Abstract
Splicing regulatory proteins often have distinct activities when bound to exons versus introns. However, less clear is whether variables besides location can influence activity. HnRNP L binds to a motif present in both CD45 variable exons 4 and 5 to affect their coordinate repression. Here we show that, in contrast to its direct repression of exon 4, hnRNP L represses exon 5 by countering the activity of a neighboring splicing enhancer. In the absence of the enhancer hnRNP L unexpectedly activates exon inclusion. As the splice sites flanking exon 4 and 5 are distinct, we directly examined the effect of varying splice site strength on the mechanism of hnRNP L function. Remarkably, binding of hnRNP L to an exon represses strong splice sites but enhances weak splice sites. A model in which hnRNP L stabilizes snRNP binding can explain both effects in a manner determined by the inherent snRNP-substrate affinity.
Keywords: hnRNP L, splicing regulation, alternative splicing, mechanism of regulation, CD45
Introduction
Large-scale analysis of gene expression data has revealed that most human genes have the capacity to encode multiple proteins through the process of alternative splicing (Pan et al., 2008; Wang et al., 2008). Importantly, variant proteins expressed from a single gene via alternative splicing often act in competition or opposition to one another, such that even small changes in the ratio of protein isoforms expressed from a given gene can have a dramatic physiologic effect (Matlin et al., 2005). Therefore, the proteins and mechanisms that control alternative splicing play a critical role in determining protein expression and cellular function.
The catalysis of pre-mRNA splicing is mediated by the “spliceosome” — a macromolecular machine comprised of five small nuclear RNAs (U1, U2, U4, U5, U6 snRNA) and associated proteins that interact with sequences at the exon/intron boundaries (“splice sites”) to direct the excision of introns and ligation of exons (Wahl et al., 2009). The catalytic spliceosome (C complex) is not a pre-formed enzyme, but rather assembles on the pre-mRNA in a stepwise pathway that involves several distinct intermediates (E-A-B complexes; Wahl et al. 2009). In higher eukaryotes, the splice site sequences are highly degenerate and alone do not typically contain sufficient information to accurately determine the sites of cleavage and ligation (Black, 1995; Matlin et al., 2005). It is now widely established that the binding of an exon by the spliceosome is typically controlled by various proteins bound to auxiliary sequences located within exons or flanking introns (Matlin et al., 2005). Interestingly, in many cases the same splicing regulatory protein can enhance inclusion of some exons while promoting skipping of others, although the mechanisms by which such dual effects are conferred remain poorly understood in most cases.
An emerging theme in alternative splicing is that of networks of co-regulated events, in which a single protein coordinates the inclusion or exclusion of exons in multiple genes. For example, coordinate regulation has been demonstrated for genes involved in controlling synaptic plasticity via the neural-specific protein Nova (Ule et al., 2006). Similarly, the neural and muscle-specific proteins Fox-1/2 regulate the splicing of multiple genes involved in neuromuscular function (Zhang et al., 2008). These studies, among others, have introduced the notion of regulatory “maps” that predict the effect of a protein based on location of binding. Critically, however, two inherent assumptions of these maps have not yet been tested. First, does a given protein always functions by the same mechanism when bound to a particular location relative to an exon and, second, is location the sole determinant of mechanism, or can other variables influence how a particular protein functions?
A well studied example of regulated alternative splicing is the CD45 gene, which has three variable exons (4, 5 and 6) that are coordinately skipped upon antigen-induced activation of T cells (Hermiston et al., 2002). Skipping of the CD45 variable exons is regulated, at least in part, by binding of hnRNP L to an activation-responsive sequence (ARS) that is located within each variable exon (Rothrock et al., 2003; Rothrock et al., 2005; Tong et al., 2005). For exons 4 and 6 the ARS motif is embedded within a 60 nt exonic splicing silencer (ESS1) element that is both necessary and sufficient for regulation (Rothrock et al., 2003; Tong et al., 2005). In contrast, the ARS motif in exon 5 is split across two regions (S1 and S2) that are separated by an exonic splicing enhancer sequence (ESE) (Tong et al., 2005; Figure 1). Therefore comparison of the regulation of CD45 exons 4 and 5 provides a powerful system for determining whether the broader sequence context of an exon can influence the mechanisms by which a particular regulatory element and/or associated proteins functions.
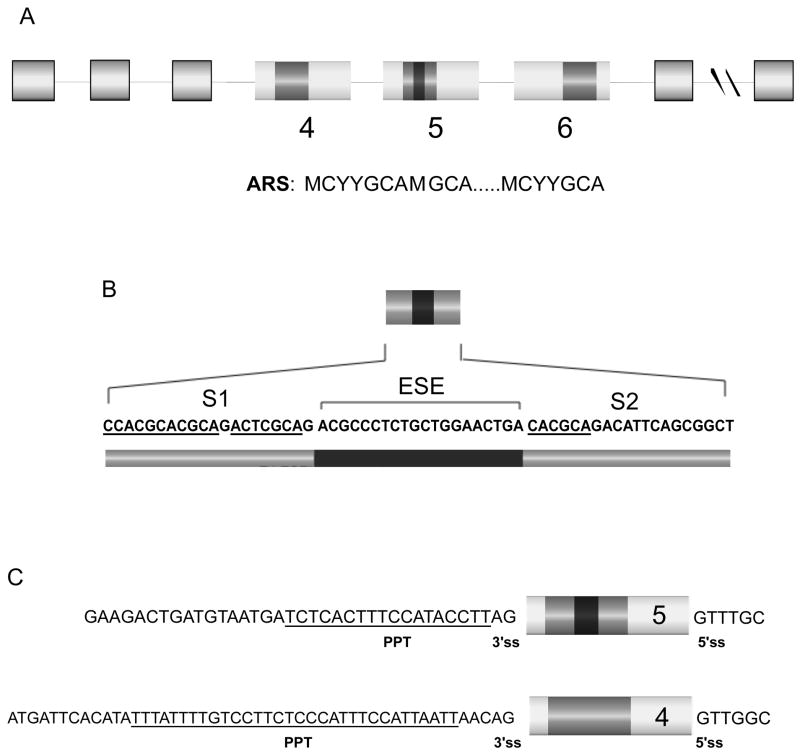
Figure 1. Differential arrangement of the ARS regulatory element in the three variable exons (4, 5, and 6) of the CD45 gene.
(A) Schematic of the human CD45 gene and its three variable exons. Exons and introns are represented by boxes or lines, respectively. The ARS-containing element (darker grey square) is embedded within a single region in exons 4 and 6, however in exon 5 the ARS is divided into two regions by an exonic splicing enhancer (ESE) (black square). The ARS consensus sequence is shown below. (B) Sequence of the regions important for regulation of exon 5: the two ARS-containing sequences, labeled S1 and S2, and the ESE. The ARS-core motif is underlined in both the S1 and S2 elements. (C) Comparison of intronic sequence flanking exons 4 and 5 with polypyrimidine tract underlined.
