Abstract
HBO1, an H4-specific histone acetylase, is a co-activator of the DNA replication licensing factor Cdt1. HBO1 acetylase activity is required for licensing, because a HAT-defective mutant of HBO1 bound at origins is unable to load the MCM complex. H4 acetylation at origins is cell-cycle regulated, with maximal activity at the G1/S transition, and co-expression of HBO1 and Jade1 increases histone acetylation and MCM complex loading. Overexpression of the Set8 histone H4 tail-binding domain specifically inhibits MCM loading, suggesting that histones are a physiologically relevant target for licensing. Lastly, Geminin, inhibits HBO1 acetylase activity in the context of a Cdt1-HBO1 complex, and it associates with origins and inhibits H4 acetylation and licensing in vivo. Thus, H4 acetylation at origins by HBO1 is critical for replication licensing by Cdt1, and negative regulation of licensing by Geminin is likely to involve inhibition of HBO1 histone acetylase activity.
Keywords: HBO1, DNA replication, origin licensing, histone acetylation, Cdt1, geminin
Introduction
In eukaryotes, DNA replication is controlled so that the genome is replicated only once per cell cycle. The first step of DNA replication is the formation of the pre-replication complex (pre-RC) on origins of replication distributed throughout the genome. The pre-RC contains the origin recognition complex (ORC), Cdt1, Cdc6, and the MCM (mini-chromosome maintenance) complex that are sequentially assembled onto replication origins in the context of chromatin. ORC associates with DNA replication origins throughout the entire cell cycle. When cells exit mitosis, the Cdt1 and Cdc6 licensing factors are loaded onto origins, followed by the MCM complex, the putative replicative helicase. The resulting pre-replication (pre-RC) complex is “licensed” for replication that will occur in the subsequent S phase (Thommes and Blow, 1997; Bell and Dutta, 2002). After replication origins fire and DNA synthesis is initiated, the pre-RC disassembles, and new pre-RC formation is prevented in S phase, thereby restricting DNA synthesis to once per cell cycle.
Regulation of Cdt1 is the key event in replication licensing that permits the ordered assembly and disassembly of the pre-RC (Arias and Walter, 2007). Cdt1 function is inhibited in S phase by ubiquitin-dependent proteolysis, thereby restricting expression to the G1 phase of the cell cycle (Nishitani et al., 2001; Zhong et al., 2003). In addition, Cdt1 activity is inhibited by Geminin in S phase via a direct interaction between these two proteins (Wohlschlegel et al., 2000; Tada et al., 2001). These mechanisms of Cdt1 regulation permit licensing, and hence subsequent DNA replication, to occur only once per cell cycle. Misregulation of Cdt1, by overexpression or by a mutant derivative that is insensitive to proteolysis, results in re-replication and genome instability (Vaziri et al., 2003; Saxena and Dutta, 2005; Tatsumi et al., 2006).
Histone acetylation is linked to pre-RC assembly and the control of initiation of DNA replication. Early-firing origins are typically localized in genomic regions that are transcribed and contain hyper-acetylated chromatin, whereas late-firing origins lie in silenced heterochromatic domains (Kemp et al., 2005; Zhou et al., 2005; Karnani et al., 2007; Lucas et al., 2007; Goren et al., 2008). In addition, histone acetylation is involved in origin activation during early development in Xenopus (Danis et al., 2004) and at the chorion gene loci in Drosophila follicle cells (Aggarwal and Calvi, 2004; Hartl et al., 2007). More generally, diverse chromatin-modifying activities can associate with pre-RC components in protein binding assays and/or genetically alter replication initiation (Takei et al., 2001; Takei et al., 2002; Vogelauer et al., 2002; Aggarwal and Calvi, 2004; Danis et al., 2004; Pappas et al., 2004; Doyon et al., 2006; Iizuka et al., 2006; Sugimoto et al., 2007; Crampton et al., 2008; Yin et al., 2008). For example ORC-dependent chromatin remodeling contributes to optimal loading of the MCM complex onto origins in yeast (Lipford and Bell, 2001), and alteration of the ordered nucleosome arrangement at the human c-Myc replicator selectively decreases MCM complex loading (Ghosh et al., 2006).
HBO1 (human acetylase binding to ORC1; also known as KAT7 and MYST2) is an H4-specific histone acetylase that interacts with transcriptional activator proteins (Georgiakaki et al., 2006; Miotto et al., 2006; Miotto and Struhl, 2006), mRNA coding regions (Saksouk et al., 2009), and with MCM2 and ORC1 (Iizuka and Stillman, 1999; Burke et al., 2001). HBO1 is required for licensing and DNA replication (Doyon et al., 2006; Iizuka et al., 2006), and in Drosophila follicle cells it increases origin activity when artificially recruited to a synthetic replication origin (Aggarwal and Calvi, 2004). In previous work, we showed that HBO1 associates with replication origins specifically during the G1 phase of the cell cycle (Miotto and Struhl, 2008). HBO1 association with origins depends on Cdt1, but is independent of Geminin. HBO1 directly interacts with Cdt1, and it enhances Cdt1-dependent re-replication. Thus, HBO1 plays a direct role at replication origins as a co-activator of the Cdt1 licensing factor, although the mechanism is unknown.
