Viable cells were counted and expressed relative to the control si-GL2 sample. Mean ± S.D. of 3 experiments.
We next performed the experiment without USP1 depletion and/or polymerase eta depletion (Figure 4C). Under control conditions (when USP1 or polymerase eta were intact) Cdt2 depletion significantly decreased the mutation frequency much like si-Rad18, demonstrating that effect of CRL4Cdt2 on TLS reported above was not an artifact of USP1 or polymerase eta depletion. Similar results were obtained when USP1 or polymerase eta were individually depleted (Figure 4C). Together, these findings demonstrate that CRL4Cdt2 cooperates with Rad18 to promote PCNA-dependent TLS activity in unperturbed proliferating cells.
It has been shown that TLS activity and PCNA monoubiquitination are required for cell survival after DNA damage (Hoege et al., 2002; Kannouche et al., 2004; Simpson et al., 2006). We therefore, tested the sensitivity of the Cdt2-depleted DT40 cells to cisplatin treatment. Cdt2-depleted cell were significantly more sensitive than the control DT40 cells (Figure 4E). The sensitization was also seen in DT40Rad18−/−, as expected from the Rad18-independent role of Cdt2 in monoubiquitinating PCNA. Collectively, these results demonstrate that CRL4Cdt2 monoubiquitinates PCNA to promote translesion DNA synthesis and is required for cell survival after DNA damage.
Discussion
CRL4Cdt2 and Rad18 promote PCNA monoubiquitination synergistically in the absence of extrinsic DNA damage
It is well established that Rad18 is essential for PCNA monoubiquitination in response to DNA damage (Shiomi et al., 2007; Watanabe et al., 2004; Yamashita et al., 2002). Our results support this conclusion. Several reports however, demonstrated that in the absence of Rad18, PCNA continues to be monoubiquitinated (albeit to a lower extent) in a variety of cell lines (Brun et al., 2008; Huang et al., 2006; Simpson et al., 2006). We demonstrated here that CRL4Cdt2 monoubiquitinates PCNA independent of Rad18 in non-DNA damaged cells. In vitro, CRL4Cdt2 was sufficient to promote PCNA monoubiquitination specifically at lys164, the same residue that is ubiquitinated via Rad18. Although CRL4Cdt2 inactivation in mammalian cells did not prevent the induction of PCNA monoubiquitination after DNA damage, it prevented the monoubiquitination in normally cycling cells and delayed the kinetics of damage-induced monoubiquitination in several cells. Intriguingly, the depletion of Rad18 also decreased basal monoubiquitination of PCNA in some cell lines and this is further decreased by CRL4Cdt2 inactivation in other cells. Thus, Rad18 and CRL4Cdt2 seem to function independently to promote basal PCNA monoubiquitination in normal cycling cells. It is important to note that the term “basal” used in this study refers to cells that are not subjected to extrinsic DNA damage, but of course, the basal monoubiquitination could result from intrinsic DNA damage caused by and/or sensed by replication forks.
CRL4Cdt2 monoubiquitinates PCNA in DNA damaged cells independent of Rad18
In several human cell lines, the requirement of CRL4Cdt2 for PCNA monoubiquitination was observed also after treating the cells with UV or cisplatin as it delayed, but did not prevent, the induction of PCNA monoubiquitination (Figure 2). Furthermore, Cdt2 is required not only for monoubiquitinating PCNA in undamaged cells, but also in response to DNA damage in the Rad18-deficient cells DT40. Thus it seems likely that although Rad18 is essential for PCNA monoubiquitination in all human cell lines tested, CRL4Cdt2 plays a supportive role to ensure optimal PCNA monoubiquitination after DNA damage.
Whereas CRL4Cdt2 appears to be required for monoubiquitinating PCNA in 293T, ES2 and MCF7 cells following DNA damage, it is dispensable in HeLa cells following UV irradiation (supplementary Fig. 3). Additionally, although basal PCNA monoubiquitination in most of the cells we examined are dependent on Cdt2, there are rare exceptions like HCT116 colon cancer cells, where Cdt2 does not appear to be required for PCNA monoubiquitination in undamaged cells (data not shown). We hypothesize that differences in levels (or activation) of PCNA, Rad6/Rad18 or CRL4Cdt2 in different cell lines account for the variable requirement of CRL4Cdt2 in basal or damage-induced monoubiquitination of PCNA in different cell lines.
