Abstract
Saliva is a readily accessible and informative biofluid, making it ideal for the early detection of a wide range of diseases including cardiovascular, renal, and autoimmune diseases, viral and bacterial infections and, importantly, cancers. Saliva-based diagnostics, particularly those based on metabolomics technology, are emerging and offer a promising clinical strategy, characterizing the association between salivary analytes and a particular disease. Here, we conducted a comprehensive metabolite analysis of saliva samples obtained from 215 individuals (69 oral, 18 pancreatic and 30 breast cancer patients, 11 periodontal disease patients and 87 healthy controls) using capillary electrophoresis time-of-flight mass spectrometry (CE-TOF-MS). We identified 57 principal metabolites that can be used to accurately predict the probability of being affected by each individual disease. Although small but significant correlations were found between the known patient characteristics and the quantified metabolites, the profiles manifested relatively higher concentrations of most of the metabolites detected in all three cancers in comparison with those in people with periodontal disease and control subjects. This suggests that cancer-specific signatures are embedded in saliva metabolites. Multiple logistic regression models yielded high area under the receiver-operating characteristic curves (AUCs) to discriminate healthy controls from each disease. The AUCs were 0.865 for oral cancer, 0.973 for breast cancer, 0.993 for pancreatic cancer, and 0.969 for periodontal diseases. The accuracy of the models was also high, with cross-validation AUCs of 0.810, 0.881, 0.994, and 0.954, respectively. Quantitative information for these 57 metabolites and their combinations enable us to predict disease susceptibility. These metabolites are promising biomarkers for medical screening.
Electronic supplementary material
The online version of this article (doi:10.1007/s11306-009-0178-y) contains supplementary material, which is available to authorized users.
Keywords: Salivary metabolome, Capillary electrophoresis-mass spectrometry, Oral cancer, Breast cancer, Pancreatic cancer
Introduction
Saliva is an important biological fluid that provides various functions, including lubrication for speech, digestion of food, and protection from microorganisms. It is produced by multiple salivary glands; particularly the three major salivary glands parotid, submandibular and sublingual, and several minor glands. Saliva is comprised of 99% water with minerals, mucus, electrolytes, nucleic acids and proteins such as enzymes, enzyme inhibitors, growth factors, cytokines, immunoglobulins, and other glycoproteins (de Almeida Pdel et al. 2008). Saliva is a filtration of blood, reflecting the physiological conditions of the body; thus it could be used to monitor clinical status and predict systemic diseases. Compared with blood, saliva offers distinct advantages for diagnostic or research purposes; its collection is cost-effective, safe, easy and non-invasive. Indeed, many of the characteristics of bodily fluids, such as blood and urine, are applicable to saliva including diurnal variation and the presence of diverse diagnostic analytes.
Cancer is a leading cause of death and oral cancer annually affects more than 400,000 individuals worldwide. Despite advances in treatment, the overall 5-year survival rate of patients with oral cancer is approximately 50% and has not improved over the past 30 years (Epstein et al. 2002; Mao et al. 2004). The mortality rate associated with oral cancer is particularly high because it is routinely discovered late, commonly after metastasis to the lymph nodes or neck has already occurred. Worldwide, more than 200,000 patients with pancreatic cancer are registered annually, and 98% of the patients die of the disease (Parkin et al. 2005). The high mortality rate from this cancer is thought to be due to a lack of adequate systemic therapies and the high rate of metastasis at the time of diagnosis. Therefore, novel diagnostic tests are urgently needed to detect these cancers at the premalignant stage.
Studies using molecular-based biomarkers in blood or urine to detect the progress of malignant tumors have mainly focused on altered DNA methylation or mutation, or on changes in the RNA or protein levels (Sidransky 2002). In addition, several molecular biomarker candidates have been identified by analyzing the transcriptome or proteome of saliva (Hu et al. 2007, 2008; Zimmermann and Wong 2008). However, sufficiently sensitive and reproducible saliva-based diagnostic methods are not yet available. In addition, conventional tumor markers, such as serum cancer antigen 19-9, which is widely used in the diagnosis of pancreatic cancer, are known to have less specificity for particular lesions. With the exception of breast cancer (Streckfus et al. 2008), few studies have used saliva to detect tumors remote from the oral cavity. Tumor markers that can discriminate individual cancer-specific differences and which are sensitive are required for clinical applications.
Metabolomics, the measurement of all intracellular metabolites, has become a powerful new tool to gain insight into cellular function. So far, several metabolomic approaches have been reported (Aharoni et al. 2002; Fiehn et al. 2000; Plumb et al. 2003; Reo 2002). In this marriage of methodologies, CE offers rapid analysis and efficient resolution, and MS provides excellent selectivity and sensitivity (Soga et al. 2006). A number of clinical applications of CE-MS exploring urinary or serum proteomics biomarkers, were developed to detect and identify the charged peptide content, which demonstrates their potential to assess the profiles of small molecules in biofluids (Fliser et al. 2005; Kolch et al. 2005; Metzger et al. 2009; Schiffer et al. 2006, 2008; Zurbig and Mischak 2008). Although diverse saliva analyses with CE have been proposed (Lloyd 2008), salivary metabolomic analysis to determine cancer-specific profiles for early cancer detection has not yet been conducted. In this study, we, for the first time, obtained and compared comprehensive salivary metabolic profiles of patients with oral, breast or pancreatic cancer, or periodontal disease, and healthy controls. We then identified individual cancer-specific markers with high discriminative ability, demonstrating the potential use of salivary metabolomics in cancer diagnosis.
Materials and methods
Patient selection
This study was approved by the UCLA Institutional Review Board. Patients with oral, breast or pancreatic cancer or periodontal disease and the healthy controls were recruited at the UCLA Medical Center. All patients had recently been diagnosed with primary disease and were without metastasis; none had received any prior treatment in the form of chemotherapy, radiotherapy, surgery or alternative therapy. No subjects had a history of prior malignancy, immunodeficiency, autoimmune disorders, hepatitis or HIV infection. Written, informed consent was obtained from all patients and from volunteers who agreed to serve as saliva donors.
Sample collection and sample preparation
The subjects were asked to refrain from eating, drinking, smoking or using oral hygiene products for at least 1 h prior to saliva collection. The subjects rinsed their mouth with water and, 5 min later, they were instructed to spit into 50-cc Falcon tubes, which were placed in a Styrofoam cup filled with crushed ice. The subjects were reminded not to cough up mucus. Five milliliters of unstimulated saliva could usually be collected in 5–10 min. Saliva collection was performed in a private room. The saliva samples were centrifuged at 2600×g for 15 min at 4°C and spun for another 20 min for incomplete separation. Equal amounts of supernatant were transferred to two fresh tubes and the samples were processed and frozen within 30 min. The protocols used for sample collection are described in more detail elsewhere (Li et al. 2004).
Saliva fluid samples were obtained from patients with oral (n = 69), breast (n = 30) and pancreatic cancer (n = 18), patients with periodontal diseases (n = 11) and healthy controls (n = 87). The race, ethnicity, sex and age of the subjects are summarized in Table 1. Except for age, clinical parameters were not collected for the non-oral cancer groups.
Table 1.
Subject characteristics
| Groups | Control | Oral cancer | Breast cancer | Pancreatic cancer | Periodontal diseases |
|---|---|---|---|---|---|
| Age | |||||
| Min–Max (median) | 20–75 (43) | 34–87 (59.5) | 29–77 (57) | 11–87 (67) | 23–76 (60) |
| Missing | 2 | 5 | 10 | 2 | 2 |
| Sex | |||||
| Male | 42 | 41 | N/A | ||
| Female | 27 | 23 | |||
| Missing | 18 | 5 | 30 | 18 | 11 |
| Race or ethnic group | |||||
| Total | 87 | 69 | 30 | 18 | 11 |
| Caucasian | 37 | 41 | N/A | ||
| Asia | 15 | 5 | |||
| African-American | 12 | 4 | |||
| Hispanic | 5 | 5 | |||
| Missing | 18 | 14 | 30 | 18 | 11 |
N/A not available
Frozen saliva was thawed and dissolved at room temperature, and 27 μl of each sample (69 patients with oral cancer and 70 healthy control samples) were added to a 1.5-ml Eppendorf tube, to which 3 μl of water containing 2 mM methionine sulfone and 2 mM 3-aminopyrrolidine as internal standards was added and mixed well. Similarly, individual thawed saliva samples (24 μl) from patients with breast or pancreatic cancer, and patients with periodontal disease and 17 healthy controls were admixed with 6 μl water containing internal standards (1 mM each of methionine sulfone and 3-aminopyrrolidine). These internal standards were selected because they were not included in the human endogenetic metabolites. Furthermore, they migrated to the center of the metabolite distribution, which was used to confirm the quality of the alignment results. Even though a unified dilution was preferred for the preparation of all samples, a greater dilution ratio was required for the control, breast, pancreatic cancer, and periodontal disease samples because of their high electrolyte content, which decreases the electrical current during the measurement.
Metabolite standards, instrumentation, and CE-TOF-MS conditions
The metabolite standards, instrumentation and CE-TOF-MS condition were used in this study as previously described (Soga et al. 2006), with slight modifications in the lock mass system setting. All chemical standards were of analytical or reagent grade and were obtained from commercial sources. They were dissolved in Milli-Q water (Millipore, Bedford, MA, USA), 0.1 mol/l HCl or 0.1 mol/l NaOH to obtain 1, 10 or 100 mmol/l stock solutions. The working solution was prepared prior to use by diluting with Milli-Q water to the appropriate concentration.
All CE-MS experiments were performed using an Agilent CE capillary electrophoresis system (Agilent Technologies, Waldbronn, Germany), an Agilent G3250AA LC/MSD TOF system (Agilent Technologies, Palo Alto, CA, USA), an Agilent 1100 series binary HPLC pump, and the G1603A Agilent CE-MS adapter and G1607A Agilent CE-ESI-MS sprayer kit. System control and data acquisition were done with G2201AA Agilent Chemstation software for CE and Analyst QS software for TOF-MS (ver. 1.1).
All samples were measured in single mode (see below); separation was done in fused-silica capillaries (50 μm i.d. × 100 cm total length) filled with 1 M formic acid as the background electrolyte. Sample solutions were injected at 50 mbar for 3 s and a voltage of 30 kV was applied. The capillary temperature was maintained at 20°C and the temperature of the sample tray was kept below 5°C using an external thermostatic cooler. The sheath liquid, comprising methanol/water (50% v/v) and 0.5 μM reserpine, was delivered at 10 μl/min. ESI-TOF-MS was conducted in the positive ion mode. The capillary voltage was set at 4 kV; the flow rate of nitrogen gas (heater temperature 300°C) was set at 10 psig. In TOF-MS, the fragmentor, skimmer and OCT RFV voltage were set at 75, 50 and 125 V, respectively. In the present study, we used a methanol dimer adduct ion ([2MeOH + H]+, m/z 65.059706) and hexakis phosphazene ([M + H]+, m/z 622.028963) to provide the lock mass for exact mass measurements. Exact mass data were acquired at the rate of 1.5 cycles/s over a 50–1000 m/z range.
Processing of CE-TOF-MS data
Raw data were analyzed with our proprietary software called MasterHands, which has already been used in several CE-TOF-MS-based profiling studies (Hirayama et al. 2009; Minami et al. 2009; Saito et al. 2009). The data analysis workflow starting with the raw data included noise-filtering, baseline correction, peak detection and integration of the peak area from sliced electropherograms (the width of each electropherogram was 0.02 m/z). Such functions are commonly used by data processing software such as MassHunter from Agilent Technologies, or XCMS (Smith et al. 2006) for liquid chromatography-MS or gas chromatography-MS data. The accurate m/z value for each peak detected within the time domain was calculated with Gaussian curve-fitting to the mass spectrum on the m/z domain peak. The alignment of peaks in multiple measurements was done by dynamic programming (DP)-based techniques (Baran et al. 2006; Soga et al. 2006) with slight modifications. The method picked up a few representative peaks using the Douglas-Peucker algorithm (Wallace et al. 2004) from unit m/z electropherograms, found corresponding peaks across multiple samples by DP, and optimized the numerical parameters of the normalization function for CE-migration (Reijenga et al. 2002). Instead of representative peaks, we used the detected peaks with accurate m/z values and regarded the peaks whose m/z difference was less than 20 ppm as ones that were derived from the same electropherograms.