Here, we show that H4 acetylation by HBO1 is critical for replication licensing and that Geminin inhibits HBO1 acetylase activity in the context of a Cdt1-HBO1 complex. Thus, by analogy with activator proteins targeting histone acetylases to enhancers to stimulate transcription, our results suggest that targeted histone acetylation at replication origins is a crucial and regulated step for DNA replication.
Results
HBO1 acetylase activity is essential for licensing of replication origins
HBO1 plays a critical role in replication licensing (Iizuka et al., 2006; Miotto and Struhl, 2008), but the importance of the histone acetylase activity is unknown. To address this issue, we analyzed human cells expressing HBO1G485, which contains a mutation of an invariant glycine in the HAT domain that abolishes enzymatic activity (Iizuka et al., 2008). HBO1G485 association with replication origins is comparable to that of wild-type HBO1 (Figure 1), indicating that the histone acetylase activity is not important for HBO1 association with origins. In contrast, over-expression of HBO1G485 impairs BrdU incorporation, indicating a defect in DNA replication (Figure S1).
Figure 1.
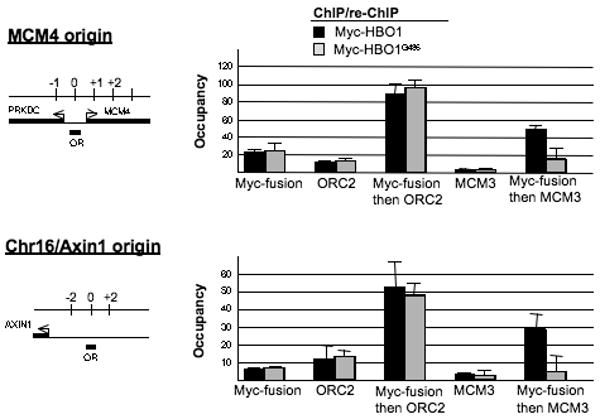
HBO1 acetylase activity is essential for DNA licensing.
DNA fragments bound by Myc-HBO1 and HAT inactive Myc-HBO1G485 were analyzed by sequential chromatin immunoprecipitation analysis for ORC and MCM complexes co-occupancy at the MCM4 and Chr16/Axin origin (n=2). The genomic regions around these replication origins (OR) are shown with neighboring genes (arrows) and positions of primer pairs (black bars) are indicated (coordinates flanking the origin are in kb). Values are expressed as relative occupancy over the control background region (mean ± SD).
To directly measure replication licensing, we used sequential ChIP to determine the association of ORC2 and MCM3 at HBO1- or HBO1G485-bound origins (Figure 1). Unlike the standard approach of monitoring licensing in crude chromatin fractions (Iizuka et al., 2006; Miotto and Struhl, 2008), sequential ChIP directly monitors MCM loading at a replication origin as a function of HBO1 activity. As HBO1 selectively associates with origins during G1 (Miotto and Struhl, 2008), MCM and ORC association detected in the second immunoprecipitation reflects MCM and ORC binding at the time of DNA licensing. As expected, HBO1-bound origins show significant co-association of ORC2 (compare lane 5 with lanes 1 and 3) and MCM3 (compare lane 9 with lanes 1 and 7); i.e. the fold-enrichments of the sequential ChIP samples are significantly higher than the fold-enrichment for HBO1 alone. In contrast, while HBO1G485-bound origins show comparable levels of ORC complex co-occupancy (lanes 5 and 6), the level of MCM complex co-occupancy is clearly reduced in comparison to that observed at HBO1-bound origins (lanes 9 and 10). The fold-enrichment of the HBO1G485 + MCM3 sequential ChIP sample is comparable to that of the individual HBO1G485 ChIP sample, indicating that little or no MCM3 associates with HBO1G485-bound origins. Therefore, recruitment of a HAT-inactive HBO1 derivative at origins selectively blocks MCM complex loading in G1, demonstrating that HBO1 acetylase activity is directly involved in DNA licensing prior to MCM complex loading.
H4 acetylation at replication origins depends on HBO1 and is cell-cycle regulated
H4 acetylation plays an important role in controlling chorion origin activity in Drosophila follicle cells (Aggarwal and Calvi, 2004; Hartl et al., 2007), and HBO1 is recruited to origins by Cdt1 (Miotto and Struhl, 2008). We therefore monitored the profile of H4 acetylation on genomic regions encompassing well-characterized origins. A peak of H4 acetylation is observed on all origins tested, with lower levels of acetylation at flanking regions. Consistent with the selectivity of HBO1 acetylation for lysine residues (Figure 2A; (Doyon et al., 2006), H4 acetylation at K5 and K12, but not K16, is specifically enriched at origins (Figure 2B).