In this study we show that hnRNP L can repress or activate an exon by distinct mechanisms due, at least in part, to differences in splice site strength. Binding and functional studies demonstrate that hnRNP L bound to the silencers in exon 5 directly competes with SF2/ASF bound to an ESE, inhibiting the ability of the ESE-complex to recruit the U2snRNP to the weak upstream 3’ss. This mechanism is markedly distinct from the previously reported mechanism of direct repression of exon 4 by hnRNP L (House and Lynch, 2006). Because the splice sites flanking exon 5 are weak compared to those of exon 4, we directly examined the effect of hnRNP L binding to exons with varying splice site strengths. Remarkably, in multiple distinct exon contexts we find that hnRNP L represses strong splice sites but enhances weak splice sites. These data provide direct evidence that a given protein can function through different mechanisms in a manner independent of location but constrained by the local sequence context. We propose a unifying model for hnRNP L function in which stabilization of U1 and U2 snRNP binding promotes assembly on weak splice sites or across an intron, but traps these snRNPs in a inactive complex when hnRNP L is bound to an exon flanked by strong splice sites.
Results
HnRNP L binds to the ARS core of exon 5 in the absence of other co-associated proteins observed on exon 4
The ARS-containing ESS1 regulatory element from CD45 exon 4 associates, in resting cells, with several members of the hnRNP family of RNA binding proteins, including hnRNPs L, E2, K, D and PTB (Rothrock et al., 2005; Melton et al., 2007; Figure 2A). Of these multiple hnRNPs, the binding of hnRNP L is most sensitive to mutations of the ARS core motif. Moreover, both in vitro and in vivo studies have confirmed that hnRNP L is the primary mediator of ESS1-dependent repression in resting cells, with the other hnRNPs having little if any functional effect (Rothrock et al., 2005; Melton et al., 2007). Upon cellular stimulation, hnRNP L-like (hnRNP LL) and the hnRNP-related protein PSF join the exon 4 ESS1-associated complex and function in combination with hnRNP L to achieve maximal exon repression (Melton et al., 2007; Oberdoerffer et al., 2008; Topp et al., 2008; Figure 2A).
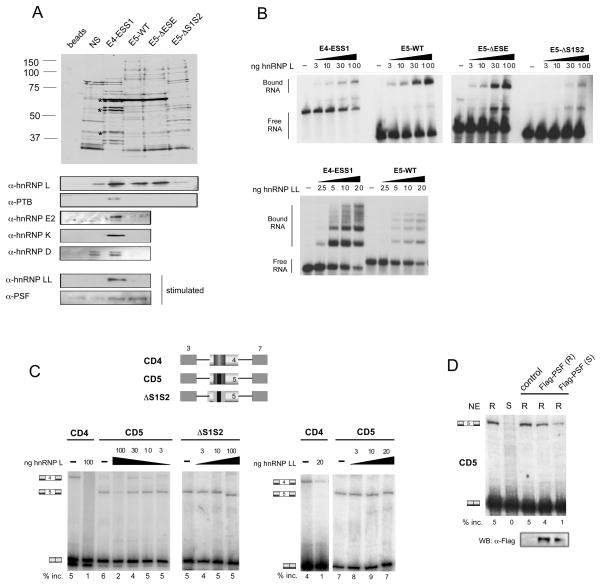
Figure 2. HnRNP L binds to the ARS core of exons 4 and 5 with different co-associated proteins.
(A) Top, silver stain of RNA-affinity pulldowns done with exon 4 (ESS1) and exon 5 (WT, ΔESE, ΔS1S2) probes. Asterisk indicates hnRNP L, PTB, and hnRNP E2. Bottom, Western blot analysis of the same RNA-pulldown samples using antibodies against previously characterized ESS1-binding proteins. (B) RNA mobility-shift experiments of radiolabeled versions of the probes from panel A, incubated with increasing amounts of recombinant hnRNP L (top) or hnRNP LL (bottom) proteins. (C) RT-PCR of in vitro splicing reactions in resting JSL1 nuclear extract supplemented with recombinant hnRNP L (left panel) or hnRNP LL (right panel). Schematics of the minigenes used in these experiments are shown at the top. Hatched boxes correspond to substitution mutation of regulatory sequences. (D) RT-PCR of in vitro splicing reactions of CD5-derived RNA incubated in JSL1 nuclear extract supplemented with Flag-tagged PSF protein purified from resting (R) or stimulated (S) JSL1 cells. Western blot with anti-Flag antibody of protein fractions added to the reactions above.
To compare the function of the ARS motif in exons 4 and 5 we performed RNA-affinity experiments to determine whether the ARS motif in exon 5 recruits a similar or overlapping set of proteins as compared to exon 4. We first determined the proteins that associate with the exon 5 RNA sequence (E5-WT) compared to RNAs that contained substitutions in the enhancer sequence (E5-ΔESE) or ARS motifs (E5-ΔS1S2). As controls, we included the 60-nt ESS1 element from exon 4 (E4-ESS1) and a nonspecific RNA (NS) that we have previously demonstrated to have unrelated silencing activity (Melton et al., 2007). The RNAs were chemically coupled to beads and incubated in nuclear extract from JSL1 T cells. This nuclear extract recapitulates ARS-mediated exon repression in in vitro splicing assays and therefore contains all functionally relevant repressor proteins (Melton et al., 2007; Rothrock et al., 2005; and see below).
Following incubation with extract, the beads were washed extensively, and the RNA-associated proteins were eluted and visualized by silver stain. Consistent with our previous studies, we observed binding of hnRNP L, PTB and hnRNP E2 to E4-ESS1 (Figure 2A). We also detected a strong signal for hnRNP L binding to the E5-WT RNA (Figure 2A). Importantly, replacement of the ARS motifs in exon 5 abolished binding of hnRNP L, while mutation of the ESE in exon 5 had little effect on the association of hnRNP L with the RNA (Figure 2A). The ARS-dependence of hnRNP L binding to exon 5 was further confirmed by RNA mobility shift assays. Titration of purified recombinant hnRNP L with 32P-labeled RNAs (Figures 2B, S1B) or competition between WT and mutant RNAs (Figure S1C) demonstrates that the inherent affinity of hnRNP L is similar for E4-ESS1, E5-WT and E5-ΔESE but was reduced by ~ 10-fold upon mutation of the ARS elements.
Interestingly, neither PTB nor hnRNP E2 were observed to associate with the E5-WT RNA by either silver stain or Western blot nor were the more weakly ESS1-associated proteins hnRNP K and D (Figure 2A). Indeed, of these four proteins only PTB showed any ability to bind exon 5 in mobility shift assays with recombinant protein, and even in this case the affinity of PTB for exon 5 is at least 10-fold lower than for exon 4 (Figure S1D and data not shown). The silver stain as well as UV crosslinking (see below) does suggest that a few proteins in addition to hnRNP L associate with E5-WT, however these proteins are in lower abundance than hnRNP L (Figure 2A) and do not exhibit specificity toward the ARS motif (Tong et al., 2005). Thus, hnRNP L is the major, if not only, protein bound to the exon 5 ARS motif in resting cells.