All peak areas were divided by the area of the internal standard (relative area) to normalize the signal intensities, and to avoid injection-volume bias and mass-spectrometry detector sensitivity bias among multiple measurements. Undetected peaks with a threshold signal-to-noise ratio of 2 were given a peak area of 0. The relative areas of the 17 healthy control samples and of the pancreatic and breast cancer, and the periodontal disease samples were multiplied by 1.25/1.1 to standardize the sample concentration.
The peaks derived from salt and neutral molecules were found in the first and the last few minutes, respectively. Then, isotopic compounds, ringing, spikes and fragment and adduct ions were eliminated and the peak data sets were compared across the sample profiles and aligned according to m/z and migration time. Although all of the metabolites were quantified separately, the sum of the quantified values of leucine and isoleucine were counted as a single marker owing to the low separation of these peaks. Peaks showing P < 0.05 in the non-parametric, multiple comparison Steel–Dwass test, between the controls and at least one disease cohort were selected as candidate markers.
Metabolite identification
The peaks were identified based on the matched m/z values and normalized migration times of the corresponding standard compounds if available. Of the peaks that did not match with any standard compounds, the concomitant peaks, such as isotopic peaks and fragment peaks, were removed based on the difference in m/z values and the normalized migration time of the two peaks with an error tolerance of 20 ppm and 0.01 min to yield only the peaks, or referred to as components, which might be derived from metabolites (Brown et al. 2009). Although CE-TOF-MS provides accurate molecular mass at the milli m/z level, the m/z alone is seldom successful to identify the metabolite (Kind and Fiehn 2006, 2007). Therefore, we used their m/z values and the migration times predicted by the Artificial Neural Networks (ANNs) (Sugimoto et al. 2005) to identify the metabolite. Briefly, the ANN model was first trained using the measured migration times of standard compounds and molecular descriptors with the net charge calculated from the pKa values. The trained ANN model then predicted the migration times of the candidate metabolites. Here, we used compounds available from the Kyoto Encyclopedia of Gene and Genomics (KEGG) database (Goto et al. 2002) and the Human Metabolome Database (HMDB) (Wishart et al. 2007) as candidates. The composition formulae obtained using the measured mass spectrometry and the matched candidates were confirmed by their isotope distribution patterns.
Statistical analysis
To evaluate the ability of the detected peaks to discriminate diseases, we conducted an unsupervised method, principal component analysis (PCA). The same analyses were also conducted to discriminate only between controls and oral samples between males and females, and between race and ethnic groups. The analyses were not performed for the other patient groups due to the unavailability of clinical parameters. Supervised classification techniques, such as partial least squares-discriminant analysis (Jonsson et al. 2005; Michell et al. 2008; Woo et al. 2009), support vector machine (SVM) (Mahadevan et al. 2008) and multiple logistic regression (MLR), are commonly used to separate subjects and to identify important features for the separation. Here, we developed independent MLR models to discriminate healthy individuals and each disease cohort using a stepwise variable selection method (backward procedure to eliminate non-predictive peaks with a threshold of P > 0.10) to construct the predictive models. The models were trained with the complete dataset and we evaluated their versatilities by tenfold cross-validation (CV). The data were randomly separated into training sets and remaining data and this process was repeated ten times for all of the values selected in the training set. The non-parametric Mann–Whitney test was used to compare two groups, e.g. comparison of metabolites in males and females.
Statistical analyses using the Steel–Dwass test were performed using the R package with the Design, Hmisc, and Lexis libraries (available at http://lib.stat.cmu.edu/R/CRAN/). Statistical analyses using the Mann–Whitney test and the heat maps were generated with TM4 software (Saeed et al. 2003). The CV data were generated using WEKA (Witten and Frank 2005). The PCA and MLR models were developed using JMP Version 7 (SAS Institute Inc., Cary, NC, USA, 1989–2007; http://www.jmp.com/software/jmp.shtml).
Results and discussion
Statistical results of discriminative metabolites
On average, CE-TOF-MS detected 3041 peaks (minimum 1585, maximum 8400, standard deviation (SD) 1137) in each saliva sample. After removing the concomitantly observed peaks such as the isotopic and fragment peaks, and noise peaks including spike and ringing peaks, an average of 90 peaks were derived from the metabolites (minimum 48, maximum 128, SD 15). The standard deviation of the relative peak areas of the metabolite-derived peaks was 1.14 (no unit), and the SDs of the migration times before and after the time normalization procedure were 1.75 min and 3.02 × 10−3 min, respectively. Of the remaining peaks, we identified 57 metabolites that were significantly different between the patients and healthy controls (P < 0.05; Steel–Dwass test).
The marker pool used to discriminate between individuals with oral cancer and healthy controls revealed 28 metabolites; namely pyrroline hydroxycarboxylic acid, leucine plus isoleucine, choline, tryptophan, valine, threonine, histidine, pipecolic acid, glutamic acid, carnitine, alanine, piperidine, taurine, and two other metabolites with a significance of P < 0.001 (Steel–Dwass test); piperidine, alpha-aminobutyric acid, phenylalanine and an additional metabolite with a significance of P < 0.01 (Steel–Dwass test); and betaine, serine, tyrosine, glutamine, beta-alanine, cadaverine, and two other metabolite with a significance of P < 0.05 (Steel–Dwass test). The overlaid electropherograms of these CE-TOF-MS peaks with a 2-dimensional map (migration time and m/z) visualizing the difference in intensity between the averaged control and oral cancer samples are shown in Fig. 1. The vertical smear lines in the first few minutes (5–7 min) and those at a later time (at 19 min) were derived from salt ions and neutral molecules, respectively, and most of the peaks derived from charged metabolites were distributed between these times. Using a similar strategy, we identified 28 metabolites for breast cancer, 48 for pancreatic cancer and 27 for periodontal disease (P < 0.05; Steel–Dwass test) as biomarker candidates. The detected markers and the statistical results are listed in Table 2; dot plots of the quantified peak areas are shown in Fig. 2 and Supplementary Fig. S1. Although, several metabolites in the dot plots achieved a statistically significant difference, individual metabolites could not separate any two groups with high sensitivity and specificity. The score plots of the PCA analyses for all individuals are shown in Fig. 3 and in Supplementary Fig. S2. Although the PCA developed using the metabolite profiles of all subjects showed no unequivocal group-specific clusters, PCAs developed individually for the control and each disease group showed partial discriminative separation of the subjects, which might be attributed to the reduced complexity of the given datasets, or the extinction in the overlap between the distribution of the score plots for all disease groups.
Fig. 1.
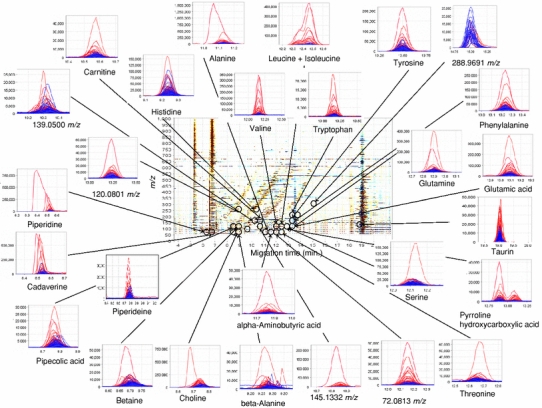
A summary of the different metabolome profiles of cations obtained from CE-TOF-MS analyses of salivary metabolites from control (n = 87) and oral cancer samples (n = 69). The X and Y axes represent the migration time and the m/z value, respectively. The color density reflects the difference in intensity between the averaged control and oral cancer samples. Black circles indicate peaks that are significantly different between healthy control and oral cancer samples (P < 0.05; Steel–Dwass test). The small linked figures include overlaid electropherograms of control (blue) and oral cancer samples (red)
Table 2.