Figure 2.
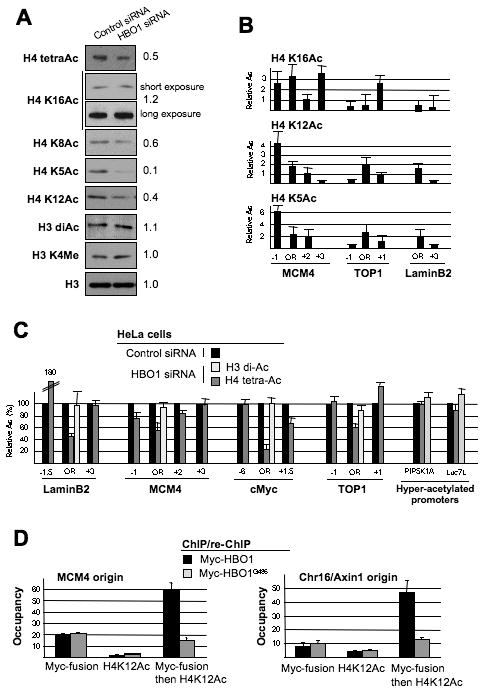
HBO1 controls H4 acetylation at origins.
(A) Bulk histones from control and HBO1 depleted HeLa cells were analyzed by Western blots with antibodies directed against the indicated modifications of H4 and H3. Total H3 serves as a loading control. The relative acetylation level in HBO1-depleted cells as compared to control cells is indicated to the right of each panel.
(B) Acetylation levels (mean ± SD) of the indicated H4 lysine at origins and surrounding regions (n=3).
(C) Acetylation levels (mean ± SD) of H3 and H4 at the indicated origins and constitutively hyper-acetylated promoters in HeLa cells depleted of HBO1 expressed as a percent of the levels in control cells (n=3).
(D) DNA fragments bound by Myc-HBO1 and HAT inactive Myc-HBO1G485 were analyzed by sequential chromatin immunoprecipitation analysis for H4-K12 acetylation (H4-K12Ac) co-occupancy at the MCM4 and Chr16/Axin origin (n=2).
HBO1 is responsible for most H4 acetylation in human cells, because depletion of HBO1 substantially reduces the overall level of H4 acetylation (Figure 2A; (Doyon et al., 2006). This loss of H4, but not H3, acetylation upon HBO1 depletion is also observed at origins (Figures 2C and S2). HBO1 depletion does not affect the expression of genes localized in the vicinity of origins (Figure S3), indicating that loss of H4 acetylation at origins is not an indirect effect of transcription. Interestingly, some promoter regions hyper-acetylated at H4 are not affected by HBO1 depletion (Figure 2C), presumably due to targeted recruitment of an H4 acetylase distinct from HBO1. Thus, recruitment of HBO1 to origins results in a peak of H4 acetylation.
We confirmed that HBO1 acetylase activity is responsible for H4 acetylation at origins by performing a sequential ChIP experiment (Figure 2D). As expected, HBO1-bound origins show significant co-association of H4-K12 acetylation. In contrast, at HBO1G485-bound origins, the level of H4-K12 acetylation co-occupancy is clearly reduced in comparison to that observed at HBO1-bound origins. Thus, the peak of H4 acetylation at origins is induced by HBO1 activity
The above results and the fact that HBO1 associates with origins in a cell-cycle-dependent manner (Miotto and Struhl, 2008) suggest that licensing is associated with a transient increase in H4 acetylation at origins. Indeed, H4 acetylation at all origins tested is 3-fold lower in cells staged in G2/M than in G1 (Figure 3A). Unexpectedly, H4 acetylation levels at the TOP1, LaminB2, and MCM4 origins are similar in cells staged in G1 and G1/S (Figure 3A), even though HBO1 association at G1/S is significantly reduced in comparison to G1 (Miotto and Struhl, 2008). However, these origins are in close proximity to an active promoter or coding sequence (Figure 2A), suggesting that transcriptional activity may maintain H4 acetylation during S phase. Indeed, H4 acetylation at isolated origins not near annotated promoter and coding sequences (Cadoret et al., 2008)(Figure S4) rises during G1 and decreases when cells are staged in S phase (Figure 3B). Thus, on isolated origins, G1-specific H4 acetylation concomitant with HBO1 binding is required for MCM complex loading. In addition, H4 deacetylation is not required for initiation of replication and MCM helicase activation, as some origins have significant level of H4 acetylation during S phase.
Figure 3.