We also investigated whether PSF and/or hnRNP LL would bind to exon 5 under activated conditions as previously determined for exon 4. Neither PSF nor hnRNP LL are readily detected by silver stain (Melton et al., 2007; data not shown). Nevertheless, Western blot confirms that both PSF and hnRNP LL associate with E4-ESS1 in extracts from cells grown under activated conditions (Figure 2A bottom). Strikingly, PSF also associates with the E5-WT RNA in stimulated extract, however there was no detectable association of hnRNP LL with E5-WT RNA in either resting or stimulated nuclear extracts (Figure 2A, S1). RNA mobility shift assays further confirm that hnRNP LL has markedly reduced affinity for exon 5 relative to exon 4 (Figure 2B, S1B). Given that the molar concentration of hnRNP L is 30–100 times that of hnRNP LL in JSL1 cells (Topp et al., 2008), the reduced affinity of hnRNP LL for exon 5 versus exon 4 is sufficient to explain why we only observe hnRNP L, and not hnRNP LL, binding to exon 5 in nuclear extract. Taken together, these results demonstrate that hnRNP L associates with the exon 5 ARS sequence in isolation under resting conditions, and together with PSF upon stimulation, without the additional binding proteins that associate with the ESS1 sequence from exon 4.
To confirm that the binding of hnRNP L and PSF to E5-WT RNA is functionally related to the regulated repression of exon 5, we performed in vitro splicing assays (Figure 2C). Pre-mRNA transcribed in vitro from a minigene containing exon 4 or exon 5 flanked by constitutive exons 3 and 7 from the CD45 gene was incubated in nuclear extract derived from resting JSL1 cells, and spliced products were detected and quantified by RT-PCR (see Experimental Procedures). Splicing of both the CD45 exon 4 and 5 minigenes in nuclear extract shows a low but detectable level of 3-exon product in the absence of additional recombinant proteins, indicative of the inclusion of exon 4 or 5 (Figure 2C, –hnRNP L). Interestingly, the differential inclusion efficiency between exons 4 and 5 in vitro is not as large as observed in vivo (Rothrock et al., 2003), suggesting that the exon 5 enhancer activity is limiting in extracts.
Addition of hnRNP L to the in vitro splicing reaction results in a decrease in exon 5 inclusion, as observed for exon 4 (Figure 2C). Importantly, this repressive effect of hnRNP L is dependent on the presence of the ARS-containing S1S2 silencers within the exon, since mutation of these sequences abolishes any effect of hnRNP L on exon inclusion (Figure 2C, CD5 vs. ΔS1S2). The repressive effect of hnRNP L is specific, as addition of PTB did not decrease inclusion of exon 5 (Figure S1E). Furthermore, addition of purified hnRNP LL has little to no effect on exon 5 inclusion at concentrations of protein that are sufficient to strongly repress exon 4 (Figure 2C right).
PSF can repress exon 4 only when purified from stimulated cells (Melton et al., 2007). Similarly, recombinant PSF purified from stimulated, but not resting, cells represses inclusion of exon 5 consistent with the repression of exon 5 observed in total nuclear extracts derived from stimulated cells (Figure 2D). Taken together, the binding and functional data demonstrate that PSF participates with hnRNP L in the stimulation-induced silencing of CD45 exon 5, as it does for exon 4, whereas hnRNP LL has little or no effect on the regulation of CD45 exon 5. While the role of PSF in the repression of exon 5 is ultimately of interest, in the remainder of this study we focus solely on the role of hnRNP L in the repression of exon 5 under resting conditions.
SF2/ASF enhances splicing of exon 5 via the ESE
The experiments above demonstrate that hnRNP L binding to the ARS motifs is the primary mediator of basal repression of CD45 exon 5, as it is for exon 4. We know however that the enhancer sequence in exon 5 is also important for the recognition and regulation of this exon (Tong et al. 2005 and see below). Therefore, our next step was to identify the trans-acting factor(s) that bind to, and function on, the exon 5 ESE. As the ESE activity is limiting in our nuclear extracts relative to hnRNP L, RNA-affinity approaches to identify the ESE-binding protein were unsuccessful. Therefore, we used computational methods to identify candidate ESE-binding protein(s). Interestingly, we found that the strongest enhancer motifs within exon 5 predicted by RESCUE-ESE (http://genes.mit.edu/burgelab/rescue-ese/, Fairbrother et al., 2002) overlapped with a binding site of SF2/ASF predicted by ESEfinder (http://rulai.cshl.edu/tools/ESE2/, Cartegni et al., 2003). Therefore we sought to determine if SF2/ASF enhanced exon 5 splicing through the E5-ESE.
Consistent with the ESEFinder results, mobility shift assays demonstrate that recombinant SF2/ASF does bind to exon 5 in an ESE-dependent manner (Figure 3A top). Endogenous SF2/ASF in JSL1 nuclear extract also binds to the exon 5 ESE as demonstrated by the ability of anti-SF2/ASF antibody to retard RNA-associated complexes in a mobility shift assay (Figure 3A bottom), as well as by western blot of RNA affinity experiments (Figure 3B). Notably, in both of these experiments, binding of SF2/ASF is observed on both E5-WT and E5-ΔS1S2 RNAs; however, substitution of the ESE essentially abolished binding. Furthermore, a second 30 kD SR protein, 9G8, displayed no binding in any of our assays (Figure 3A and data not shown).
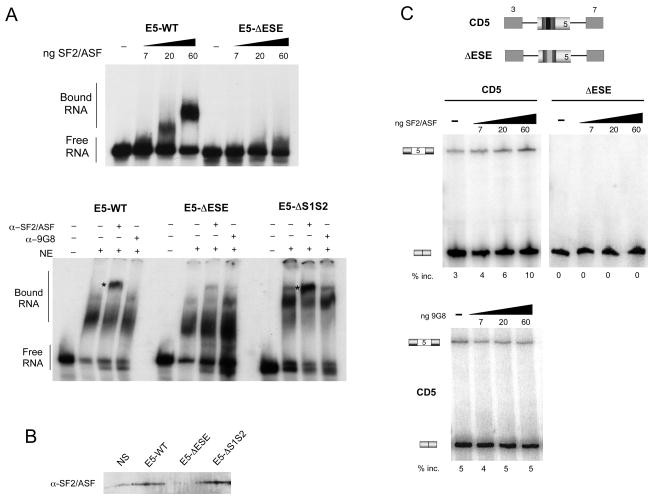
Figure 3. SF2/ASF is a candidate ESE-binding protein of CD45 exon 5.
(A) Top, RNA-mobility shift experiment using radiolabeled E5-WT and ESE RNAs and recombinant SF2/ASF. Bottom, mobility shift assay done with indicated RNAs in JSL1 nuclear extract, in the absence (−) or presence (+) of anti-SF2/ASF (α-SF2/ASF) or anti-9G8 (α-9G8) antibody. Super-shifted complexes are indicated with asterisk. (B) Western blot with anti-SF2/ASF of RNA-affinity pulldowns done with nonspecific (NS) and exon 5 (E5, ΔESE, ΔS1S2) probes as in Figure 2A. (C) RT-PCR of in vitro splicing reactions done with indicated RNAs in JSL1 nuclear extract supplemented with recombinant SF2/ASF (top) or 9G8 (bottom). The numbers shown below each panel represent the mean exon inclusion, n=3.
The binding of SF2/ASF to the ESE within exon 5 is functionally significant, as predicted from our binding data, since addition of recombinant SF/ASF protein to in vitro splicing reactions increased the level of exon 5 inclusion in a dose-dependent manner (Figure 3C, CD5). Importantly, SF2/ASF has no enhancement activity on a substrate that lacks the ESE (Figure 3C, ΔESE). In addition, 9G8, which does not bind exon 5, also has no effect on splicing (Figure 3C bottom). Therefore, we conclude that SF2/ASF binds specifically to the ESE element within exon 5 and functions to enhance CD45 exon 5 splicing.