Salivary metabolite marker candidates (P < 0.05 Steel–Dwass test) and the ratio of the relative area of oral, breast and pancreatic cancers, and periodontal diseases to controls (n = 215)
| Metabolite | m/z | Identification (*1) | HMDB (*2) | p- Value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Theoretical | Measured | Error (ppm) | Control vs oral cancer | Control vs breast cancer | Control vs pancreatic cancer | Control vs periodontal desease | Oral cancer vs breast cancer | |||
| C2H6N2 | 59.0604 | 59.0616 | 21 | 0.260 | 4.17 × 10−6*** | 2.67 × 10−4*** | 3.46 × 10−4*** | 1.51 × 10−8*** | ||
| C32H48O13 (*4) | 214.4438 | 214.4440 | 1.0 | 0.834 | 0.00385** | 0.271 | 0.907 | 4.75 × 10−4*** | ||
| C3H7NO2 | 90.0550 | 90.0553 | 3.7 | 0.955 | 0.167 | 1.40 × 10−6*** | 0.0242* | 0.154 | ||
| C4H12N5 | 131.1165 | 131.1174 | 6.8 | 0.686 | 0.00290** | 3.37 × 10−6*** | 0.0251* | 0.323 | ||
| C4H9NO2 | 104.0706 | 104.0706 | −0.35 | 0.620 | 0.913 | 5.78 × 10−6*** | 0.779 | 1.00 | ||
| Cadaverine | 103.1230 | 103.1231 | 1.4 | Standard | HMDB02322 | 0.0422* | 0.00889** | 0.00104** | 0.00100** | 0.993 |
| C5H11NO2 (10.05 min.) | 118.0863 | 118.0864 | 0.86 | 0.910 | 0.408 | 0.192 | 0.0308* | 0.416 | ||
| C30H62N19O2S3 (*3) | 409.2321 | 409.2312 | −2.2 | 0.367 | 2.39 × 10−5*** | 4.64 × 10−4*** | 0.0141* | 0.0354* | ||
| C30H55N27O3S (*3) | 437.7424 | 437.7442 | 4.2 | 0.428 | 0.0305* | 0.0114* | 0.672 | 0.532 | ||
| C18H32N6O6 (*3) | 215.1264 | 215.1269 | 2.2 | 0.891 | 0.386 | 0.0387* | 0.997 | 0.143 | ||
| Alpha-Aminobutyric acid | 104.0706 | 104.0705 | −1.0 | Standard | HMDB00650 | 0.00256** | 0.0269* | 4.37 × 10−6*** | 0.00796** | 1.00 |
| Alanine | 90.0550 | 90.0551 | 1.1 | Standard | HMDB00161 | 2.45 × 10−4*** | 0.0277* | 4.57 × 10−6*** | 0.00647** | 0.956 |
| Putrescine | 89.1073 | 89.1075 | 1.5 | Standard | HMDB01414 | 0.890 | 0.00108** | 1.91 × 10−4*** | 0.0227* | 0.138 |
| Methylimidazoleacetic acid (*5) | 141.0659 | 141.0660 | 0.02 | Predicted | HMDB02820 | 0.183 | 0.390 | 5.32 × 10−4*** | 0.213 | 1.00 |
| Trimethylamine (*5) | 60.0808 | 60.0814 | 11 | Predicted | HMDB00906 | 0.609 | 0.921 | 4.97 × 10−4*** | 1.99 × 10−4*** | 1.00 |
| C5H14N5 | 145.1322 | 145.1332 | 6.9 | 0.0212* | 0.0700 | 2.28 × 10−4*** | 0.0825 | 0.998 | ||
| Piperidine | 86.0964 | 86.0966 | 2.4 | Standard | 0.00119** | 0.934 | 0.0347* | 0.137 | 0.165 | |
| Taurine | 126.0219 | 126.0221 | 1.0 | Standard | HMDB00251 | 3.57 × 10−5*** | 5.94 × 10−6*** | 2.36 × 10−4*** | 6.70 × 10−5*** | 6.82 × 10−10*** |
| Piperideine (*5) | 84.0808 | 84.0807 | −0.41 | Predicted | 2.83 × 10−4*** | 0.0128* | 0.569 | 3.19 × 10−4*** | 2.79 × 10−6*** | |
| Pipecolic acid | 130.0863 | 130.0861 | −0.93 | Standard | HMDB00070 | 1.87 × 10−4*** | 0.264 | 0.943 | 0.00948** | 3.47 × 10−4*** |
| C4H9N | 72.0808 | 72.0813 | 7.8 | 2.02 × 10−7*** | 0.729 | 3.37 × 10−6*** | 0.949 | 5.62 × 10−4*** | ||
| C8H9N | 120.0808 | 120.0801 | −5.9 | 2.64 × 10−5*** | 4.60 × 10−4*** | 1.00 | 0.0385* | 1.02 × 10−8*** | ||
| Pyrroline hydroxycarboxylic acid (*5) | 130.0499 | 130.0498 | −0.49 | Predicted | HMDB01369 | 1.28 × 10−5*** | 0.141 | 0.228 | 0.176 | 1.36 × 10−5*** |
| Betaine | 118.0863 | 118.0864 | 0.86 | Standard | HMDB00043 | 0.0162* | 0.719 | 0.987 | 0.576 | 0.0183* |
| C4H7N | 70.0651 | 70.0655 | 5.8 | 0.101 | 0.637 | 0.997 | 0.502 | 0.0349* | ||
| C6H6N2O2 | 139.0502 | 139.0500 | −1.8 | 0.00270** | 0.265 | 1.65 × 10−10−4*** | 0.127 | 0.948 | ||
| Leucine + Isoleucine | 132.1019 | 132.1019 | −0.30 | Standard | HMDB00687 + HMDB00172 | 1.56 × 10−5*** | 0.00136** | 7.47 × 10−9*** | 0.00150** | 1.00 |
| Phenylalanine | 166.0863 | 166.0863 | 0.46 | Standard | HMDB00159 | 0.00333** | 0.0149* | 2.95 × 10−7*** | 0.0198* | 1.00 |
| Tyrosine | 182.0812 | 182.0812 | 0.16 | Standard | HMDB00158 | 0.0253* | 0.00908** | 1.00 × 10−5*** | 0.0279* | 0.926 |
| Histidine | 156.0768 | 156.0766 | −1.2 | Standard | HMDB00177 | 6.87 × 10−4*** | 0.350 | 0.00141** | 0.246 | 0.805 |
| Proline | 116.0705 | 116.0705 | −0.19 | Standard | HMDB00162 | 0.968 | 0.00165** | 0.00115** | 0.0598 | 0.0299* |
| Lysine | 147.1128 | 147.1127 | −1.0 | Standard | HMDB00182 | 0.0779 | 9.56 × 10−4*** | 5.22 × 10−6*** | 0.459 | 0.426 |
| Glycine | 76.0393 | 76.0397 | 4.8 | Standard | HMDB00123 | 1.00 | 0.0140* | 0.00216** | 0.193 | 0.0753 |
| Ornithine | 133.0972 | 133.0971 | −0.22 | Standard | HMDB00214 | 0.293 | 0.0114* | 0.0220* | 0.0801 | 0.652 |
| C17H26N4O5 | 367.1976 | 367.1977 | 0.21 | 0.981 | 0.0990 | 0.0309* | 0.0561 | 0.430 | ||
| Pro-Gly-Pro or Pro-Pro-Gly (*6) | 270.1449 | 270.1452 | 1.1 | Predicted | 0.143 | 0.635 | 0.00165** | 0.425 | 0.990 | |
| C7H12N2O3 | 173.0921 | 173.0919 | −1.1 | 0.892 | 0.854 | 0.00289** | 0.902 | 0.999 | ||
| Burimamide (*5) | 213.1168 | 213.1233 | 30 | Predicted | 0.999 | 0.266 | 0.0439* | 0.950 | 0.224 | |
| Ethanolamine | 62.0600 | 62.0601 | 1.1 | Predicted | HMDB00149 | 0.684 | 0.0483* | 2.43 × 10−7*** | 0.00187** | 0.597 |
| Gamma-Aminobutyric acid | 104.0706 | 104.0706 | −0.35 | Standard | HMDB00112 | 0.833 | 0.139 | 2.10 × 10−7*** | 0.0133 | 0.897 |
| Aspartic acid | 134.0448 | 134.0447 | −0.34 | Standard | HMDB00191 | 0.287 | 0.00564** | 1.56 × 10−7*** | 0.403 | 0.416 |
| Valine | 118.0863 | 118.0862 | −0.72 | Standard | HMDB00883 | 7.31 × 10−5*** | 0.0204* | 1.94 × 10−8*** | 0.0138 | 0.990 |
| Tryptophan | 205.0972 | 205.0972 | −0.091 | Standard | HMDB00929 | 6.13 × 10−5*** | 0.402 | 2.99 × 10−8*** | 0.0113* | 0.229 |
| Beta-Alanine | 90.0550 | 90.0551 | 1.1 | Standard | HMDB00056 | 0.0407* | 0.722 | 3.56 × 10−6*** | 0.268 | 0.842 |
| Citrulline | 176.1030 | 176.1028 | −0.80 | Standard | HMDB00904 | 0.148 | 0.0509 | 0.00420** | 0.130 | 0.960 |
| Glutamic acid | 148.0604 | 148.0603 | −1.1 | Standard | HMDB00148 | 4.95 × 10−4*** | 0.00528** | 1.49 × 10−8*** | 0.0757 | 1.00 |
| Threonine | 120.0655 | 120.0654 | −0.64 | Standard | HMDB00167 | 1.18 × 10−4*** | 0.00790** | 5.72 × 10−8*** | 1.80 × 10−4*** | 1.00 |
| Serine | 106.0499 | 106.0499 | 0.33 | Standard | HMDB00187 | 0.0197* | 0.0119* | 3.42 × 10−7*** | 0.00699** | 0.846 |
| Glutamine | 147.0764 | 147.0764 | −0.43 | Standard | HMDB00641 | 0.0327* | 0.368 | 1.07 × 10−6*** | 0.111 | 0.975 |
| Hypoxanthine | 137.0458 | 137.0457 | −0.96 | Standard | HMDB00157 | 0.207 | 0.616 | 4.93 × 10−6*** | 0.00279** | 0.107 |
| Choline (*5) | 104.1070 | 104.1070 | 0.22 | Predicted | HMDB00097 | 2.30 × 10−5*** | 1.00 | 1.91 × 10−4*** | 0.0580 | 0.0115* |
| Carnitine | 162.1125 | 162.1124 | −0.38 | Standard | HMDB00062 | 7.60 × 10−4*** | 0.956 | 0.00341** | 0.996 | 0.247 |
| C5H11NO2 (13.42 min.) | 118.0863 | 118.0863 | 0.33 | 0.0606 | 0.958 | 3.73 × 10−4*** | 0.915 | 0.754 | ||
| Glycerophosphocholine | 258.1107 | 258.1121 | 5.5 | Standard | HMDB00086 | 0.287 | 4.53 × 10−5*** | 0.00263** | 0.0322* | 7.05 × 10−6*** |
| C7H8O3S | 173.0267 | 173.0285 | 10 | 0.962 | 1.88 × 10−5*** | 0.00553** | 0.0154* | 2.71 × 10−4*** | ||
| C4H5N2O11P | 288.9704 | 288.9691 | −4.3 | 0.0421* | 0.0815 | 0.0776 | 0.256 | 5.17 × 10−6*** | ||
| p-Value | Ratio of relative average to controls | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Metabolite | Oral cancer vs pancreatic cancer | Oral cancer vs periodontal desease | Breast cancer vs pancreatic cancer | Breast cancer vs periodontal desease | Pancreatic cancer vs periodontal desease | Oral cancer | Breast cancer | Pancreatic cancer | Periodontal desease |
| C2H6N2 | 2.34 × 10−6*** | 6.26 × 10−6*** | 0.995 | 0.936 | 0.999 | 0.577 | 3.96 | 4.42 | 5.28 |
| C32H48O13 (*4) | 0.137 | 0.444 | 0.951 | 0.467 | 0.970 | 0.954 | 2.21 | 2.03 | 1.11 |
| C3H7NO2 | 3.68 × 10−4*** | 0.0662 | 0.0350* | 0.748 | 0.744 | 1.24 | 1.62 | 3.00 | 2.16 |
| C4H12N5 | 0.00636** | 0.469 | 0.132 | 0.928 | 0.661 | 2.69 | 2.46 | 6.98 | 2.34 |
| C4H9NO2 | 0.00504** | 0.995 | 0.0138* | 0.995 | 0.214 | 1.73 | 1.88 | 5.16 | 1.71 |
| Cadaverine | 0.449 | 0.488 | 0.569 | 0.781 | 1.00 | 5.20 | 2.21 | 6.15 | 2.71 |
| C5H11NO2 (10.05 min.) | 0.270 | 0.169 | 0.976 | 0.543 | 0.897 | 1.51 | 1.73 | 2.10 | 2.35 |
| C30H62N19O2S3 (*3) | 0.0812 | 0.420 | 1.00 | 0.992 | 0.973 | 2.18 | 10.3 | 6.65 | 3.40 |
| C30H55N27O3S (*3) | 0.225 | 0.993 | 0.954 | 0.980 | 0.790 | 2.06 | 5.47 | 5.56 | 2.11 |
| C18H32N6O6 (*3) | 0.0247* | 0.914 | 0.657 | 0.917 | 0.578 | 1.34 | 2.39 | 6.70 | 0.942 |
| Alpha-Aminobutyric acid | 0.0543 | 0.811 | 0.0321* | 0.885 | 0.299 | 3.44 | 2.26 | 4.01 | 2.46 |
| Alanine | 0.0968 | 0.945 | 0.0447* | 0.543 | 0.573 | 3.91 | 1.94 | 3.67 | 1.92 |
| Putrescine | 0.0399* | 0.444 | 0.652 | 0.999 | 0.717 | 3.53 | 2.80 | 3.98 | 2.10 |
| Methylimidazoleacetic acid (*5) | 0.286 | 0.973 | 0.365 | 0.903 | 0.632 | 4.19 | 2.46 | 4.64 | 2.18 |
| Trimethylamine (*5) | 0.146 | 0.0643 | 0.0256* | 0.00788** | 0.977 | 4.80 | 1.36 | 4.13 | 3.75 |
| C5H14N5 | 0.471 | 0.994 | 0.211 | 0.889 | 0.839 | 8.31 | 2.20 | 5.35 | 2.69 |