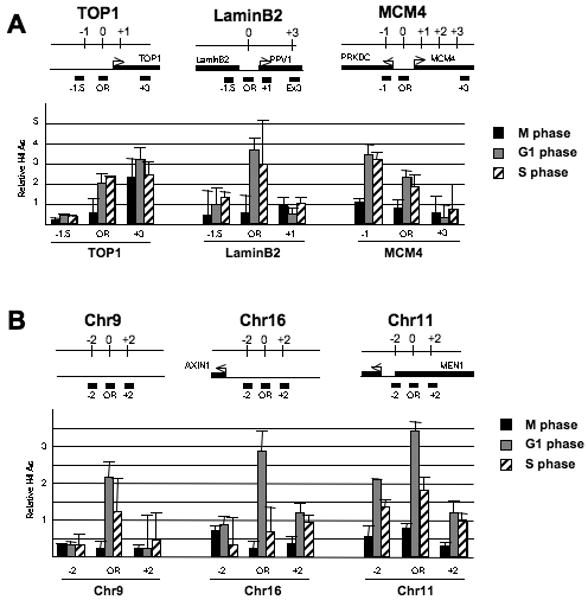
H4 acetylation at origins is cell cycle regulated.
(A) H4 acetylation levels (mean ± SD) at replication origins and flanking regions (indicated in kb from the origin) in cells at the indicated stages of the cell cycle (n=3). The genomic regions around these replication origins (OR) are shown with neighboring genes (arrows) and positions of primer pairs (black bars) are indicated (coordinates flanking the origin are in kb).
(B) Same as above, except that the origins were isolated from annotated promoters.
Histone H4 acetylation at origins influences MCM complex loading
HBO1 mediates targeted H4 acetylation at origins, and its HAT activity is required for licensing, suggesting that H4 acetylation is required for loading of the MCM complex. However, as is typical for histone acetylases, HBO1 acetylates non-histone substrates in vitro including ORC2, Geminin, MCM2 and Cdc6 (Iizuka et al., 2006). To establish that H4 acetylation is important for MCM complex loading, we utilized two independent approaches in which proteins presumed to specifically affect acetylation of H4 but not non-histone substrates were introduced into cells.
First, we analyzed the HBO1 co-factor Jade-1/PHF17. Jade-1 and HBO1 are components of two ING complexes implicated in DNA replication (Doyon et al., 2006), and Jade-1 stabilizes HBO1 in the nucleus and increases HBO1 association with chromatin through its PHD domains (Foy et al., 2008). Over-expression of HBO1 alone barely increases H4 acetylation, whereas co-expression of HBO1 and Jade-1 increases H4 acetylation levels (Foy et al., 2008). Co-expression of Jade-1 and HBO1 strongly enhances MCM loading, whereas over-expression of HBO1 alone does not (Figure 4A). Importantly, the synergy of HBO1 and Jade-1 for MCM complex loading is not observed in parallel experiments involving the catalytically inactive HBO1G485 derivative (Figure 4B). Therefore, increased HBO1 association with chromatin and H4 hyperacetylation is sufficient to promote excess MCM loading. Nevertheless, cells over-expressing Jade-1 and HBO1 show no detectable re-replication (data not shown), as also observed for cells treated with HDAC inhibitors (data not shown) or expressing HBO1 alone (Miotto and Struhl, 2008).
Figure 4.
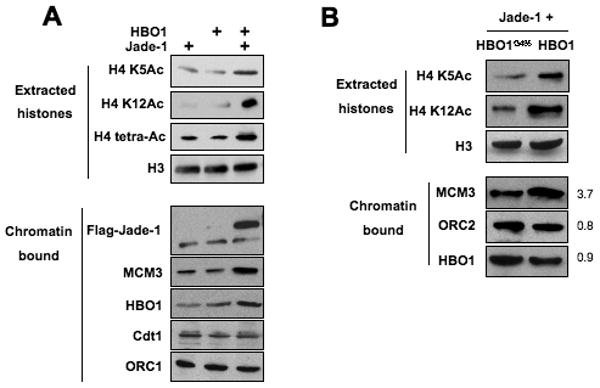
Co-expression of HBO1 and Jade-1 stimulates H4 acetylation and MCM complex loading.
(A) Western blot analysis showing H4 acetylation status (extracted histones; H3 levels serves as the internal control) and presence of the indicated proteins with in chromatin in cells over-expressing Jade-1L or HBO1 or both.
(B) Similar experiment in cells co-expressing Jade-1L with either HBO1 or HAT-defective derivative HBO1G485.