PCNA monoubiquitination via the CRL4Cdt2 E3 ubiquitin ligase complex; a surveillance mechanism to monitor and respond to DNA lesions associated with replication stress
Because CRL4Cdt2-dependent monoubiquitination of PCNA is constitutively counteracted by USP1, our data support the hypothesis that in normally cycling cells, PCNA monoubiquitination is regulated by the balanced activity of CRL4Cdt2 and USP1. We propose that stress associated with DNA replication may interfere with the activity of USP1 thus allowing CRL4Cdt2 to promote PCNA monoubiquitination necessary for bypassing DNA lesions. This hypothesis is supported by the observation that CRL4Cdt2 inactivation specifically impairs TLS activity in undamaged cells (Figure 4). Recently, it has been shown that polymerase eta is required for surveillance of the genome and S phase progression in normally cycling cells (Rey et al., 2009), demonstrating the functional significance of regulating basal PCNA monoubiquitination in normally proliferating cells in the absence of external DNA damage and highlighting the significant role of CRL4Cdt2 in promoting such an activity. Since TLS is required for cell survival but increases mutation frequency, it is tempting to speculate that the over-expression of Cul4A or Cdt2 observed in some human cancer (Pan et al., 2006; Schindl et al., 2007) may promote malignancy by enhancing PCNA-dependent and TLS-mediated mutagenesis.
CRL4Cdt2 induces mono- and polyubiquitination
Although CRL4Cdt2 complex promotes the polyubiquitination and degradation of several substrates, polyubiquitination of PCNA by CRL4Cdt2 was not detected in vitro. Monoubiquitination versus polyubiquitination by the same E3 ligase has been observed in other contexts. For example, the CRL4DDB2 E3 ubiquitin ligase not only monoubiquitinates histone H3 and H4 (Wang et al., 2006) and promotes the monoubiquitination of Histone H2A at UV-induced DNA damage sites (Kapetanaki, 2006; Guerrero-Santoro, 2008), it also promotes the polyubiquitination of XPC protein (Sugasawa et al., 2005). Since monoubiquitinated PCNA has a high affinity interaction with Y-family polymerases through an ubiqutin binding domain and a PIP box, it is possible that the bound polymerase sterically hinders the continued interaction of PCNA with CRL4Cdt2. In such a model, it is also possible that PCNA monoubiqutination could inhibit CRL4Cdt2-dependent polyubiquitination of other substrates by displacing the CRL4Cdt2 from the substrate-bound-PCNA ring.
Experimental Procedures
Cell lines and reagents
HeLa, 293T, and U2OS cell lines were obtained from ATCC. Wild type DT40 and DT40Rad18−/− cells were kindly provided by Shunichi Takeda (Kyoto University). The antibodies were purchased as indicated: against PCNA (PC10), Cul1 (D-5), Cul4 (C-19) (Santa Cruz Biotechnologies), against Rad18, USP1, DDB2 (Rockland Immunochemicals) and against DDB1 (Invitrogen).
Gene silencing by RNAi
siRNA tranfections into human cell lines were performed using oligofectamine (Invitrogen). For introducing siRNA into DT40 cells, Amaxa Nucleofector Kit T was used. The following siRNA oligonucleotides were purchased from Invitrogen: polymerase eta, 5′-GUGGAGCAGCGGCAAAAUCdTdT-3′ human USP1, 5′-AGCUACAAGUGAUACAUUAdTdT-3′; chicken USP1, 5′-AGUGAAAGUUACAGAGGAAdTdT-3′; chicken Cdt2, 5′-CCAAAGAGCUUGUGAUGAAdTdT-3′. Other siRNA oligonucleotides were described before (Abbas et al., 2008). mRNA of beta-actin, Cdt2, and USP1 in DT40 cells were measured by using the following primers: USP1, 5′-GCTTTGTGCATCTCCAACACAGG-3′ and 5′-CACACCTATTTACACACTTTGC-3′; Cdt2, 5′-CCTTACAGATTCAGCCTGAAGC-3′ and 5′-GCTAGTGATCTTGTTCTACC-3′; beta-actin. 5′-CATTGCTGACAGGATGCAGAAGG-3′ and 5′-TGCTTGCTGATCCACATCTGCTGG-3′.
In vitro ubiquitination assays
The purification of CRL4Cdt2 complex was previously described (Abbas et al., 2008). Briefly, Cul4A-myc, DDB1-myc, flag-Roc1 and flag-Cdt2 were transiently overexpressed in 293T cells and purified by immunoprecipitation with anti-myc antibody. Purified complexes were mixed with recombinant wild type or a K164A mutant of PCNA in ubiquitination reaction buffer. The ubiquitination reactions were incubated at 30°C for one hour and terminated by adding Laemmli sample buffer. Proteins were resolved by SDS-PAGE and immunoblotted with anti-PCNA or anti-ubiquitin antibody (Sigma).
Mutation frequency assays
293T cells were transfected with siRNAs as described above. Twenty-four hours later, pSP189 plasmids (Parris and Seidman, 1992) were irradiated with UV (1000 J/m2) and transfected into the cells using Lipofectamine 2000 (Invitrogen). Forty-eight hours after, cells were harvested for plasmid purification using DNA miniprep kit (Qiagen). Purified plasmids were Dpn1-digested and introduced by electroporation into the MBM7070 bacterial strain. The mutation frequency in the SupF coding region was analyzed by counting the ratio between blue (wild type) and white (mutant) colonies. Mutations in the SupF coding region were confirmed by sequencing.