| Piperidine | 1.00 | 0.972 | 0.190 | 0.730 | 0.992 | 18.4 | 1.20 | 3.73 | 1.46 |
| Taurine | 5.82 × 10−7*** | 1.10 × 10−6*** | 0.994 | 0.612 | 0.793 | 2.35 | 0.358 | 0.376 | 0.00 |
| Piperideine (*5) | 0.00226** | 2.91 × 10−5*** | 0.740 | 0.304 | 0.0296* | 2.43 | 0.509 | 0.665 | 0.113 |
| Pipecolic acid | 0.175 | 1.97 × 10−4*** | 0.330 | 0.529 | 0.0161* | 2.25 | 0.833 | 1.11 | 0.358 |
| C4H9N | 0.996 | 0.00766** | 0.00286** | 0.992 | 0.00416** | 4.81 | 1.18 | 2.70 | 0.846 |
| C8H9N | 0.0123* | 2.94 × 10−4*** | 0.00358** | 0.995 | 0.0379* | 3.23 | 0.209 | 0.817 | 0.168 |
| Pyrroline hydroxycarboxylic acid (*5) | 0.867 | 0.00267** | 0.0170* | 0.947 | 0.0400* | 4.15 | 0.454 | 2.28 | 0.184 |
| Betaine | 0.133 | 0.0668 | 0.998 | 0.999 | 0.913 | 2.04 | 0.864 | 0.822 | 0.637 |
| C4H7N | 0.853 | 0.0858 | 0.825 | 0.998 | 0.632 | 2.21 | 0.742 | 1.14 | 0.517 |
| C6H6N2O2 | 0.0715 | 0.974 | 0.0460* | 0.827 | 0.429 | 1.61 | 1.44 | 2.89 | 1.42 |
| Leucine + Isoleucine | 7.44 × 10−4*** | 0.868 | 0.00272** | 0.855 | 0.00574** | 4.65 | 3.05 | 7.71 | 2.19 |
| Phenylalanine | 0.00351** | 0.961 | 0.0122* | 0.967 | 0.0165* | 2.25 | 1.78 | 3.54 | 1.66 |
| Tyrosine | 0.0286* | 0.969 | 0.235 | 1.00 | 0.215 | 1.84 | 1.99 | 2.90 | 1.49 |
| Histidine | 0.698 | 0.997 | 0.275 | 0.990 | 0.717 | 1.70 | 1.35 | 2.02 | 1.29 |
| Proline | 0.0171* | 0.291 | 0.825 | 0.995 | 0.689 | 1.63 | 2.48 | 3.99 | 1.58 |
| Lysine | 0.00513** | 1.00 | 0.226 | 0.730 | 0.0190* | 1.84 | 2.96 | 3.97 | 1.22 |
| Glycine | 0.0122* | 0.352 | 0.731 | 0.998 | 0.689 | 1.38 | 2.32 | 3.10 | 1.67 |
| Ornithine | 0.516 | 0.912 | 0.986 | 1.00 | 0.941 | 1.69 | 2.13 | 1.97 | 1.43 |
| C17H26N4O5 | 0.248 | 0.357 | 0.917 | 0.997 | 0.913 | 2.19 | 2.63 | 2.69 | 1.52 |
| Pro-Gly-Pro or Pro-Pro-Gly (*6) | 0.413 | 1.00 | 0.205 | 0.972 | 0.660 | 4.25 | 2.65 | 4.27 | 2.00 |
| C7H12N2O3 | 0.0627 | 0.995 | 0.238 | 1.00 | 0.602 | 1.80 | 1.65 | 2.48 | 1.48 |
| Burimamide (*5) | 0.0675 | 0.841 | 0.779 | 0.961 | 0.631 | 1.68 | 2.32 | 2.56 | 1.42 |
| Ethanolamine | 2.34 × 10−4*** | 0.132 | 0.00404** | 0.486 | 0.401 | 1.66 | 1.84 | 4.35 | 2.23 |
| Gamma-Aminobutyric acid | 0.00108** | 0.367 | 0.0140 | 0.712 | 0.215 | 1.82 | 1.62 | 3.31 | 2.07 |
| Aspartic acid | 4.37 × 10−5*** | 0.980 | 0.0114* | 0.932 | 0.00672** | 1.53 | 1.70 | 4.10 | 1.26 |
| Valine | 0.00325** | 0.999 | 0.00549** | 0.967 | 0.00785** | 4.53 | 2.64 | 5.92 | 1.75 |
| Tryptophan | 0.0461* | 1.00 | 1.83 × 10−4*** | 0.319 | 0.0424* | 4.26 | 1.59 | 6.47 | 2.44 |
| Beta-Alanine | 0.156 | 0.999 | 0.00471** | 0.730 | 0.375 | 2.22 | 1.51 | 3.04 | 1.74 |
| Citrulline | 0.291 | 0.880 | 0.865 | 0.987 | 0.992 | 1.98 | 2.30 | 3.10 | 1.65 |
| Glutamic acid | 0.00312** | 1.00 | 0.00214** | 1.00 | 0.00574** | 2.87 | 2.12 | 4.80 | 1.54 |
| Threonine | 3.08 × 10−4*** | 0.0829 | 0.00141** | 0.166 | 0.162 | 2.15 | 1.71 | 4.75 | 2.19 |
| Serine | 1.16 × 10−4*** | 0.435 | 0.00920** | 0.868 | 0.0868 | 1.74 | 1.66 | 4.34 | 1.63 |
| Glutamine | 0.00228** | 0.998 | 0.00167** | 0.903 | 0.0251* | 2.35 | 1.59 | 4.96 | 1.39 |
| Hypoxanthine | 0.0917 | 0.452 | 1.94 × 10−4*** | 0.00237** | 0.952 | 2.97 | 0.839 | 3.35 | 2.30 |
| Choline (*5) | 0.871 | 0.983 | 0.00374** | 0.265 | 0.429 | 2.98 | 1.05 | 2.63 | 1.51 |
| Carnitine | 0.652 | 0.670 | 0.0895 | 1.00 | 0.335 | 2.10 | 1.21 | 1.88 | 1.11 |
| C5H11NO2 (13.42 min.) | 0.0687 | 0.930 | 0.0161* | 0.999 | 0.0544 | 2.13 | 1.06 | 2.17 | 1.03 |
| Glycerophosphocholine | 7.33 × 10−4*** | 0.0138* | 1.00 | 1.00 | 1.00 | 2.78 | 0.00 | 0.00 | 0.00 |
| C7H8O3S | 0.0176* | 0.0307* | 0.864 | 1.00 | 0.860 | 0.972 | 0.494 | 0.592 | 0.467 |
| C4H5N2O11P | 0.628 | 0.808 | 6.92 × 10−4*** | 0.0116* | 1.00 | 0.418 | 1.96 | 0.00 | 0.00 |
* P < 0.05; ** P < 0.01; *** P < 0.001 (Steel–Dwass test)
(*1) The peaks marked “standard” were identified based on the matched m/z values and normalized migration times of the corresponding standard compounds. The peaks marked “predicted” were obtained from Kyoto Encyclopedia of Gene and Genomics (KEGG) database (http://www.genome.jp/kegg/) and HMDB (HMDB; http://www.hmdb.ca/) by matching the composition formula and migration times that were calculated from the isotope distribution and molecular structure, respectively
(*2) HMDB: accession numbers for the Human Metabolome Database
(*3) The peaks were detected as bivalent ions
(*4) The peaks were detected as trivalent ions
(*5) The normalized measured and predicted migration times were 8.16 min and 7.78 min (0.38 min) for ethanolamine, 11.61 min and 10.42 min (1.19 min) for burimamide, 8.75 min and 7.78 min (0.97 min) for choline, 8.16 min and 7.78 min (0.38 min) for ethanolamine, 10.012 min and 10.10 min (0.98 min) for methylimidazoleacetic acid, 8.78 min and 7.84 min (0.94 min) for piperidine, 12.90 min and 13.09 min (0.19 min) for pyrroline hydroxycarboxylic acid, and 7.61 min and 8.26 min (0.65 min) for trimethylamine. The parenthetic values are differences between the predicted and the measured time
(*6) Because we did not find any candidate compounds for the 270.1454 m/z and 12.23 min peaks in the databases, we considered Pro-Gly-Pro, Pro-Pro-Gly or Gly-Pro-Pro as possible candidates ([M + H]+ = 270.3044 m/z in all cases). The predicted migration time of Pro-Gly-Pro and Pro-Pro-Gly was 11.33 min and that of Gly-Pro-Pro was 10.98 min; therefore, Gly-Pro-Pro was excluded as a candidate
Fig. 2.
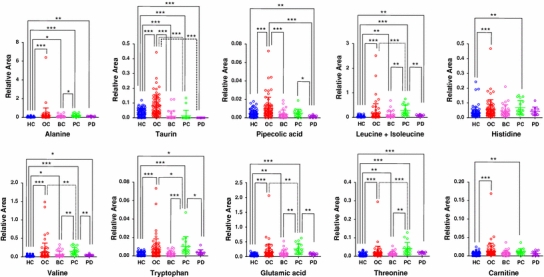
Representative dot plots for the relative area of detected metabolites in samples from all groups. The colored dots denote healthy controls (blue), oral (red), breast (pink), pancreatic cancer (green), and periodontal disease (purple). The Y- and X-axes denote the relative peak area (no units) and the group name, respectively. The horizontal, center long bars and the short top/bottom bars indicate the means and standard deviations, respectively. The stars indicates * P < 0.05, ** P < 0.01, and *** P < 0.001 (Steel–Dwass test). Only metabolites showing a significant difference between oral cancer and controls at P < 0.001 and matched with standard library are displayed. The dot plots of the other metabolites are shown in Supplementary Fig. S1
Fig. 3.
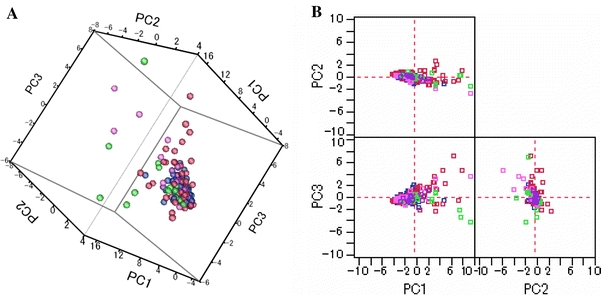
Score plots of principal components (PC) analyses. The subjects in all groups are shown in 3-dimensional (a) and 2-dimensional (b) plots without outliers. The cumulative proportions of the first, second and third PCs (PC1, PC2, and PC3) were 44.8, 57.6 and 67.0%. The same analyses presented for all datasets are shown in Supplementary Fig. S2
The MLR model developed for oral cancer yielded a high AUC (0.865), and the trained models also showed high separation ability in the CV (AUC = 0.810). The receiver operating characteristic (ROC) curves and selected parameters of the MLR models for each disease are shown in Fig. 4 and Supplementary Table S1, respectively. The MLR models for pancreatic cancer and periodontal disease yielded high AUCs in the CV test (0.944 and 0.954, respectively), using only five and two metabolic markers, respectively; while oral and breast cancers (0.810 and 0.881, respectively) used 9 and 14 metabolites, respectively, with lower AUCs. On the metabolite heat map (Fig. 5), the control group and the periodontal disease group were relatively lower and the pancreatic cancer group tended to be homologically higher, while the oral and breast cancers exhibited more diverse profiles compared with the other groups. This suggests that our MLR models for oral and breast cancer require additional parameters for accurate classification. The heterogeneous nature of oral cancers, including oral squamous cell carcinoma (OSCC), oropharyngeal, tongue and neck cancer, may produce different profiles; this diminishes the discriminative capability of a single classification model. The diverse profiles associated with breast cancer may result in a similar situation because breast cancer comprises structurally differing types according to the expression of hormone receptors such as estrogen and progesterone, and is affected by clinical parameters, such as the patient’s age or menopause status. Three metabolites, taurine, piperidine, and a peak at 120.0801 m/z, were oral cancer-specific markers (different from all of the other groups at P < 0.05; Steel–Dwass test) and eight metabolites (leucine with isoleucine, tryptophan, valine, glutamic acid, phenylalanine, glutamine, and aspartic acid) were pancreatic cancer-specific markers. Although several metabolites in breast cancer patients yielded a statistically significant difference between breast cancer and healthy controls, including taurine and lysine (P < 0.001 for both; Steel–Dwass test), there were no differences in metabolites between breast cancer and other cancer, and they were not unique for breast cancer.