HnRNP L represses CD45 exon 5 by blocking the activity of the ESE
Having identified hnRNP L as the primary repressor of exon 5 splicing in resting cells and SF2/ASF as an enhancer of this exon, we investigated if the interplay of these two activities might influence the mechanism of hnRNP L repression. To this end we carried out a systematic deletion analysis of the exon 5 regulatory sequences (ESE and S1S2) alone or in combination. In these cell-based assays we used minigenes in which exon 5, or derivatives thereof, is flanked by exons from the human β-globin gene (SC5, Figure 4A). These minigenes were stably expressed in our JSL1 cell line, RNA was isolated from resting (−PMA) or activated (+PMA) cells, and the spliced mRNA products from these minigenes were assayed by RT-PCR. Deletion of the ARS-containing motifs decreased the level of exon skipping compared to that seen with the WT minigene (Figure 4A, SC5 8.5% vs. S1S2 1.5% skipping). Importantly, the signal-induced decrease in exon inclusion was particularly dependent on the presence of the silencers within exon 5, consistent with the ARS motif functioning to confer both basal and activation-induced silencing in the wildtype context (Figure 4A).
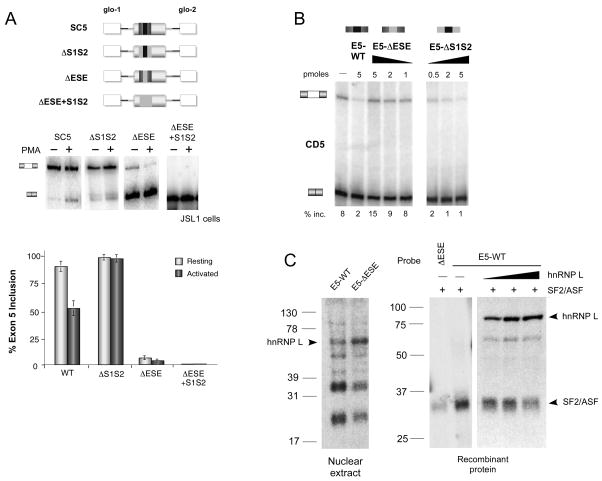
Figure 4. The ARS motifs in exon 5 repress the exon by antagonizing the activity of the ESE.
(A) RT-PCR analysis of RNA derived from resting (−PMA) or stimulated (+PMA) JSL1 clones that stably express WT (SC5) and mutant (ΔS1S2, ΔESE, ΔESE+S1S2) exon 5 minigenes, schematics of which are shown at the top. White boxes and black lines correspond to sequence from the human β-globin gene. Rest of coloration is consistent with Figures 1 and 2. Bottom, mean percent inclusion of exon 5 +/− SD, n>6. (B) RT-PCR of in vitro splicing reactions using WT CD5 substrate in the absence (−) or presence of increasing amounts of various exogenous RNA competitors. Mean % inclusion is shown below, n>3. (C) Left, UV crosslinking of radiolabeled exon 5 probes (WT and ESE) with JSL1 nuclear extract or recombinant proteins as indicated.
In contrast to the data shown above, substitution of the ESE in exon 5 almost entirely abolished exon inclusion in resting cells (Figure 4A, ΔESE; Tong et al., 2005). One of two possible models explains these data. First, the increase in exon 5 repression observed with the ΔESE minigene could be due to the S1S2 sequences directly repressing exon 5, in which case we would expect an increase in the level of exon 5 inclusion if the S1S2 sequences were removed in the ΔESE background. Alternatively, the drop in exon 5 inclusion from ~80% to ~10% upon substitution of the ESE could also be due simply to the loss of the enhancer element which could be crucial for recognition of exon 5 by the spliceosome. This model would predict that deletion of the adjacent S1S2 sequences would result in no change in the inclusion of exon 5. Our data supports the second of these two models, as we observed no increase in the level of exon inclusion between the ΔESE and ΔESE+S1S2 minigenes (Figure 4A). This result suggests that the “silencers” have no silencer activity on their own in the absence of the ESE and that the decrease in exon inclusion in the ΔESE construct is due solely to the loss of the ESE in the exon. Interestingly, we actually note a decrease in exon inclusion with ΔESE+S1S2 minigene, compared to ΔESE alone, consistent with some residual enhancer activity from the S1S2 sequence (see below). However, comparison of the ΔS1S2 minigene to the ΔESE+S1S2 minigene clearly indicates that the ESE alone has the vast majority of the normal enhancer activity for this exon and functions completely independently of the silencers (Figure 4A, 99% inc. ΔS1S2 vs. 1% inc. ΔESE+S1S2).
The fact that the S1S2 sequences have no repressive activity in exon 5 in the absence of the ESE suggests that the silencers function to directly counter the ESE activity. To determine if this interplay between the enhancer and silencers is due to competition of binding by hnRNP L and SF2/ASF, we next tested the effects of titrating various exogenous competitor RNAs into an in vitro splicing assay with the standard CD5 minigene. Upon addition of exogenous E5-WT RNA we observed a notable decrease in the level of exon 5 inclusion, suggesting that the E5-WT RNA titrates more SF2/ASF than hnRNP L away from the substrate RNA, leading to a loss of exon enhancement (Figure 4B). Addition of the competitor lacking the silencers (E5-ΔS1S2) reduced exon inclusion at even lower concentrations than the E5-WT competitor (Figure 4B, 0.5 pmol ΔS1S2 vs. 5 pmol E5-WT), consistent with the lack of hnRNP L binding to the ΔS1S2 RNA allowing more efficient recruitment and sequestration of SF2/ASF. In contrast, addition of the E5-ΔESE RNA competitor increases the level of exon inclusion in a dose dependent manner (Figure 4B) as we would expect if this E5-ΔESE RNA primarily sequesters the repressive hnRNP L via the remaining S1S2 sequences. Importantly, in control experiments the ΔESE competitor does not increase exon use in a CD5 ΔESE substrate (Figure S2A), confirming that binding of hnRNP L to the ARS sequences in exon 5 has no inherent silencing activity in the absence of the ESE.
Together these data strongly argue that there is direct competition between SF2/ASF and hnRNP L to bind exon 5; and that the balance of these competing activities ultimately determines the extent of exon inclusion. This model is also supported by gel shift assays in which the E5-WT complex migrates at a diffuse midpoint between that observed for the E5-ΔESE and E5-ΔS1S2 RNAs (Figure 3A, S2B) suggesting that the E5-WT RNA binds a mixture of the “enhancer-complex” and the “silencer-complex”. We further characterized these complexes by supershifting with antibodies to hnRNP L and SF2/ASF. As we predicted, the complex assembled on the E5-WT RNA contains both proteins (Figure 3A, S2C). In contrast, mutation of the ESE abolished binding of SF2 (Figure 3A, E5-ΔESE) while mutation of the S1S2 abolished binding of hnRNP L (Figure S2C, E5-ΔS1S2).