Second, we analyzed MCM complex loading in cells where H4 acetylation is presumed to be specifically blocked in G1 via the histone-binding domain (HBD) of the Set8 H4-K20 histone methylase. The Set8 HBD interacts with H4 tails, lacks proteolytic destruction sites, and accumulates in G1 (Yin et al., 2008). As previously reported (Yin et al., 2008), over-expression of Set8-HBD, but not Set8, during G1 reduces bulk H4 acetylation on histone H4 residues K5, K8 and K12 but not K16 and blocks the cell cycle progression prior to S phase entry (Figure 5A). Importantly, over-expression of Set8-HBD, but not full-length Set8, also blocks MCM complex association at origins (Figure 5B) without affecting loading of HBO1, Cdt1, Cdc6, and the ORC complex or expression of MCM components (Figure 5C). As over-expression of Set8-HBD is unlikely to inhibit acetylation of non-histone substrates, this observation strongly argues that histones are the physiological substrate for HBO1 that is required for efficient MCM complex loading.
Figure 5.
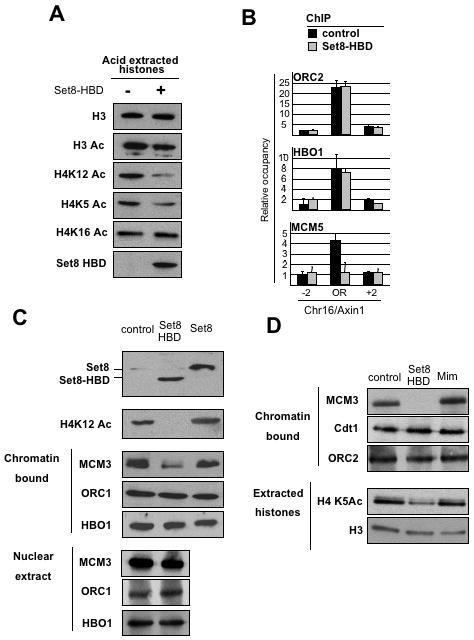
Expression of the Set8 histone tail-binding domain inhibits H4 acetylation and MCM complex loading.
(A) Western blot analysis to measure acetylation at the indicated residues in acid extracted histones prepared from HeLa cells that do or do not express the histone-binding domain (HBD) of Set8.
(B) Association of ORC2, HBO1 and MCM5 (mean ± SD) at the Chr16/Axin1 origin detected by ChIP in cell that do or do not express Set8-HBD (n=3).
(C) Comparison of full-length Set8 and Set8-HBD effects on H4 acetylation and levels of MCM3, ORC1, and HBO1 in chromatin and nuclear extract.
(D) Western blot analysis to examine the effect of mimosine and Set8-HBD (blocks in early S-phase) on H4 acetylation and levels of MCM3, Cdt1 and ORC2 in the chromatin.
To exclude the possibility that Set8-HBD indirectly affects H4 acetylation and MCM complex recruitment due to the block in G1, we examined cells treated with mimosine, a drug used to block the cell cycle in late G1 (Miotto and Struhl, 2008). Cells staged in G1 by mimosine have an equivalent amount of H4 acetylation and association of ORC, Cdt1, and the MCM complex as compared to untreated cells (Fig. 5D). The observation that Set8-HBD and mimosine treatment affect different steps during replication initiation, even though they both block the cell cycle in late G1, strongly emphasizes the functional connection between H4 acetylation and MCM complex loading during G1.
Importance of H4 acetylation for licensing is not an indirect consequence of transcriptional effects
Set8-HBD-expressing or HBO1-depleted cells have a global defect in H4 acetylation, and such low levels of H4 acetylation could indirectly affect MCM complex loading via an effect on transcription by RNA polymerase (Pol) II. Three lines of evidence exclude this possibility. First, as observed in HBO1-depleted cells (Iizuka et al., 2006; Miotto and Struhl, 2008), Set8-HBD expression does not alter the accumulation of pre-RC subunits (Figure 5C). Second, in contrast to MCM complex loading, chromatin association of Pol II, TBP, TBP-associated factors (TAFs), TFIIB, TFIIH (CCNH subunit), and Mediator (Med26 subunit) are not affected by Set8-HBD expression (Figure S5A) or HBO1 depletion (Figure S5B). Third, Set8-HBD expression and HBO1 depletion do not affect mRNA levels (Figure S3 and data not shown) or Pol II occupancy in the coding sequence of all genes tested (Figure S5C), indicating that low levels of H4 acetylation do not have a general effect on Pol II transcription. These observations are consistent with transcription-independent replication assays in Xenopus extracts showing the involvement of HBO1 in pre-RC assembly prior to MCM complex loading (Iizuka et al., 2006). Therefore it is highly likely that targeted HBO1-dependent H4 acetylation controls MCM complex loading at origins, independently of global effect at the transcriptional level.
Geminin, a Cdt1 repressor, inhibits HBO1 acetylase activity in a Cdt1-dependent manner
Geminin interacts directly with Cdt1, and it is a potent inhibitor of Cdt1 licensing activity (Wohlschlegel et al., 2000; Tada et al., 2001). However, Geminin does not block the interaction of Cdt1 with HBO1 in vitro or Cdt1-dependent recruitment of HBO1 to replication origins in vivo (Miotto and Struhl, 2008). In addition, the enzymatic activity of HBO1 immunoprecipitates is enhanced from mitosis to late G1 phase (Iizuka et al., 2006), suggesting the possibility that HBO1 acetylase activity is regulated by a repressor. As our results indicate that H4 acetylation by HBO1 is required for licensing, we considered the possibility that Geminin inhibits HBO1 histone acetylase activity.