Fig. 4.
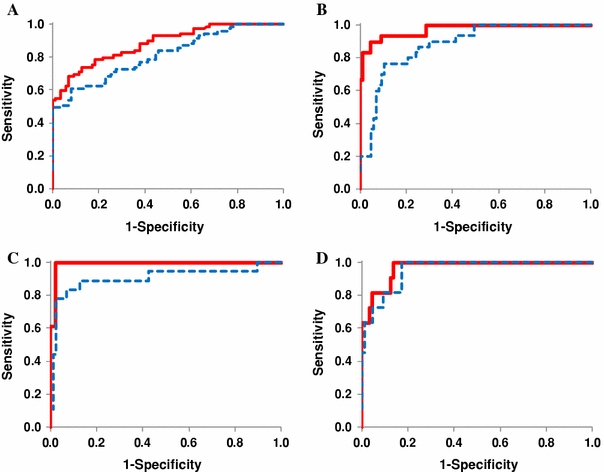
ROC curve analysis of the ability of salivary metabolites to discriminate between samples from patients with a oral (n = 69), b breast (n = 30) or c pancreatic cancer (n = 18), and d samples from patients with periodontal diseases (n = 11) and the controls (n = 87). The solid (red) and dotted (blue) ROC curves were obtained using the complete data as a training set and with a tenfold cross-validation, respectively. Using a cut-off probability of 50%, the calculated area under the ROC curves were 0.865 (0.810) for oral, 0.973 (0.881) for breast and 0.993 (0.944) for pancreatic cancer, and 0.969 (0.954) for periodontal diseases. The non-parenthetic values were obtained with the full-training data and parenthetic values by tenfold cross-validation
Fig. 5.
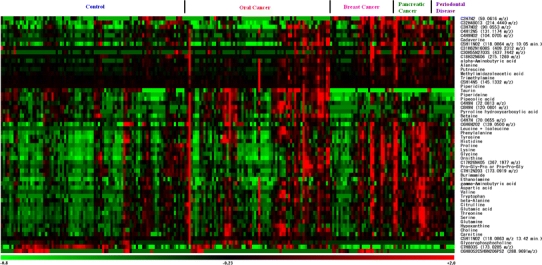
Heat map of 57 peaks showing significantly different levels (P < 0.05; Steel–Dwass test) between control samples (n = 87) and samples from patients with at least one disease (n = 128). Each row shows data for a specific metabolite and each column shows an individual. The colors correspond to the relative metabolite areas that were converted to Z-scores
Comparison of the obtained metabolites with previous studies
Of the metabolite profiles obtained, the annotated metabolites included carnitines (betaine, choline, carnitine, glycerophosphocholine), polyamines (cadaverine and putrescine), a purine (hypoxanthine), amino alcohols (ethanolamine), aliphatic and aromatic amine (trimethylamine), and amino acids (the others), in accordance with the defined chemical class category in HMDB. Because each MLR model developed to discriminate between control and patient groups reached high accuracy by incorporating quantified multiple metabolites, the quantitative associations between the multiple metabolites and the individual markers are important. Changes in the individual metabolites were generally consistent with those of earlier studies. For example, polyamines are correlated with cell growth and proliferation (Casero and Marton 2007; Gerner and Meyskens 2004; Tabor and Tabor 1984), and with tumor growth in oral cancer (Dimery et al. 1987), while putrescine is used to monitor the effect of chemotherapy on oral cancer cells (Okamura et al. 2007). The serum concentration of putrescine and cadaverine are decreased in cancer patients undergoing radiotherapy but remain higher than those in healthy individuals (Khuhawar et al. 1999). There were no significant differences in urinary polyamine levels between the healthy individuals and breast cancer patients; however, the levels of putrescine, spermine and other metabolites were significantly higher in patients with breast cancer (Byun et al. 2008). Oral polyamine levels are also affected by periodontitis and gum healing (Silwood et al. 2002). We found that the levels of ornithine and putrescine were higher in patients with breast or pancreatic cancer, and were markedly higher in patients with oral cancer, than in our healthy controls, while there was no significant difference between patients with periodontal disease and the controls. Although the quantitative level of polyamines is associated with regulation of tumor growth and with periodontitis, our results indicate that salivary polyamines are affected by the cancer type and by periodontitis, and that their levels were markedly higher in patients with oral cancer.
In addition to polyamines, the level of tryptophan (Carlin et al. 1989), which is increased in oral and pancreatic cancer, is a direct marker for tumor development. In terms of an indirect connection between the detected metabolites and human cancer, the repeat peptide Pro-Pro-Gly, which is expressed at high levels in breast cancer, is an inhibitor of matrix metalloproteinase-2 (MMP-2, gelatinase A), which plays an important role in tumor invasion and metastasis (Jani et al. 2005). The expression levels of the amino acid transporters ACST2 and LAT1 are elevated in primary human cancers, and cancer cells optimize their metabolic pathways by activating the extra- to intracellular exchange of amino acids. Peptides and acids are derived from various sources, such as fragmented proteins, and the saliva metabolome profiles comprising these compounds may reflect the integrated results.
A significantly decreased level of arginine was observed in plasma samples from several cancers including breast, colonic and pancreatic cancer, which might be due to increased uptake of arginine by tumor tissues with high arginase activity (Vissers et al. 2005). However, salivary arginine was hardly changed, and there were no differences among the groups (Supplementary Fig. S3 and Table S2). A trend for decreasing levels of amino acids, including leucine, isoleucine, valine and alanine, has been reported in pancreatic cancer samples (Fang et al. 2007). The levels of amino acids in breast cancer tissue samples showed similar patterns, with low levels of isoleucine, leucine, lysine and valine (Yang et al. 2007). The decreased amino acid levels appear to be the result of enhanced energy metabolism or upregulation of the appropriate biosynthetic pathways, and required cell proliferation in cancer tissues. However, the observed salivary amino acid levels showing significant differences in the cancer groups (in Table 2) were higher than in the controls. The heterogeneous systems that transport amino acids from blood to saliva via the salivary gland, such as kinetic differences, or the dependence or independence of small ions such as potassium and sodium (Mann and Yudilevich 1987), altered the concentration of these ions because of water movement through the paracellular route (Melvin 1999) or channels (Ishikawa and Ishida 2000). Metabolism in the salivary gland itself might also play a major contribution to the differences in profiles between saliva and blood. Further validation of these findings by comparing saliva profiles with blood and tissue profiles is needed to understand the reason for the different saliva amino acid profiles.
Choline, a quaternary amine, is an essential nutrient that is predominantly supplied by the diet, and choline-containing metabolites are important constituents of phospholipid metabolism of cell membranes and are associated with malignant transformation, such as breast, brain and prostate cancers (Ackerstaff et al. 2003). Magnetic resonance spectroscopy (MRS) is routinely used to quantify choline-based metabolism in malignancies such as head and neck cancer and breast cancers (Bolan et al. 2003). Choline is highly metabolized in tumors to phosphocholine and is also highly oxidized to betanine; hence, the low concentration of choline and high concentrations of phosphocholine and betaine (Katz-Brull et al. 2002) were observed. Furthermore, the levels of choline metabolites were higher in tumors than in benign lesions or normal tissues (reviewed in Haddadin et al. 2009). In tumor cells, an excessive increase in plasma choline levels in patients with breast cancer was also shown (Katz-Brull et al. 2001). Aberrant choline metabolism can be explained as a result of enhanced membrane synthesis and degradation, which represent excessive proliferation of cancer cells. Pancreatic cancer tissue had a unique profile showing decreased levels of phosphocholine and glycerophosphocholine, but not choline (Fang et al. 2007). We found that the levels of phosphocholine (Supplementary Fig. S3 and Table S2) and glycerophosphocholine (Table 2) were increased in the saliva samples from oral cancer patients and were decreased in the other groups.
Creatine phosphate acts as a store for high-energy phosphates. Therefore, its concentration might be altered in energy-demanding tissues (Maheshwari et al. 2000). Previous studies showed an increase in the choline-creatinine ratio in tumor tissues and in the serum of patients with OSCC (Maheshwari et al. 2000; Tiziani et al. 2009). Creatine is converted to creatine phosphate by creatine kinase. Increased creatine phosphate levels were also found in other tumors, such as breast and gastrointestinal tract tumors. In our study, the salivary choline level was significantly higher in subjects with oral and pancreatic cancers (P = 2.30 × 10−5 and P = 1.91 × 10−4, respectively; Steel–Dwass test), but not in the other groups. Therefore, the salivary choline–creatinine ratio showed oral cancer-specific elevation (Supplementary Table S2 and Fig. S3). However, this finding needs to be interpreted with care because choline is a nutrient present in most foods.
Compared with oral cancer, breast and pancreatic tumors are physically remote from the oral cavity. Therefore, it can be questioned why salivary metabolite profiles reflect the aberrant localized tumour metabolism. Systemic biofluids, such as blood and lymph fluid, are one of the routes that readily bypass these tumors and the salivary gland, which blends saliva with contaminating blood. Several metabolites in tumor tissues, such as lactate, which is derived from tumor exposed to hypoxia, were altered both with and without metastasis (Hirayama et al. 2009; Walenta et al. 2000). Although abnormal arginine levels in breast cancer without metastasis were observed, the same metabolic changes were shown in a pooled group of patients with colonic and pancreatic cancer with/without metastasis (Vissers et al. 2005). In OSCC patients without metastasis from the primary tumor, cancer-specific changes in serum and salivary mRNA levels (Li et al. 2006; Pickering et al. 2007) and blood metabolome levels (Tiziani et al. 2009; Zhou et al. 2009) were shown. Although this does not constitute direct proof that the aberration in salivary metabolites is attributed to a remote tumor, evidence that the salivary metabolite profiles reflects the systemic and localized tumor status or its response to chemotherapies, such as breast and lung cancer, has accumulated (Emekli-Alturfan et al. 2008; Gao et al. 2009; Harrison et al. 1998; Streckfus et al. 2006, 2008). Although previous studies have demonstrated an increase in choline metabolites in blood in various cancers, the increase in choline metabolites in oral cancer patients in study indicate that the transportation of these metabolites from the blood to the saliva through the salivary gland is low, even though their levels in blood are elevated. Alternatively, these metabolites were diffused from the oral malignancy to the salivary gland via a route other than the blood vessel. We acknowledge that the current study merely mined the data and showed that the changes in salivary metabolites had cancer-specific features. Further biological studies to compare the metabolite profiles obtained concurrently from saliva, blood and cancer tissue is needed to provide rational evidence for the systemic metabolite links.
Bias derived from clinical parameters
We evaluated the metabolite bias introduced by relevant clinical parameters (age, gender, race and ethnicity). The PCA score plots showed poor separation between male and female subjects among healthy controls and patients with oral cancer (Supplementary Fig. S4). Statistical comparisons of the relative area are presented in Supplementary Table S3. Takeda et al. (2009) measured the gender-specific differences in salivary metabolites and found that formate, lactate, propionate and taurine were significantly higher in males. Compared with these metabolites, the gender-specific level of taurine, which was the only metabolite observed in our measurement condition, showed little difference between the subjects in the control and oral cancer groups. By contrast, in the control group, tyrosine and a metabolite at 214.4440 m/z were significantly higher in females than in males (P = 0.0492 and p = 0.0261, respectively; Mann–Whitney test). In the oral cancer group, threonine and serine were significantly higher in males and piperidine was higher in females (P = 0.0340, P = 0.0462, and P = 0.0221, respectively; Mann–Whitney test). Takeda et al. (2009) discussed that these gender-specific differences might be attributed to dental care, hormones such as estrogen, and oral pathogenesis carriers such as microflora. Indeed, infection of the oral environment with viruses such as human papillomavirus or micro-organisms is known to be a risk factor for the development of oral cancer (Meurman and Uittamo 2008). Although we found that the gender-specific differences in metabolic profiles differed between the tumor types, the number of metabolites showing significant differences was low, which implies that the disease-specific variation is predominantly embedded in the 57 metabolites identified here.