To provide direct evident for a competition model, we carried out UV crosslinking assays. Crosslinking with JSL1 nuclear extract showed that a 70 kDa protein, which we have previously shown by immunoprecipitation to be hnRNP L (Tong et al. 2005), associates more strongly with the E5 RNA when the ESE is mutated (Figure 4C left). In contrast, two additional bands are markedly reduced upon mutation of the ESE, demonstrating differential association of exon 5 by these ESE binding proteins versus hnRNP L. As the available SF2/ASF antibody is not adequate to conclusively demonstrate that SF2/ASF is among the proteins that associate in an ESE-specific manner, we also carried out UV crosslinking assays with recombinant hnRNP L and SF2/ASF proteins (Figure 4C right). This experiment confirms that SF2/ASF crosslinks to exon 5 in a largely ESE-dependent manner. More importantly, titration of an increasing concentration hnRNP L to the reaction resulted in reduced SF2/ASF binding to the E5-WT substrate, thus confirming direct competition between hnRNP L and SF2/ASF for binding to exon 5.
HnRNP L repression of CD45 exons 4 and 5 occurs by distinct mechanisms
Taken together, the data in Figure 4 demonstrate that hnRNP L bound to the silencer sequences in exon 5 represses inclusion by directly competing with the activity of a critical enhancer element bound by SF2/ASF. This mechanism is surprisingly distinct from the repression of exon 4 in which hnRNP L blocks inclusion by directly stalling spliceosome assembly after the ATP-dependent addition of the U1 and U2 snRNPs (House and Lynch, 2006). Therefore, to better define the mechanism by which hnRNP L functions on exon 5 we next investigated what step in spliceosome assembly is regulated by the ESE and S1S2 silencers.
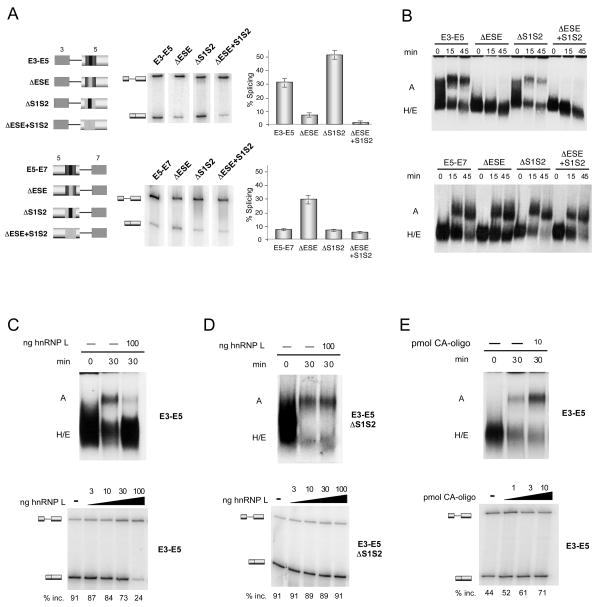
To facilitate analysis of spliceosome assembly we made single intron minigene constructs that consist of the upstream exon 3 and exon 5 (E3–E5) or exon 5 and the downstream exon 7 (E5–E7) (Figure 5A). Intron retention in these minigenes corresponds to exon 5 repression. Splicing of the WT E3–E5 RNA is efficient in nuclear extract from resting JSL1 cells. However, deletion of the ESE from the E3–E5 construct greatly abrogates the efficiency of splicing compared to the wildtype version (Figure 5A top; ~30% to ~6%, p<0.0002), demonstrating that the enhancer element is functional in this single-intron minigene. Consistent with regulation of the complete exon 5 minigene, mutation of the silencers increases splicing efficiency in the E3–E5 background, but only of a construct that contains the enhancer (Figure 5A top, 30% to 53%, p<0.001), indicating once again that the silencers in exon 5 function specifically to counter the enhancer.
Figure 5. The ESE in exon 5 activates the formation of A-complex on its upstream intron.
(A) RT-PCR of in vitro splicing reactions. Schematics of each of the minigenes used are shown on the left. Graph represents mean +/− SD from 3 independent experiments. (B) Radiolabeled RNA substrates derived from each of the minigenes shown in panel A were incubated in nuclear extract for the times indicated and the resulting spliceosome complexes were resolved on native agarose gels. (C) Assembly and RT-PCR analysis done in the absence (−) or presence (+) of 100 ng of recombinant hnRNP L protein. (D) Same as panel C except with ΔS1S2 substrate (E) Same as in panel C, except reactions were incubated in the absence (−) or presence (+) of 10 pmol of CA-oligo.
By contrast, we find that the exon 5 enhancer is not functional on the downstream intron, as removal of the ESE from the E5–E7 construct does not decrease splicing efficiency either in the presence or absence of the silencers (Figure 5A bottom). The increase in splicing efficiency upon removal of the ESE in the wildtype context appears to be due a spurious context effect as this is not seen in the absence of the silencers and is counter to the effect of deleting the ESE in the full minigene. Removal of the silencer sequences also has no effect on the splicing efficiency of the E5–E7 substrate (Figure 5A bottom). Together these data indicate that the intron downstream of exon 5 is essentially refractory to control by the core exonic regulatory sequences and that the ESE, and thus the S1S2 silencers, function primarily on the upstream intron to regulate inclusion of exon 5. Importantly, the fact that the E3–E5 minigene is regulated in a manner consistent with the full CD5 minigene also provides further proof that the silencing of exon 5 is mechanistically distinct from that of exon 4, as single-intron constructs are unable to form the exon-defined complex required for repression of exon 4 and thus do not recapitulate the silencing of exon 4 (House and Lynch, 2006).
We next analyzed the single-intron splicing reactions from Figure 5A on nondenaturing agarose gels to separate the different spliceosome intermediates. Spliceosome assembly on E3–E5 progresses efficiently in resting nuclear extract, as a complex, confirmed to be the pre-spliceosomal A complex by its dependence on ATP and the U2 snRNP, is readily detected after a 30 minute incubation (Figure 5B, S3). We are unable to resolve the subsequent B and C complexes on these gels, most probably due to the limited resolution capacity of the agarose, however these complexes must also form efficiently since up to 30% of the E3–E5 pre-mRNA is converted to spliced product (Figure 5A top). Remarkably, no detectable A complex is formed on the E3–E5ΔESE substrate, consistent with the significant loss of splicing we observe upon substitution of the ESE (Figure 5B, S3A). By contrast, the efficiency of A complex formation on an E3–E5 substrate lacking the S1S2 sequences is the same or greater than for the WT construct. Importantly, deletion of the silencers in the background of the ESE mutation does not restore A complex formation (Figure 5B, S3A) demonstrating that the loss of A complex formation on the E3–E5ΔESE substrate is again a direct result of loss of the enhancer and not a result of residual silencer activity. We also observe efficient A complex formation on the E5–E7 substrate, however, formation of this complex is not dependent on the presence of the ESE or S1S2 silencers alone or in combination (Figure 5B bottom, S3A). These results are consistent with the splicing of the E5–E7 construct not decreasing upon deletion of the ESE or the S1S2 sequences.