Geminin does not inhibit acetylation of H4-K5 or H4-K12 by the purified yeast piccolo NuA4 complex (Figure 6A), indicating that Geminin does not interact with the histone H4 tail and non-specifically mask H4 acetylation sites. In addition, acetylation of an H4 peptide by immunoprecipitated Flag-HBO1 is not inhibited significantly by Geminin (Figure 6A). The observed activity is due to HBO1, because immunoprecipitated Flag-HBO1G485 has no detectable HAT activity. In contrast, when Flag-HBO1 is immunoprecipitated from cells also expressing HA-Cdt1, the bound material is enriched in HBO1/Cdt1 complex whose ability to acetylate the H4 tail is inhibited by Geminin in a concentration dependent manner (Figure 6B, C).
Figure 6.
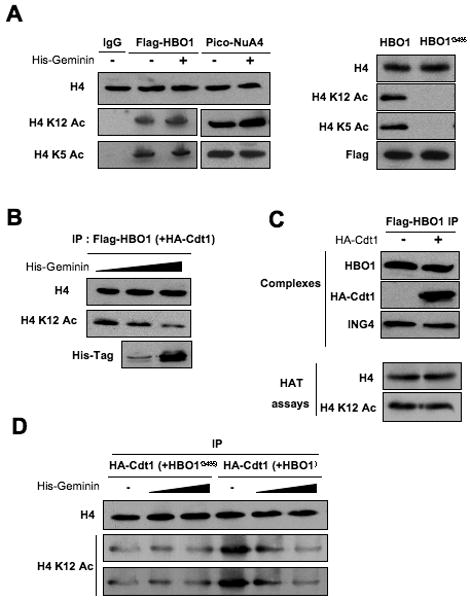
Geminin inhibits H4 acetylase activity of HBO1 in vitro.
(A) Effect of recombinant full-length Geminin on the H4 acetylase activity of immunoprecipitated Flag-HBO1, Flag-HBO1G485, and purified yeast piccolo NuA4 complex.
(B) Same experiment as in (A) except that Flag-HBO1 was purified from cells over-expressing Flag-HBO1 and HA-Cdt1.
(C) Flag-HBO1 immunoprecipitates prepared from cells that do or do not co-express HA-Cdt1 were analyzed by Western blotting with antibodies against HBO1, HA-Cdt1, and ING4. HBO1 complexes in the presence or absence of Cdt1 were also tested for their HAT activity on H4-K12.
(D) Effect of Geminin on the H4 activity of HA-Cdt1 complexes purified from cells co-expressing either HA-Cdt1+Flag-HBO1 or HA-Cdt1+Flag-HBO1G485. Numbers under the panels indicate the relative amount of acetylated H4-K12 HAT as compared to cells in the absence of Geminin. Note that weak H4-K12 acetylase activity observed in the HA-Cdt1+Flag-HBO1G485 immunoprecipitates is insensitive to Geminin.
To verify that Geminin inhibits H4 acetylation by the Cdt1-HBO1 complex, we purified HA-Cdt1 from cells co-expressing either Flag-HBO1 or Flag-HBO1G485, and assessed the activity of the resulting complexes in presence of recombinant Geminin (Figure 6D). The Cdt1/HBO1G485 complex has weak H4 activity and acetylates H4 in presence of recombinant Geminin, presumably due to weak association of a histone acetylase other than HBO1. Importantly, the Cdt1/HBO1 complex has strong H4 acetylase activity that is inhibited by recombinant Geminin in a concentration dependent manner (Figure 6D). Therefore, Geminin inhibits HBO1 HAT activity in a Cdt1-dependent manner, consistent with the observation that Geminin interacts with HBO1 only in the presence of Cdt1 (Miotto and Struhl, 2008).
Geminin associates with replication origins and inhibits H4 acetylation in vivo
The above biochemical observations predict that Geminin interferes with HBO1-dependent H4 acetylation at origins in vivo. To test this hypothesis, we over-expressed a non-degradable derivative of Geminin, GemL26A (Wohlschlegel et al., 2002), that blocks MCM complex loading but does not interfere with binding of ORC, Cdt1, Cdc6 and HBO1 (Miotto and Struhl, 2008). As shown in Figure 7A, GemL26A inhibits H4 acetylation at the Myc and Chr16 replication origin, but not at flanking regions or at control hyper-acetylated loci. Furthermore, GemL26A associates with all four replication origins tested (2-4 fold-enrichments), but not with control hyperacetylated regions (Figure 7B), suggesting that Geminin directly affects H4 acetylation at origins via its interaction with Cdt1 and HBO1. Taken together, these biochemical and genetic observations further support the crucial role of HBO1 acetylase activity during replication licensing, and they strongly suggest a mechanism (not necessarily exclusive) for how Geminin represses licensing.