In the control and oral cancer groups, the PCA based on race and ethnicity were visualized using score plots (Supplementary Fig. S5) and the statistical analytical results are presented in Supplementary Table S4. In the control group, there were no significant differences between African-Americans and Caucasians, or African-Americans and Hispanics. Meanwhile, 11 and 12 significantly different (P < 0.05; Steel–Dwass test) metabolites were observed between African-Americans and Asians, and Asians and Caucasians, respectively. Similarly, the profiles between Asians and Hispanics, and Caucasians and Hispanics revealed three and seven significantly different metabolites (P < 0.05; Steel–Dwass test). Of particular note, levels of putrescine, proline, glycine and unannotated metabolites at 118.0864 m/z and 10.05 min were low in Asians, while the level of burimamide was high in African-Americans. A country-dependant bias in human urinary metabolite profiles has also been reported elsewhere (Holmes et al. 2008). In their study, positively charged metabolites, such as alanine-related metabolites, showed discriminative characteristics and were correlated with several dietary factors such as energy intake, dietary cholesterol and alcohol intake. However, in our study, there were no differences in alanine levels in either the control or the oral cancer subjects. In the control group, there were no differences in 34 out of 57 marker candidates among the race or ethnic groups. In subjects with oral cancer, only a metabolite at 211.4440 m/z showed a significant difference (P = 0.0386; Steel–Dwass test). Although biases based on race or ethnicity-were found in the 57 metabolic profiles, the number of the metabolites showing significant differences were less than the number of peaks showing significant differences in cancer-specific profiles, which implies that this bias might be more moderate than disease-specific differences.
Age-related differences have been reported in a transcriptome study of the salivary gland (Srivastava et al. 2008). The coefficients of regression lines for age and relative area for all 57 metabolite markers are presented in Supplementary Table S5. It has been reported that other commonly used methods for standardization of metabolites in biofluid yield different statistical results (Schnackenberg et al. 2007); therefore, consistent decreases or increases in levels of metabolites among subjects with correlated clinical parameters should be accounted for. In the control subjects and patients with pancreatic cancer, there was a positive correlation between metabolites and age, whereas the opposite was true for patients with oral or breast cancer or periodontal diseases. Accordingly, it is unlikely that age is correlated with the concentrations of salivary metabolites.
Several limitations in this study need to be acknowledged. First, the metabolite profiles in saliva might fluctuate to similar or greater levels compared with other omics profiles, such as the proteome and transcriptome, in response to systemic conditions such as stress, and oral conditions including gingival crevicular fluid and oral microbiota (reviewed in Fabian et al. 2008). Therefore, the reproducibility of the sample collection protocol used in this study should be rigorously verified under various conditions. Circadian rhythms in salivary flow rate and components have been reported (Dawes 1972). Levels of putrescine and cadaverine, which correlate with oral malodor, were markedly altered during waking time, even in healthy donors (Cooke et al. 2003). Although, the samples were collected within a limited period of time in the morning, levels of these metabolites were generally higher in patients with most types of cancer in the present study. The variance in the concentrations of these metabolites should be validated in future studies. Another external factor that alters saliva contents is the time-course of fluoride concentration, which has been tracked, and the changes in concentrations continued for 30 min after eating food (Hedman et al. 2006). Therefore, the 1-h period before sample collection should be evaluated in terms of food intake. Smoking is also known to affect salivary metabolites such as citrate lactate, pyruvate and sucrose (Takeda et al. 2009). The metabolites identified in this study could not be compared with these metabolites because they were not positively charged in our measurement condition. Therefore, the profiles of positively charged metabolites should be explored in further analyses.
Second, the sample sizes, particularly the number of patients with breast or pancreatic cancer or periodontal diseases, were relatively small. A larger cohort, including samples from an independent institute, would allow for statistical comparisons with greater power and a more rigorous validation. In addition, samples from patients with systemic diseases showing similar symptoms, such as oral leukoplakia and oral cancer (Zhou et al. 2009), chronic pancreatitis and pancreatic cancer (Fang et al. 2007; Kojima et al. 2008), should be compared with evaluate the sensitivity and specificity of the detected metabolites. In this study, the patients’ age was collected for all samples and only a few additional parameters, namely sex and race, or ethnic group, were collected for the control and oral cancer group. Analyses and validation studies taking into account the complete clinical and pathological parameters, including menopausal status, estrogen and progesterone receptors for breast cancer, and risk factors including smoking and alcohol drinking for oral cancers are essential before actual diagnostic application of the classification model obtained in this study. In this study, although we used stepwise feature selection and an MLR model to identify classifiers, other feature selection and classification methods are also applicable, such as regression tree models (Li et al. 2004, 2006) and concurrent use of ANN with SVM (Ayers et al. 2004). Instead of developing a classification model only based on the salivary metabolome profiles of matched subjects, the construction of a marker model incorporating related clinical features or risk factors and biomarkers can be used to visualize the probability of a specific diseases status; for example, nomograms are a commonly used strategy (Brennan et al. 2004; Gross et al. 2008; Katz et al. 2008).
A metabolomic study using serum samples from patients with oral cancer showed stage-specific profiles (Tiziani et al. 2009). The profiles obtained in this study were simply categorized into the type of cancer. Therefore, future studies are needed that integrate histological and clinical features. Simultaneous analyses of the metabolic profiles in blood and tissue collected from the same patients are also needed to track the biological sources of the disease-specific signatures in salivary metabolite profiles. Although there are still several limitations to be addressed, the methodology used in this study to detect salivary metabolite profiles are not limited to early diagnosis but offer the potential to aid the characterization of malignant neoplasms or tumors by integrating histological or clinical features, such as staging.
Concluding remarks
This is the first study to comprehensively analyze salivary metabolites and to identify metabolic profiles specific to oral, breast and pancreatic cancers. A larger number of patient samples, particularly those from different institutes, and additional clinical variables are needed for further validation and future clinical application of our method. In addition, integrating the knowledge obtained from other omics studies may help us to understand the biological basis of these disease-specific metabolic profiles.
In conclusion, our study has demonstrated that CE-TOF-MS can readily and effectively be applied to salivary metabolomics. We have proposed an alternative use for salivary diagnosis to be applied for the detection of oral, breast and pancreatic cancers.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Dot plots for the relative area of detected metabolites in samples from all groups. The colored dots denote healthy controls (blue), oral (red), breast (pink), pancreatic cancer (green), and periodontal disease (purple). The Y- and X-axes denote the relative peak area (no units) and the group name, respectively. The horizontal, center long bars and short top/bottom bars indicate the means and standard deviations, respectively. * p < 0.05, ** p < 0.01 and *** p < 0.001 (Steel–Dwass test). Metabolites showing a significant difference between oral cancer and controls at p < 0.001 (Steel–Dwass test) and matched with the standard library are shown in Figure 2. (PPT 2514 kb)
Score plots of individually developed principal components (PC) analyses. The subjects in all groups are shown in 3-dimensional (a) and 2-dimensional (b) plots. The same data presented without three outliers are depicted in Figure 3. The score plots yielded by independent PC analysis are shown for control subjects versus patients with oral cancer (c), breast cancer (d), pancreatic cancer (e), and periodontal diseases (f). The colored dots indicate control (blue), oral cancer (red), breast cancer (pink), pancreatic cancer (green) and periodontal diseases (purple). The cumulative proportions of the first, second and third PCs (PC1, PC2, and PC3) were 49.6%, 64.0% and 74.6% for (c), 48.9%, 57.4% and 63.5% for (d), 47.5%, 57.6% and 65.1% for (e), and 38.7%, 49.6% and 58.0% for (f), respectively. (PPT 437 kb)
Dot plots for the relative area of creatinine, arginine, phosphocholine, and choline-creatinine ratio. The colored dots denote healthy controls (blue), oral (red), breast (pink), pancreatic cancer (green), and periodontal disease (purple). The Y- and X-axes denote the relative peak area (no units) or ratio of the relative peak area and the group name, respectively. The horizontal, center long bars and short top/bottom bars indicate the means and standard deviations, respectively. * p < 0.05, ** p < 0.01 and *** p < 0.001 (Steel–Dwass test). (PPT 255 kb)
Score plots of principal components (PC) analysis according to sex. The values for males and females are colored blue and red, respectively. Three-dimensional and 2-dimensional PC plots for healthy controls (a) and (c), respectively, and for subjects with oral cancer (b) and (d), respectively, are shown. A total of 42 and 27 male and female control subjects and 41 and 23 male and female patients with oral cancer were included in this analysis. Eighteen control subjects, five patients with oral cancer and all of the patients with other diseases were excluded because of unavailable sex information. The cumulative proportions of PC1, PC2, and PC3 for the control subjects were 43.0%, 52.9% and 60.6%, respectively, and those for patients with oral cancer were 50.2%, 65.7% and 77.5%, respectively. (PPT 218 kb)
Principal components (PC) analysis score plots for race and ethnic groups. The data for African-American, Asian, Caucasian and Hispanic subjects are colored red, blue, green, and orange, respectively. Three-dimensional and 2-dimensional PC plots for healthy controls (a) and (c), respectively, and subjects with oral cancer (b) and (d), respectively, are shown. The race or ethnic groups included 12 and four African-Americans, 15 and five Asians, 37 and 41 Caucasians, and five and five Hispanics as healthy controls and subjects with oral cancer, respectively. Eighteen control subjects, five patients with oral cancer, and all of the patients with other diseases were excluded because of unavailable race or ethnic information. The cumulative proportions of PC1, PC2 and PC3 for the control subjects were 43.0%, 52.9% and 60.9%, respectively, and those for subjects with oral cancer were 42.0%, 63.2% and 70.2%, respectively.(PPT 240 kb)
Acknowledgments
This work was supported by NIH grant DE017170 (to D.T.W.), a grant from the Global COE Program entitled “Human Metabolomic Systems Biology,” and “Grant-in-Aid” research grants from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan for Scientific Research on Priority Areas “Systems Genomes”, as well as research funds from Yamagata prefectural government and the city of Tsuruoka (to M.S., A.H., T.S., and M.T.). D.T.W. acknowledges the colleagues who provided the clinical samples for this study: Elliot Abemayor, Marilene Wang, Mai Brooks, James Ferrell, and Perry Klokkevoid and members of his laboratory at UCLA: Bradley Henson, Jianghua Wang, Xinmin Yang and David Akin. M.S., A.H., T.S., and M.T. thank Shinobu Abe and Kenjiro Kami of IAB for technical support and valuable discussions, and Dr. Ursula Petralia for editing the manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