The loss of A complex upon removal of the ESE in the E3–E5 substrate could be due to a direct block in A complex formation, or alternatively, to inhibition of the earlier E complex. Formation of E complex involves the initial ATP-independent recognition of the 5′ and 3′ splice sites by the U1 snRNP and U2AF protein respectively, and precedes the ATP-dependent loading of U2 snRNP in A complex. To directly assess formation of E complex we incubated the E3–E5 and E5–E7 substrates in the absence of ATP, and then resolved the assembly reactions on agarose gels optimized to resolve H and E complexes. The identity of E complex was confirmed by its dependence on U1 snRNA and sensitivity to heparin, two well established hallmarks of E complex (Figure S3D, E). Notably, we observed no significant difference in the efficiency of E complex formation in the absence or presence of ESE in exon 5 with both the E3–E5 or E5–E7 substrates (Figure S3D). Thus the ESE within exon 5 is not required for the initial recognition of this exon during E complex formation. Rather we conclude that the ESE within exon 5 promotes A complex formation on the upstream intron, presumably by recruiting the U2 snRNP to the 3′ splice site region upstream of exon 5.
The data above suggest that hnRNP L represses exon 5 by inhibiting the ability of the enhancer complex to recruit the U2snRNP. To directly test this model, we analyzed the effect of modulating levels of hnRNP L on formation of A complex on the E3–E5 minigene. Strikingly, addition of excess recombinant hnRNP L to an assembly reaction specifically inhibited A complex formation, coincident with decreasing splicing efficiency (Figure 5C). Importantly, this inhibitory effect of hnRNP L is dependent on the presence of the ARS-containing silencer sequences as hnRNP L has no effect on the formation of A complex for the E3–E5 ΔS1S2 substrate (Figure 5D). In a reverse experiment, we used a poly-CA oligo to functionally deplete hnRNP L, as has been done in previous studies (Hui et al., 2003). Such depletion of hnRNP L results in an increase in both splicing and A complex formation (Figure 5E), thereby confirming that hnRNP L normally represses A complex formation on the intron upstream of exon 5.
Importantly, this mechanism is entirely distinct from that by which hnRNP L represses exon 4, in which U1 and U2 snRNPs are not blocked from assembling around the exon but rather are stalled after binding to the substrate in an A-like exon-defined complex (AEC) (House and Lynch, 2006). Therefore, we propose that hnRNP L regulates exon 5 by a distinct mechanism from that of exon 4 due to inherent differences in the efficiency of snRNP assembly on the flanking splice sites. In particular, as U2 snRNP is not stably bound upstream of exon 5 in the absence of the ESE activity, we propose that preventing U2 association by inhibiting enhancer activity is a more efficient mechanism of regulation than suppressing assembly after U2 association (see discussion).
HnRNP L binding represses exons with strong splice sites but activates weak splice sites
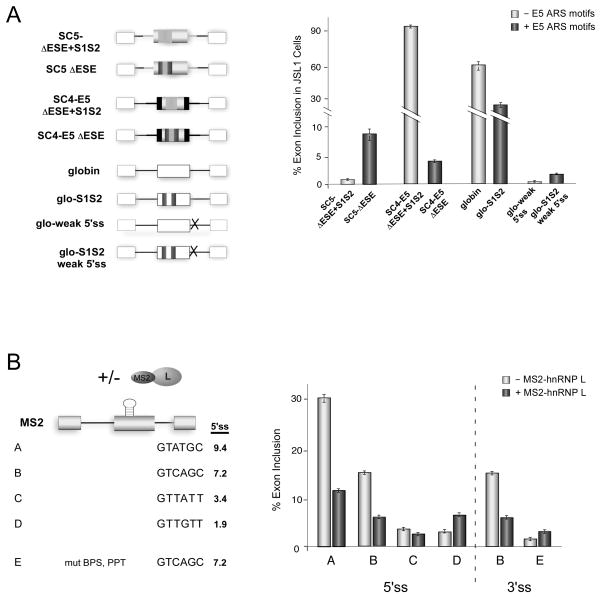
Closer inspection of the exon 4 and exon 5 sequences pointed to a difference in flanking 3’ss strength. We noticed that compared to exon 4, exon 5 has a noticeably shorter polypyrimidine tract making it an intrinsically weaker 3’ss (Figure 1C). This initial observation offered a rationale for the differential mechanism of regulation of exon 4 and 5, wherein we would predict that hnRNP L can only stall an AEC complex when the binding of U1 and/or U2 snRNPs to the flanking splice sites is inherently strong. Consistent with this idea, is our observation that in the absence of the ESE element not only do the S1S2 sequences not silence exon 5, they in fact have a significant enhancing effect on an otherwise unused exon (Figures 3A, 5A and S4A, 1.1% to 8.8%). This enhancing activity is in striking contrast to the repressive activity of the exon 5 S1S2 sequence when placed in the background of exon 4 (Figure 6A, S4A; SC4-E5ΔESE+S1S2 vs SC4-E5ΔESE). These data demonstrate that the hnRNP L-binding sequence from exon 5 can have either a positive or negative effect on exon inclusion depending on context. The most notable distinction between the exon 5 and exon 4 backgrounds is the overall level of exon inclusion in the absence of the S1S2 sequence (Figure 6A, S4A, SC5ΔESE+S1S2 1% vs. SC4-E5ΔESE+S1S2 93.7%), which is consistent with the strength of both the 5′ splice site and length of the polypyrimidine track flanking these exons differing markedly (Figure 1C).
Figure 6. HnRNP L represses strong splice sites but activates weak splice sites.
(A) Mean exon inclusion +/− SD from RT-PCR of stable cell lines expressing the minigenes shown, done in triplicate. Black boxes and bold black lines represent exonic and intronic sequence from CD45 exon 4 respectively, rest of coloration is consistent with other figures. Glo-weak and glo-weak S1S2 minigenes carry mutations in the 5’ss downstream of the central exon. (B) Mean exon inclusion +/− SD from triplicate in vitro splicing reactions, done in the absence or presence of MS2-hRNP L, using RNAs transcribed from minigenes shown. Numbers shown for 5’ss represent score for 5′ splice site strength (http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html).
To determine if splice site strength is a determinant in whether the S1S2 sequences function as an enhancer or silencer we inserted them into a β-globin test exon and then weakened the 5′ splice site (Figure 6A, globin and glo-S1S2). The globin minigene alone has a relatively high level of exon inclusion, and the presence of the S1S2 sequence causes exon repression (Figure 6A, S4A). Remarkably, however, we find that when the 5’ss flanking the β-globin central exon is weakened the presence of the S1S2 sequences enhanced exon inclusion by ~ 6 fold (0.3 % to 1.9% exon inclusion, Figure 6A, S4A).
To test if the above correlation between directionality of hnRNP L regulation and splice site strength is a general phenomena, and to isolate affects of hnRNP L away from potential co-binding proteins, we engineered a single-hairpin binding site for the MS2 coat protein into a chimeric exon consisting of β-globin splice sites fused to splicing-inert sequence from CD45 exon 9 used to lengthen the exon to ~ 200 nts (Rothrock et al., 2003). Inclusion of this MS2 test exon was highly efficient in vitro, as expected since the splicing sites flanking this exon are strong (Figure 6B, construct A). Notably, addition of partially purified MS2-hnRNP L to the in vitro splicing reactions markedly repressed exon inclusion, consistent with hnRNP L functioning as a silencer of exons with strong splice sites. MS2-hnRNP L had no effect on the splicing of a related minigene lacking the MS2 binding site, and MS2 protein alone had no silencing activity on the MS2 hairpin-containing substrate, confirming that repression is due to the hnRNP L component of the fusion protein and that exon binding is required for repression (Figure S4B and C). We also confirmed that the binding of MS2-hnRNP L to the RNA does not alter message stability (Figure S4D).