Figure 7.
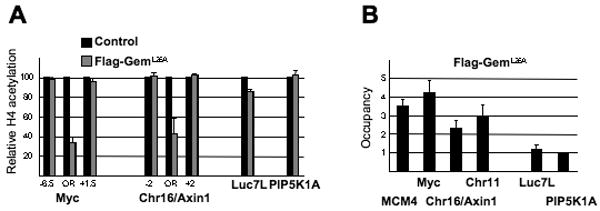
Geminin associates with replication origins and inhibits H4 acetylation in vivo.
(A) Relative H4 tetra-acetylation level (mean ± SD) at origins in cells expressing the non-degradable Flag-GemL26A derivative (n=3).
(B) Association of Flag-GemL26A (mean ± SD) at origins (MCM4, Myc, Chr16, Chr11) and control regions (Luc7L, PIP5K1A)(n=3).
Discussion
HBO1 acetylase activity is important for licensing
Although the Cdt1 licensing factor directly interacts with the MCM complex (Tanaka and Diffley, 2002; You and Masai, 2008), recruitment of HBO1 to origins by Cdt1 is required for MCM complex loading in human cells. Here, we demonstrate that HBO1 function at origins requires its acetylase activity, indicating that MCM loading is enhanced by a Cdt1-dependent acetylation event. Specifically, as assayed by sequential ChIP, a replication origin simultaneously bound by ORC and HBO1G485 is defective in recruiting the MCM complex, (Figure 1). Thus, HBO1 acetylase activity is required prior to MCM complex loading for pre-RC assembly at origins. The importance of HBO1 acetylase activity for licensing is also supported by the observation that modulators of this enzymatic activity such as Geminin (Figure 7), Jade-1 (Figure 4), p53 (Iizuka et al., 2008), and Polo-like kinase 1 (Wu and Liu, 2008) are linked to MCM loading under specific conditions. More generally, regulation of chromatin structure regulation can affect MCM complex loading (Lipford and Bell, 2001; Ghosh et al., 2006), and our results provide an additional mechanism by which this occurs.
H4 acetylation by HBO1 is critical for licensing
Several observations suggest the importance of H4 acetylation for licensing. First, HBO1 is the predominant H4 acetylase in human cells (Figure 2A (Iizuka et al., 2008), and it is recruited to origins at the time of licensing, where it is important for loading the MCM complex (Miotto and Struhl, 2008). Second, treatments of cells that either increase (overexpression of HBO1 and Jade-1) or decrease (Set8-HBD overexpression) H4 acetylation reveal a correlation between levels of bulk H4 acetylation and MCM complex loading (Figures 4, 5). Third, in accord with the HBO1 association profile, H4 acetylation peaks at replication origins in comparison to most flanking sequences (Figure 2B), and it is cell-cycle regulated (Figure 3).
The strong correlation between H4 acetylation and replication licensing does not necessarily indicate that histones are the physiologically relevant substrate for HBO1. As is typical for histone acetylases, HBO1 acetylates non-histone substrates in vitro including ORC2, Geminin, MCM2 and Cdc6 (Iizuka et al., 2006). Thus, the requirement for the HBO1 acetylase activity might involve a non-histone substrate, and the increased H4 acetylation at origins might be a consequence of recruitment, rather than having a direct effect on licensing Establishing that histones are the physiological substrate is difficult in human cells, unlike the case in yeast where it is possible to examine strains with genetically modified histones.
Although formal proof is lacking, two observations strongly suggest that H4 is the physiologically relevant substrate for MCM complex loading. First, overexpression of Jade-1 and wild-type (but not mutant) HBO1 increases H4 acetylation and MCM complex loading (Figure 4). The PHD fingers of Jade-1 are required for histone binding and stimulation of H4 acetylation, but not for the association of Jade-1 and HBO1 (Foy et al., 2008). The requirement for the PHD fingers suggests that Jade-1 can stimulate acetylation of H4, but not non-histone substrates. Second, the histone tail-binding domain of Set8, a histone methylase, blocks H4 acetylation and MCM complex loading (Figure 5). As this Set8 domain is unlikely to interact with non-histone substrates, the observation strongly suggests that the observed effects on MCM complex loading involve an interaction with H4.