M. Sugimoto and D. T. Wong contributed equally to this work.
References
- Ackerstaff E, Glunde K, Bhujwalla ZM. Choline phospholipid metabolism: A target in cancer cells? Journal of Cellular Biochemistry. 2003;90:525–533. doi: 10.1002/jcb.10659. [DOI] [PubMed] [Google Scholar]
- Aharoni A, Ric de Vos CH, Verhoeven HA, et al. Nontargeted metabolome analysis by use of Fourier transform ion cyclotron mass spectrometry. OMICS. 2002;6:217–234. doi: 10.1089/15362310260256882. [DOI] [PubMed] [Google Scholar]
- Ayers M, Symmans WF, Stec J, et al. Gene expression profiles predict complete pathologic response to neoadjuvant paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide chemotherapy in breast cancer. Journal of Clinical Oncology. 2004;22:2284–2293. doi: 10.1200/JCO.2004.05.166. [DOI] [PubMed] [Google Scholar]
- Baran R, Kochi H, Saito N, et al. MathDAMP: A package for differential analysis of metabolite profiles. BMC Bioinformatics. 2006;7:530. doi: 10.1186/1471-2105-7-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolan PJ, Meisamy S, Baker EH, et al. In vivo quantification of choline compounds in the breast with 1H MR spectroscopy. Magnetic Resonance in Medicine. 2003;50:1134–1143. doi: 10.1002/mrm.10654. [DOI] [PubMed] [Google Scholar]
- Brennan MF, Kattan MW, Klimstra D, Conlon K. Prognostic nomogram for patients undergoing resection for adenocarcinoma of the pancreas. Annals of Surgery. 2004;240:293–298. doi: 10.1097/01.sla.0000133125.85489.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M, Dunn WB, Dobson P, et al. Mass spectrometry tools and metabolite-specific databases for molecular identification in metabolomics. Analyst. 2009;134:1322–1332. doi: 10.1039/b901179j. [DOI] [PubMed] [Google Scholar]
- Byun JA, Lee SH, Jung BH, et al. Analysis of polyamines as carbamoyl derivatives in urine and serum by liquid chromatography-tandem mass spectrometry. Biomedical Chromatography. 2008;22:73–80. doi: 10.1002/bmc.898. [DOI] [PubMed] [Google Scholar]
- Carlin JM, Ozaki Y, Byrne GI, Brown RR, Borden EC. Interferons and indoleamine 2, 3-dioxygenase: role in antimicrobial and antitumor effects. Experientia. 1989;45:535–541. doi: 10.1007/BF01990503. [DOI] [PubMed] [Google Scholar]
- Casero RA, Jr, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nature Reviews. Drug Discovery. 2007;6:373–390. doi: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- Cooke M, Leeves N, White C. Time profile of putrescine, cadaverine, indole and skatole in human saliva. Archives of Oral Biology. 2003;48:323–327. doi: 10.1016/S0003-9969(03)00015-3. [DOI] [PubMed] [Google Scholar]
- Dawes C. Circadian rhythms in human salivary flow rate and composition. Journal of Physiology. 1972;220:529–545. doi: 10.1113/jphysiol.1972.sp009721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida Pdel V, Gregio AM, Machado MA, de Lima AA, Azevedo LR. Saliva composition and functions: A comprehensive review. Journal of Contemporary Dental Practice. 2008;9:72–80. [PubMed] [Google Scholar]
- Dimery IW, Nishioka K, Grossie VB, Jr, et al. Polyamine metabolism in carcinoma of the oral cavity compared with adjacent and normal oral mucosa. American Journal of Surgery. 1987;154:429–433. doi: 10.1016/0002-9610(89)90018-4. [DOI] [PubMed] [Google Scholar]
- Emekli-Alturfan E, Demir G, Kasikci E, et al. Altered biochemical parameters in the saliva of patients with breast cancer. Tohoku Journal of Experimental Medicine. 2008;214:89–96. doi: 10.1620/tjem.214.89. [DOI] [PubMed] [Google Scholar]
- Epstein JB, Zhang L, Rosin M. Advances in the diagnosis of oral premalignant and malignant lesions. Journal Canadian Dental Association. 2002;68:617–621. [PubMed] [Google Scholar]
- Fabian TK, Fejerdy P, Csermely P. Salivary Genomics, transcriptomics and proteomics: The emerging concept of the oral ecosystem and their use in the early diagnosis of cancer and other diseases. Current Genomics. 2008;9:11–21. doi: 10.2174/138920208783884900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F, He X, Deng H, et al. Discrimination of metabolic profiles of pancreatic cancer from chronic pancreatitis by high-resolution magic angle spinning 1H nuclear magnetic resonance and principal components analysis. Cancer Science. 2007;98:1678–1682. doi: 10.1111/j.1349-7006.2007.00589.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O, Kopka J, Dormann P, et al. Metabolite profiling for plant functional genomics. Nature Biotechnology. 2000;18:1157–1161. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- Fliser D, Wittke S, Mischak H. Capillary electrophoresis coupled to mass spectrometry for clinical diagnostic purposes. Electrophoresis. 2005;26:2708–2716. doi: 10.1002/elps.200500187. [DOI] [PubMed] [Google Scholar]
- Gao K, Zhou H, Zhang L, et al. Systemic disease-induced salivary biomarker profiles in mouse models of melanoma and non-small cell lung cancer. PLoS ONE. 2009;4:e5875. doi: 10.1371/journal.pone.0005875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerner EW, Meyskens FL., Jr Polyamines and cancer: Old molecules, new understanding. Nature Reviews Cancer. 2004;4:781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- Goto S, Okuno Y, Hattori M, Nishioka T, Kanehisa M. LIGAND: Database of chemical compounds and reactions in biological pathways. Nucleic Acids Research. 2002;30:402–404. doi: 10.1093/nar/30.1.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross ND, Patel SG, Carvalho AL, et al. Nomogram for deciding adjuvant treatment after surgery for oral cavity squamous cell carcinoma. Head and Neck. 2008;30:1352–1360. doi: 10.1002/hed.20879. [DOI] [PubMed] [Google Scholar]
- Haddadin IS, McIntosh A, Meisamy S, et al. Metabolite quantification and high-field MRS in breast cancer. NMR in Biomedicine. 2009;22:65–76. doi: 10.1002/nbm.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison T, Bigler L, Tucci M, et al. Salivary sIgA concentrations and stimulated whole saliva flow rates among women undergoing chemotherapy for breast cancer: An exploratory study. Special Care in Dentistry. 1998;18:109–112. doi: 10.1111/j.1754-4505.1998.tb00914.x. [DOI] [PubMed] [Google Scholar]
- Hedman J, Sjoman R, Sjostrom I, Twetman S. Fluoride concentration in saliva after consumption of a dinner meal prepared with fluoridated salt. Caries Research. 2006;40:158–162. doi: 10.1159/000091064. [DOI] [PubMed] [Google Scholar]
- Hirayama A, Kami K, Sugimoto M, et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Research. 2009;69:4918–4925. doi: 10.1158/0008-5472.CAN-08-4806. [DOI] [PubMed] [Google Scholar]
- Holmes E, Loo RL, Stamler J, et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453:396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Arellano M, Boontheung P, et al. Salivary proteomics for oral cancer biomarker discovery. Clinical Cancer Research. 2008;14:6246–6252. doi: 10.1158/1078-0432.CCR-07-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Yu T, Xie Y, et al. Discovery of oral fluid biomarkers for human oral cancer by mass spectrometry. Cancer Genomics and Proteomics. 2007;4:55–64. [PubMed] [Google Scholar]
- Ishikawa Y, Ishida H. Aquaporin water channel in salivary glands. Japanese Journal of Pharmacology. 2000;83:95–101. doi: 10.1254/jjp.83.95. [DOI] [PubMed] [Google Scholar]
- Jani M, Tordai H, Trexler M, Banyai L, Patthy L. Hydroxamate-based peptide inhibitors of matrix metalloprotease 2. Biochimie. 2005;87:385–392. doi: 10.1016/j.biochi.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Jonsson P, Johansson AI, Gullberg J, et al. High-throughput data analysis for detecting and identifying differences between samples in GC/MS-based metabolomic analyses. Analytical Chemistry. 2005;77:5635–5642. doi: 10.1021/ac050601e. [DOI] [PubMed] [Google Scholar]
- Katz A, Smith BL, Golshan M, et al. Nomogram for the prediction of having four or more involved nodes for sentinel lymph node-positive breast cancer. Journal of Clinical Oncology. 2008;26:2093–2098. doi: 10.1200/JCO.2007.11.9479. [DOI] [PubMed] [Google Scholar]
- Katz-Brull R, Margalit R, Degani H. Differential routing of choline in implanted breast cancer and normal organs. Magnetic Resonance in Medicine. 2001;46:31–38. doi: 10.1002/mrm.1157. [DOI] [PubMed] [Google Scholar]
- Katz-Brull R, Seger D, Rivenson-Segal D, Rushkin E, Degani H. Metabolic markers of breast cancer: Enhanced choline metabolism and reduced choline-ether-phospholipid synthesis. Cancer Research. 2002;62:1966–1970. [PubMed] [Google Scholar]
- Khuhawar MY, Memon AA, Jaipal PD, Bhanger MI. Capillary gas chromatographic determination of putrescine and cadaverine in serum of cancer patients using trifluoroacetylacetone as derivatizing reagent. Journal of Chromatography. B, Biomedical Science and Applications. 1999;723:17–24. doi: 10.1016/S0378-4347(98)00478-2. [DOI] [PubMed] [Google Scholar]
- Kind T, Fiehn O. Metabolomic database annotations via query of elemental compositions: Mass accuracy is insufficient even at less than 1 ppm. BMC Bioinformatics. 2006;7:234. doi: 10.1186/1471-2105-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind T, Fiehn O. Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinformatics. 2007;8:105. doi: 10.1186/1471-2105-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K, Asmellash S, Klug CA, et al. Applying proteomic-based biomarker tools for the accurate diagnosis of pancreatic cancer. Journal of Gastrointestinal Surgery. 2008;12:1683–1690. doi: 10.1007/s11605-008-0632-6. [DOI] [PubMed] [Google Scholar]
- Kolch W, Neususs C, Pelzing M, Mischak H. Capillary electrophoresis-mass spectrometry as a powerful tool in clinical diagnosis and biomarker discovery. Mass Spectrometry Reviews. 2005;24:959–977. doi: 10.1002/mas.20051. [DOI] [PubMed] [Google Scholar]
- Li Y, Elashoff D, Oh M, et al. Serum circulating human mRNA profiling and its utility for oral cancer detection. Journal of Clinical Oncology. 2006;24:1754–1760. doi: 10.1200/JCO.2005.03.7598. [DOI] [PubMed] [Google Scholar]
- Li Y, St. John MA, Zhou X, et al. Salivary transcriptome diagnostics for oral cancer detection. Clinical Cancer Research. 2004;10:8442–8450. doi: 10.1158/1078-0432.CCR-04-1167. [DOI] [PubMed] [Google Scholar]
- Lloyd DK. Capillary electrophoresis analysis of biofluids with a focus on less commonly analyzed matrices. Journal of Chromatography. B, Analytical Technologies in the Biomedical and Life Sciences. 2008;866:154–166. doi: 10.1016/j.jchromb.2008.01.058. [DOI] [PubMed] [Google Scholar]
- Mahadevan S, Shah SL, Marrie TJ, Slupsky CM. Analysis of metabolomic data using support vector machines. Analytical Chemistry. 2008;80:7562–7570. doi: 10.1021/ac800954c. [DOI] [PubMed] [Google Scholar]
- Maheshwari SR, Mukherji SK, Neelon B, et al. The choline/creatine ratio in five benign neoplasms: Comparison with squamous cell carcinoma by use of in vitro MR spectroscopy. AJNR American Journal of Neuroradiology. 2000;21:1930–1935. [PMC free article] [PubMed] [Google Scholar]
- Mann GE, Yudilevich DL. Regulation of amino acid influx and efflux at the basolateral plasma membrane of the salivary gland epithelium: Effects of parasympathetic nerve stimulation. Journal of Dental Research. 1987;66:569–575. doi: 10.1177/00220345870660023101. [DOI] [PubMed] [Google Scholar]
- Mao L, Hong WK, Papadimitrakopoulou VA. Focus on head and neck cancer. Cancer Cell. 2004;5:311–316. doi: 10.1016/S1535-6108(04)00090-X. [DOI] [PubMed] [Google Scholar]
- Melvin JE. Chloride channels and salivary gland function. Critical Reviews in Oral Biology and Medicine. 1999;10:199–209. doi: 10.1177/10454411990100020601. [DOI] [PubMed] [Google Scholar]