We next systematically weakened the 5′ or 3′ splice site signals flanking the MS2 test exon. As shown in Figure 6B, partial weakening of exon efficiency results in a loss of responsiveness to hnRNP L binding (Figure 6B, construct C). Strikingly, however, when the 5′ splice site was rendered weakest, binding of MS2-hnRNP L actually enhanced exon inclusion by two to three fold (Figure 6B, S4E, construct D, 2.6% to 6.9%). This result was not solely specific for weak 5′ splice sites as weakening of the upstream branch point sequence and polypyrimidine track also resulted in MS2-hnRNP L functioning as an activator (Figure 6B, constructs E, 1.6% to 3.4%). Therefore, we present here three distinct systems in which hnRNP L functions as a repressor of efficient exons; however, once the absolute level of exon inclusion is below ~2–3 %, hnRNP L functions as an enhancer. These data emphasize that variables in addition to binding location can alter the effect a given regulatory protein has on the assembly pathway of the spliceosome.
Discussion
Previous studies have shown that CD45 variable exons 4 and 5 are independently repressed through the activity of the ARS core motif sequence and its cognate binding protein hnRNP L (Rothrock et al., 2005; Tong et al., 2005). Here we demonstrate that, remarkably, unique features of the sequence context of the ARS element in exon 5 result in hnRNP L functioning to repress this exon by a distinct mechanism from that described for exon 4. Moreover, by extending the analysis of the role of splice site strength in determining regulatory mechanism, we find that weakening of splice site strength can flip the activity of exon-bound hnRNP L binding from a repressor to an activator. These data provide direct evidence that a given protein can exert different effects on the spliceosome and that for any given exon the mechanism by which a protein functions is constrained by the rate-limiting step in spliceosome assembly.
Mechanism of repression of CD45 exon 5 by hnRNP L
Our data argue that the difference in splice site strength between exons 4 and 5 necessitates the distinct mechanisms by which hnRNP L affects coordinated repression of these exons. Specifically, exon 5 cannot efficiently recruit the U2 snRNP in the absence of the ESE, therefore hnRNP L is unlikely to be able to trap a bound U2 snRNP on constructs lacking the ESE, as occurs on exon 4. Indeed, in the absence of the ESE, hnRNP L likely promotes U2 snRNP recruitment to a limited extent (see below). However when the enhancer is present it strongly promotes A complex formation effectively strengthening the splice sites and shifting the rate-limiting step in spliceosome assembly. In this context, the presence of hnRNP L is able to repress exon usage by competing with binding of SF2/ASF to the ESE thereby removing the enhancer activity. The resulting loss of the SF2/ASF enhancer activity causes a bigger decrease in splicing than is compensated for by the “enhancer” activity of hnRNP L, therefore the net result of hnRNP L in this context is a reduction of exon inclusion.
A remaining question we have not addressed in this study is how the recruitment of PSF to exon 5 alters the mechanism of repression upon cellular stimulation. Interestingly, while the basal silencing activity of the ARS motifs in exon 5 requires the presence of the ESE, activation-induced silencing of exon 5 still occurs in the absence of this enhancer (Figure 4A), indicating that the mechanism of activation-induced repression of exon 5 is distinct from the basal repression by hnRNP L. One explanation for these results is that the addition of PSF to the ARS complex could directly inhibit the enhancement activity of hnRNP L, perhaps by blocking the ability of hnRNP L to recruit the U2 snRNP. Alternatively, PSF could be directly antagonizing spliceosome assembly formation on exon 5. This latter possibility is consistent with the fact that addition of PSF to the exon 4 silencer complex does not change the mechanism of repression (A.E. House and K.W. Lynch, unpublished), suggesting that PSF helps hnRNP L trap the U1/U2 snRNP-containing AEC complex from progressing on in assembly. However, further studies will be required to understand how the addition of PSF upon cellular activation alters the mechanism of exon 5 silencing.
A unified model for hnRNP L function
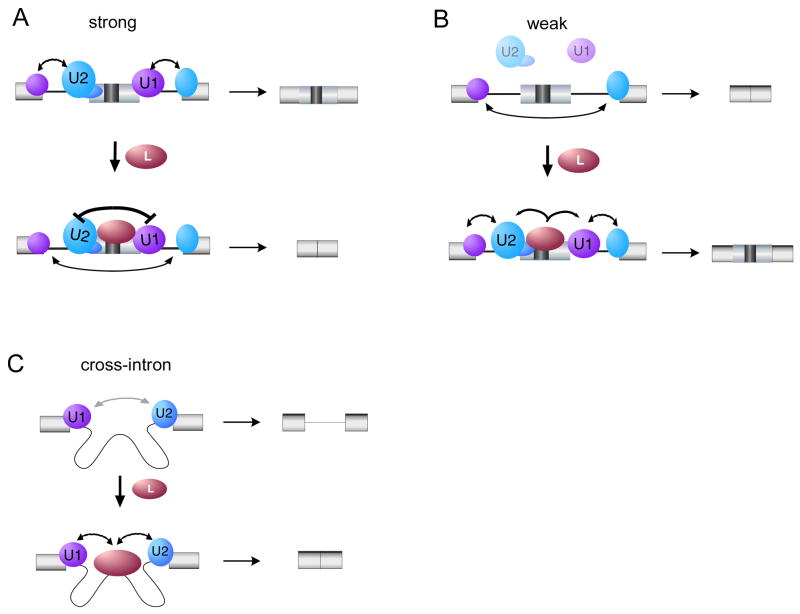
The correlation between splice site strength and mechanism of regulation can best be understood by a model in which the overall efficiency of snRNP association across an exon needs to be strong enough to promote exon definition, but not so strong as to prevent remodeling to the cross-intron interactions required for catalysis (Figure 7). In this model hnRNP L interacts directly, or indirectly, with the U1 and U2 snRNPs to promote their association with the substrate. In the case in which hnRNP L is bound to an exon flanked by strong splice sites, we predict the sum total of the interaction between U1-hnRNP L-U2 and the RNA sequesters the snRNPs in an inactive conformation that cannot progress further on in the assembly pathway (Figure 7A). This interpretation is consistent with our data regarding repression of CD45 exon 4, including the fact that the stalled AEC is more resistant to salt and heparin destabilization than is a canonical A complex. (House and Lynch, 2006; N. Chiou and K.W.L., unpublished). Moreover this model is consistent with recent data from the Nilsen group demonstrating that subtle perturbations in snRNP-splice site interactions can alter the efficiency of subsequent assembly steps (Yu et al., 2008).
Figure 7. Model for hnRNP L function.