Supplementary Material
Acknowledgments
We thank members of the Dutta lab for helpful discussions. This work was supported by grants from the NIH (R01 CA60499) to A.D. K.T. was supported by the Uehara Memorial Foundation Fellowship and the U.S. Department of Defense Breast Cancer Research Program (W81XWH-08-1-0355). T.A. was supported by the Cancer Training Grant T32CA009109. AAJ was supported by the Women’s Oncology Program of University of Virginia’s Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22:2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen PL, Xu F, Xiao W. Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res. 2008;18:162–173. doi: 10.1038/cr.2007.114. [DOI] [PubMed] [Google Scholar]
- Arias EE, Walter JC. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat Cell Biol. 2006;8:84–90. doi: 10.1038/ncb1346. [DOI] [PubMed] [Google Scholar]
- Brun J, Chiu R, Lockhart K, Xiao W, Wouters BG, Gray DA. hMMS2 serves a redundant role in human PCNA polyubiquitination. BMC Mol Biol. 2008;9:24. doi: 10.1186/1471-2199-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa LA, Banks D, Wu M, Kobayashi R, Sun H, Zhang H. L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle. 2006;5:1675–1680. doi: 10.4161/cc.5.15.3149. [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Huang J, Huen MS, Kim H, Leung CC, Glover JN, Yu X, Chen J. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol. 2009;11:592–603. doi: 10.1038/ncb1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D’Andrea AD. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell. 2006;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- Kim SH, Michael WM. Regulated proteolysis of DNA polymerase eta during the DNA-damage response in C. elegans. Mol Cell. 2008;32:757–766. doi: 10.1016/j.molcel.2008.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Starostina NG, Kipreos ET. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 2008;22:2507–2519. doi: 10.1101/gad.1703708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H, Shiomi Y, Iida H, Michishita M, Takami T, Tsurimoto T. CDK inhibitor p21 is degraded by a proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. J Biol Chem. 2008;283:29045–29052. doi: 10.1074/jbc.M806045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan HW, Chou HY, Liu SH, Peng SY, Liu CL, Hsu HC. Role of L2DTL, cell cycle-regulated nuclear and centrosome protein, in aggressive hepatocellular carcinoma. Cell Cycle. 2006;5:2676–2687. doi: 10.4161/cc.5.22.3500. [DOI] [PubMed] [Google Scholar]
- Parris CN, Seidman MM. A signature element distinguishes sibling and independent mutations in a shuttle vector plasmid. Gene. 1992;117:1–5. doi: 10.1016/0378-1119(92)90482-5. [DOI] [PubMed] [Google Scholar]
- Rey L, Sidorova JM, Puget N, Boudsocq F, Biard DS, Monnat RJ, Jr., Cazaux C, Hoffmann JS. Human DNA polymerase eta is required for common fragile site stability during unperturbed DNA replication. Mol Cell Biol. 2009;29:3344–3354. doi: 10.1128/MCB.00115-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindl M, Gnant M, Schoppmann SF, Horvat R, Birner P. Overexpression of the human homologue for Caenorhabditis elegans cul-4 gene is associated with poor outcome in node-negative breast cancer. Anticancer Res. 2007;27:949–952. [PubMed] [Google Scholar]
- Senga T, Sivaprasad U, Zhu W, Park JH, Arias EE, Walter JC, Dutta A. PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J Biol Chem. 2006;281:6246–6252. doi: 10.1074/jbc.M512705200. [DOI] [PubMed] [Google Scholar]
- Shibutani ST, de la Cruz AF, Tran V, Turbyfill WJ, 3rd, Reis T, Edgar BA, Duronio RJ. Intrinsic negative cell cycle regulation provided by PIP box- and Cul4Cdt2-mediated destruction of E2f1 during S phase. Dev Cell. 2008;15:890–900. doi: 10.1016/j.devcel.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi N, Mori M, Tsuji H, Imai T, Inoue H, Tateishi S, Yamaizumi M, Shiomi T. Human RAD18 is involved in S phase-specific single-strand break repair without PCNA monoubiquitination. Nucleic Acids Res. 2007;35:e9. doi: 10.1093/nar/gkl979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson LJ, Ross AL, Szuts D, Alviani CA, Oestergaard VH, Patel KJ, Sale JE. RAD18-independent ubiquitination of proliferating-cell nuclear antigen in the avian cell line DT40. EMBO Rep. 2006;7:927–932. doi: 10.1038/sj.embor.7400777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, Mori T, Iwai S, Tanaka K, Hanaoka F. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument-Bromage H, Tempst P, Xiong Y, Zhang Y. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell. 2006;22:383–394. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. Embo J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita YM, Okada T, Matsusaka T, Sonoda E, Zhao GY, Araki K, Tateishi S, Yamaizumi M, Takeda S. RAD18 and RAD54 cooperatively contribute to maintenance of genomic stability in vertebrate cells. Embo J. 2002;21:5558–5566. doi: 10.1093/emboj/cdf534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Chea J, Meng X, Zhou Y, Lee EY, Lee MY. PCNA is ubiquitinated by RNF8. Cell Cycle. 2008;7:3399–3404. doi: 10.4161/cc.7.21.6949. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.