- Metzger J, Luppa PB, Good DM, Mischak H. Adapting mass spectrometry-based platforms for clinical proteomics applications: The capillary electrophoresis coupled mass spectrometry paradigm. Critical Reviews in Clinical Laboratory Sciences. 2009;46:129–152. doi: 10.1080/10408360902805261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurman JH, Uittamo J. Oral micro-organisms in the etiology of cancer. Acta Odontologica Scandinavica. 2008;66:321–326. doi: 10.1080/00016350802446527. [DOI] [PubMed] [Google Scholar]
- Michell AW, Mosedale D, Grainger DJ, Barker RA. Metabolomic analysis of urine and serum in Parkinson’s disease. Metabolomics. 2008;4:191–201. doi: 10.1007/s11306-008-0111-9. [DOI] [Google Scholar]
- Minami Y, Kasukawa T, Kakazu Y, et al. Measurement of internal body time by blood metabolomics. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9890–9895. doi: 10.1073/pnas.0900617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura M, Kobayashi M, Suzuki F, Shimada J, Sakagami H. Induction of cell death by combination treatment with cisplatin and 5-fluorouracil in a human oral squamous cell carcinoma cell line. Anticancer Research. 2007;27:3331–3337. [PubMed] [Google Scholar]
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA: A Cancer Journal for Clinicians. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- Pickering V, Jordan RC, Schmidt BL. Elevated salivary endothelin levels in oral cancer patients—A pilot study. Oral Oncology. 2007;43:37–41. doi: 10.1016/j.oraloncology.2005.12.027. [DOI] [PubMed] [Google Scholar]
- Plumb R, Granger J, Stumpf C, et al. Metabonomic analysis of mouse urine by liquid-chromatography-time of flight mass spectrometry (LC-TOFMS): detection of strain, diurnal and gender differences. Analyst. 2003;128:819–823. doi: 10.1039/b304296k. [DOI] [PubMed] [Google Scholar]
- Reijenga JC, Martens JH, Giuliani A, Chiari M. Pherogram normalization in capillary electrophoresis and micellar electrokinetic chromatography analyses in cases of sample matrix-induced migration time shifts. Journal of Chromatography. B, Analytical Technologies in the Biomedical and Life Sciences. 2002;770:45–51. doi: 10.1016/S0378-4347(01)00527-8. [DOI] [PubMed] [Google Scholar]
- Reo NV. NMR-based metabolomics. Drug and Chemical Toxicology. 2002;25:375–382. doi: 10.1081/DCT-120014789. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Saito N, Robert M, Kochi H, et al. Metabolite profiling reveals YihU as a novel hydroxybutyrate dehydrogenase for alternative succinic semialdehyde metabolism in Escherichia coli. Journal of Biological Chemistry. 2009;284:16442–16451. doi: 10.1074/jbc.M109.002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer E, Mischak H, Novak J. High resolution proteome/peptidome analysis of body fluids by capillary electrophoresis coupled with MS. Proteomics. 2006;6:5615–5627. doi: 10.1002/pmic.200600230. [DOI] [PubMed] [Google Scholar]
- Schiffer E, Mischak H, Theodorescu D, Vlahou A. Challenges of using mass spectrometry as a bladder cancer biomarker discovery platform. World Journal of Urology. 2008;26:67–74. doi: 10.1007/s00345-007-0234-z. [DOI] [PubMed] [Google Scholar]
- Schnackenberg LK, Sun J, Espandiari P, et al. Metabonomics evaluations of age-related changes in the urinary compositions of male Sprague–Dawley rats and effects of data normalization methods on statistical and quantitative analysis. BMC Bioinformatics. 2007;8(Suppl 7):S3. doi: 10.1186/1471-2105-8-S7-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidransky D. Emerging molecular markers of cancer. Nature Reviews Cancer. 2002;2:210–219. doi: 10.1038/nrc755. [DOI] [PubMed] [Google Scholar]
- Silwood CJ, Lynch E, Claxson AW, Grootveld MC. 1H and (13)C NMR spectroscopic analysis of human saliva. Journal of Dental Research. 2002;81:422–427. doi: 10.1177/154405910208100613. [DOI] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Analytical Chemistry. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Soga T, Baran R, Suematsu M, et al. Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. Journal of Biological Chemistry. 2006;281:16768–16776. doi: 10.1074/jbc.M601876200. [DOI] [PubMed] [Google Scholar]
- Srivastava A, Wang J, Zhou H, Melvin JE, Wong DT. Age and gender related differences in human parotid gland gene expression. Archives of Oral Biology. 2008;53:1058–1070. doi: 10.1016/j.archoralbio.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streckfus CF, Bigler LR, Zwick M. The use of surface-enhanced laser desorption/ionization time-of-flight mass spectrometry to detect putative breast cancer markers in saliva: A feasibility study. Journal of Oral Pathology and Medicine. 2006;35:292–300. doi: 10.1111/j.1600-0714.2006.00427.x. [DOI] [PubMed] [Google Scholar]
- Streckfus CF, Mayorga-Wark O, Arreola D, et al. Breast cancer related proteins are present in saliva and are modulated secondary to ductal carcinoma in situ of the breast. Cancer Investigation. 2008;26:159–167. doi: 10.1080/07357900701783883. [DOI] [PubMed] [Google Scholar]
- Sugimoto M, Kikuchi S, Arita M, et al. Large-scale prediction of cationic metabolite identity and migration time in capillary electrophoresis mass spectrometry using artificial neural networks. Analytical Chemistry. 2005;77:78–84. doi: 10.1021/ac048950g. [DOI] [PubMed] [Google Scholar]
- Tabor CW, Tabor H. Polyamines. Annual Review of Biochemistry. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- Takeda I, Stretch C, Barnaby P, et al. Understanding the human salivary metabolome. NMR in Biomedicine. 2009;22:577–584. doi: 10.1002/nbm.1369. [DOI] [PubMed] [Google Scholar]
- Tiziani, S., Lopes, V., & Gunther, U. L. (2009) Early stage diagnosis of oral cancer using 1H NMR-based metabolomics. Neoplasia. 11, 269–276, 264–269. [DOI] [PMC free article] [PubMed]
- Vissers YL, Dejong CH, Luiking YC, et al. Plasma arginine concentrations are reduced in cancer patients: Evidence for arginine deficiency? American Journal of Clinical Nutrition. 2005;81:1142–1146. doi: 10.1093/ajcn/81.5.1142. [DOI] [PubMed] [Google Scholar]
- Walenta S, Wetterling M, Lehrke M, et al. High lactate levels predict likelihood of metastases, tumor recurrence, and restricted patient survival in human cervical cancers. Cancer Research. 2000;60:916–921. [PubMed] [Google Scholar]
- Wallace WE, Kearsley AJ, Guttman CM. An operator-independent approach to mass spectral peak identification and integration. Analytical Chemistry. 2004;76:2446–2452. doi: 10.1021/ac0354701. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Tzur D, Knox C, et al. HMDB: The human metabolome database. Nucleic Acids Research. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten IH, Frank E. Data mining: Practical machine learning tools and techniques. Boston, MA: Morgan Kaufman; 2005. [Google Scholar]
- Woo HM, Kim KM, Choi MH, et al. Mass spectrometry based metabolomic approaches in urinary biomarker study of women’s cancers. Clinica Chimica Acta. 2009;400:63–69. doi: 10.1016/j.cca.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Yang, C., Richardson, A. D., Smith, J. W., & Osterman, A. (2007). Comparative metabolomics of breast cancer. Pacific Symposium on Biocomputing, 12, 181–192. [PubMed]
- Zhou J, Xu B, Huang J, et al. 1H NMR-based metabonomic and pattern recognition analysis for detection of oral squamous cell carcinoma. Clinica Chimica Acta. 2009;401:8–13. doi: 10.1016/j.cca.2008.10.030. [DOI] [PubMed] [Google Scholar]
- Zimmermann BG, Wong DT. Salivary mRNA targets for cancer diagnostics. Oral Oncology. 2008;44:425–429. doi: 10.1016/j.oraloncology.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurbig P, Mischak H. Capillary electrophoresis coupled to mass spectrometry for biomarker discovery and diagnosis of kidney diseases. Contributions to Nephrology. 2008;160:107–126. doi: 10.1159/000125938. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dot plots for the relative area of detected metabolites in samples from all groups. The colored dots denote healthy controls (blue), oral (red), breast (pink), pancreatic cancer (green), and periodontal disease (purple). The Y- and X-axes denote the relative peak area (no units) and the group name, respectively. The horizontal, center long bars and short top/bottom bars indicate the means and standard deviations, respectively. * p < 0.05, ** p < 0.01 and *** p < 0.001 (Steel–Dwass test). Metabolites showing a significant difference between oral cancer and controls at p < 0.001 (Steel–Dwass test) and matched with the standard library are shown in Figure 2. (PPT 2514 kb)
Score plots of individually developed principal components (PC) analyses. The subjects in all groups are shown in 3-dimensional (a) and 2-dimensional (b) plots. The same data presented without three outliers are depicted in Figure 3. The score plots yielded by independent PC analysis are shown for control subjects versus patients with oral cancer (c), breast cancer (d), pancreatic cancer (e), and periodontal diseases (f). The colored dots indicate control (blue), oral cancer (red), breast cancer (pink), pancreatic cancer (green) and periodontal diseases (purple). The cumulative proportions of the first, second and third PCs (PC1, PC2, and PC3) were 49.6%, 64.0% and 74.6% for (c), 48.9%, 57.4% and 63.5% for (d), 47.5%, 57.6% and 65.1% for (e), and 38.7%, 49.6% and 58.0% for (f), respectively. (PPT 437 kb)
Dot plots for the relative area of creatinine, arginine, phosphocholine, and choline-creatinine ratio. The colored dots denote healthy controls (blue), oral (red), breast (pink), pancreatic cancer (green), and periodontal disease (purple). The Y- and X-axes denote the relative peak area (no units) or ratio of the relative peak area and the group name, respectively. The horizontal, center long bars and short top/bottom bars indicate the means and standard deviations, respectively. * p < 0.05, ** p < 0.01 and *** p < 0.001 (Steel–Dwass test). (PPT 255 kb)
Score plots of principal components (PC) analysis according to sex. The values for males and females are colored blue and red, respectively. Three-dimensional and 2-dimensional PC plots for healthy controls (a) and (c), respectively, and for subjects with oral cancer (b) and (d), respectively, are shown. A total of 42 and 27 male and female control subjects and 41 and 23 male and female patients with oral cancer were included in this analysis. Eighteen control subjects, five patients with oral cancer and all of the patients with other diseases were excluded because of unavailable sex information. The cumulative proportions of PC1, PC2, and PC3 for the control subjects were 43.0%, 52.9% and 60.6%, respectively, and those for patients with oral cancer were 50.2%, 65.7% and 77.5%, respectively. (PPT 218 kb)
Principal components (PC) analysis score plots for race and ethnic groups. The data for African-American, Asian, Caucasian and Hispanic subjects are colored red, blue, green, and orange, respectively. Three-dimensional and 2-dimensional PC plots for healthy controls (a) and (c), respectively, and subjects with oral cancer (b) and (d), respectively, are shown. The race or ethnic groups included 12 and four African-Americans, 15 and five Asians, 37 and 41 Caucasians, and five and five Hispanics as healthy controls and subjects with oral cancer, respectively. Eighteen control subjects, five patients with oral cancer, and all of the patients with other diseases were excluded because of unavailable race or ethnic information. The cumulative proportions of PC1, PC2 and PC3 for the control subjects were 43.0%, 52.9% and 60.9%, respectively, and those for subjects with oral cancer were 42.0%, 63.2% and 70.2%, respectively.(PPT 240 kb)