(A) Interaction of hnRNP L with U1 and U2 snRNPs bound to strong flanking splice sites sequesters them in an inactive conformation that cannot progress further in the spliceosome assembly pathway. (B) However, if an ARS-containing exon is flanked by weak splice sites, then the interaction between U1 and U2 snRNPs and the exon-intron boundary is highly inefficient. In such a case interaction of hnRNP L with U1 and U2 may stabilize their interaction with the splice sites thus promoting progression through assembly pathway. (C) If the hnRNP L-binding sites are located within an intron then the interaction of U1 and/or U2 with hnRNP L would be predicted to bring these snRNPs together in a productive complex.
In contrast, if the splice sites are so weak that recruitment of U1 or U2 snRNP is essentially absent (consistent with the <3% inclusion of exons E5ΔESE+S1S2, glo-weak 5’ss, MS2-construct D and E; Figure 6), then the interaction of exon-bound hnRNP L with the snRNPs likely stabilizes the otherwise transient recruitment of the snRNPs to the flanking splice site(s) (Figure 7B). This model also provides an explanation for why intermediate strength splice sites (such as MS2-C, Figure 6B) are essentially refractory to regulation by hnRNP L, as these would likely be in a range in which their interactions with the snRNPs are sufficient to not be helped by hnRNP L, but also not so strong that snRNPs can be “trapped” by hnRNP L. Interestingly, hnRNP L has been shown to increase inclusion of at least five exons that contain putative hnRNP L binding sites (MYL6, FAM48, PAPOLA (Hung et al., 2008); ERK1, GCK, A. Tong and K.W. Lynch, unpublished), although the mechanism of such enhancement has not been investigated. Interestingly, all five of these exons are flanked by short polypyrimidine tracts and/or 5′ splice sites, consistent with the model we propose here. Our data here therefore suggest a possible explanation for this activity of hnRNP L. However, further investigation will be required to determine if splice site strength is the sole determinant of directionality of hnRNP L on these exons.
An appealing aspect of the above model is that it also accommodates the enhancing effect of hnRNP L that has been observed for several genes when this protein is bound within an intron (Hui et al., 2003). It is easy to imagine that co-association of U1, hnRNP L and U2 across an intron would promote cross-intron pairing of the snRNPs, thereby promoting subsequent spliceosome assembly (Figure 7C). Taken together the data presented in this study indicate that the same protein, through the same molecular interactions, can differentially influence spliceosome assembly in a manner that is determined at least in part on the strength of the flanking splice sites.
However, this is not to say that location is not also an important arbitrator of regulation. Indeed, the data alluded to in Figure 7 demonstrates that location of hnRNP L binding (intronic versus exonic) can strongly influence regulatory outcome, and numerous other examples of location-dependent mechanism have been well-characterized (Ule et al., 2006; Zhang et al., 2008). Moreover, splice site strength is unlikely to be the only aspect of context that influences splicing mechanism. Binding of additional proteins to flanking regulatory elements (Matlin et al., 2005) and neighboring RNA motifs and/or structure (Yu et al., 2008) are just two other examples of additional context differences that have been shown to alter the sensitivity of a gene to a particular regulatory protein. Therefore, we conclude that the mechanism by which a particular protein regulates any given exon cannot be solely attributed to either location or context, but rather relates to how that protein impinges on the rate-limiting step in assembly of the spliceosome on that exon, and how this relates to the efficiency of competing assembly pathways on the same transcript.
Experimental Procedures
Minigenes
Constructs SC5, CD4 and CD5 were previously described in Tong et al. (2005) and Rothrock et al (2003). Construction of additional plasmids is described in Supplemental Materials. Oligonucleotides encoding the 100 nt E5-WT, −ΔESE, and −ΔS1S2 were cloned directly downstream of a T7 polymerase promoter and served as minigene templates for transcription of competitor RNAs and RNA probes in the absence or presence of 32P-CTP.
Nuclear extract and recombinant proteins
Nuclear extract was purified from JSL1 cells using a standard protocol previously described in Rothrock et al., (2005). Purification of recombinant proteins is described in Supplemental Materials.
Splicing and protein binding analysis
RT-PCR, in vitro splicing, RNA-affinity purification, UV crosslinking, gel shift analysis, and spliceosome assembly assays were done as described previously (Rothrock et al., 2005; House and Lynch, 2006). Additional experimental details are available online (Supplemental Methods).
Supplementary Material
Acknowledgments
We thank Nicholas Conrad and members of the Lynch laboratory for their helpful discussion and comments. This work was supported by US National Institutes of Health grant R01 GM067719 to K.W.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Black DL. Finding splice sites within a wilderness of RNA. RNA. 1995;1:763–771. [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. NAR. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- Hermiston ML, Xu Z, Majeti R, Weiss A. Reciprocal regulation of lymphocyte activation by tyrosine kinases and phosphatases. J Clin Invest. 2002;109:9–14. doi: 10.1172/JCI14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House AE, Lynch KW. An exonic splicing silencer represses spliceosome assembly after ATP-dependent exon recognition. Nat Struct Mol Biol. 2006;13:937–944. doi: 10.1038/nsmb1149. [DOI] [PubMed] [Google Scholar]
- Hui J, Stangl K, Lane WS, Bindereif A. HnRNP L stimulates splicing of the eNOS gene by binding to variable-length CA repeats. Nat Struct Biol. 2003;10:33–37. doi: 10.1038/nsb875. [DOI] [PubMed] [Google Scholar]
- Hung LH, Heiner M, Hui J, Schreiner S, Benes V, Bindereif A. Diverse roles of hnRNP L in mammalian mRNA processing: a combined microarray and RNAi analysis. RNA. 2008;14:284–296. doi: 10.1261/rna.725208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Melton AA, Jackson J, Wang J, Lynch KW. Combinatorial control of signal-induced exon repression by hnRNP L and PSF. Mol Cell Biol. 2007;27:6972–6984. doi: 10.1128/MCB.00419-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer S, Moita LF, Neems D, Freitas RP, Hacohen N, Rao A. Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science. 2008;321:686–691. doi: 10.1126/science.1157610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Gent. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Rothrock C, Cannon B, Hahm B, Lynch KW. A conserved signal-responsive sequence mediates activation-induced alternative splicing of CD45. Mol Cell. 2003;12:1317–1324. doi: 10.1016/s1097-2765(03)00434-9. [DOI] [PubMed] [Google Scholar]
- Rothrock CR, House AE, Lynch KW. HnRNP L represses exon splicing via a regulated exonic splicing silencer. EMBO J. 2005;24:2792–2802. doi: 10.1038/sj.emboj.7600745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A, Nguyen J, Lynch KW. Differential expression of CD45 isoforms is controlled by the combined activity of basal and inducible splicing-regulatory elements in each of the variable exons. J Biol Chem. 2005;280:38297–38304. doi: 10.1074/jbc.M508123200. [DOI] [PubMed] [Google Scholar]
- Topp JD, Jackson J, Melton AA, Lynch KW. A cell-based screen for splicing regulators identifies hnRNP LL as a distinct signal-induced repressor of CD45 variable exon 4. RNA. 2008;14:2038–2049. doi: 10.1261/rna.1212008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Maroney PA, Denker JA, Zhang XH, Dybkov O, Luhrmann R, Jankowsky E, Chasin LA, Nilsen TW. Dynamic regulation of alternative splicing by silencers that modulate 5′ splice site competition. Cell. 2008;135:1224–1236. doi: 10.1016/j.cell.2008.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.