In accord with HBO1 association with origins being restricted to G1, isolated origins far from annotated promoters show preferential H4 acetylation at G1. However, H4 acetylation at other origins remains high throughout S phase, as also observed at the Drosophila chorion gene origin (Hartl et al., 2007) and the Epstein Barr oriP origin (Zhou et al., 2005). This difference between H4 acetylation and HBO1 association at some origins during S phase likely reflects transcriptional-related events in the vicinity of the origin that involve other H4 acetylases such as Tip60. The effects of H4 acetylation may vary among origins, and the histone deacetylase inhibitor TSA alters DNA replication patterns in human cells (Kemp et al., 2005). Similarly, mutation of the Sir2 histone deacetylase affects licensing in yeast, but only 4 of the 25 origins tested show reduced levels of H4-K16 acetylation, possibly due to a conserved DNA element bound by a well-positioned nucleosome (Crampton et al., 2008).
While our results indicate that Cdt1-dependent H4 acetylation by HBO1 is important for MCM complex loading, the precise role of H4 acetylation during replication licensing is unknown. One possibility is that H4 acetylation increases accessibility or fluidity of chromatin, thereby facilitating association of the MCM complex. In this regard, Cdt1 interacts with SNF2H and WSTF (Sugimoto et al., 2007), which are components of nucleosome remodeling complexes that preferentially associate with acetylated histones in chromatin (Hakimi et al., 2002; Fujiki et al., 2005). Alternatively, the MCM complex or an associated factor might recognize the acetylated lysines on H4, and this interaction would stabilize the interaction of the MCM complex with chromatin. Although an HBO1 homologue does not exist in yeast. HAT1 histone acetylase interacts with the ORC complex and affects ORC function in vivo (Suter et al., 2007). Like HBO1, HAT1 preferentially acetylates H4 at K5, K8, and K12 (but not K16), suggesting that it might play an analogous role at origins.
Geminin inhibits HBO1 histone acetylase activity in the context of a Cdt1 complex
Geminin plays a key role in the down-regulation of Cdt1 activity when cells enter S phase. Although Geminin directly interacts with Cdt1, it does not block the interaction of Cdt1 and HBO1 but rather forms a ternary complex in vitro (Miotto and Struhl, 2008). This ternary complex is mediated by independent interactions of Geminin or HBO1 with different surfaces of Cdt1, because Geminin and HBO1 do not interact with each other. In accord with these observations, forcing Geminin expression in G1 does not inhibit HBO1 association with origins in vivo, even though it inhibits licensing (Miotto and Struhl, 2008). It has been suggested that Geminin binding to Cdt1 inhibits the interaction between Cdt1 and the MCM complex (Yanagi et al., 2002; Cook et al., 2004; Lee et al., 2004), although Cdt1 lacking the evolutionarily conserved region that interacts with the MCM complex is capable of re-replication (Teer and Dutta, 2008).
Here, we show that Geminin inhibits HBO1-dependent acetylation of H4 in vitro (Figure 6). This inhibition requires Cdt1, and presumably occurs in the context of the Cdt1-HBO1-geminin complex. Geminin-mediated inhibition of HBO1 histone acetylase activity might also account for the observation that enzymatic activity of HBO1 immunoprecipitates is enhanced from mitosis to late G1 phase (Iizuka et al., 2006). Most importantly, H4 acetylation is specifically impaired at origins in cells expressing the GemL26A derivative, and this is likely to be a direct effect because GemL26A associates with origins (Figure 7). Given the importance of H4 acetylation in MCM complex loading at origins, our results strongly suggest that Geminin inhibition of HBO1 histone acetylase activity contributes to replication licensing and the blockage of re-replication.
While likely to be important, the ability of Geminin to inhibit the acetylase activity of HBO1 might not fully account for how it regulates Cdt1 activity. This model is not mutually exclusive with the idea that geminin blocks the interaction between Cdt1 and the MCM complex. Geminin also protects Cdt1 from proteasome-mediated degradation by inhibiting its ubiquitination, and inhibition of Geminin during M phase impairs pre-RC formation during the following cell cycle (Ballabeni et al., 2004). In addition, a Geminin-Cdt1 complex can license origins, but block re-replication in vitro, and it has been suggested that this switch is due to increased stoichiometry of Geminin with respect to Cdt1 (Lutzmann et al., 2006). As inhibition of HBO1 histone acetylase activity by Geminin is concentration-dependent, perhaps this mechanism is more important for preventing re-replication as opposed to blocking licensing. In any event, as replication licensing and the prevention of re-replication is of paramount importance for long term genome stability, it would not be surprising that Geminin functions by multiple mechanisms.
Experimental Procedures
Plasmids and reagents
Expression vectors for Myc-HBO1G485, YFP-Set8, YFP-Set8 histone binding domain (Set8-HBD) and Flag-Jade-1 (long isoform) were described elsewhere (Contzler et al., 2006; Foy et al., 2008; Yin et al., 2008). Other materials not listed here have been previously described (Miotto and Struhl, 2008).
Antibodies
Antibodies were obtained from the following sources: Santa Cruz biotechnology for HBO1, CCNH, MED26, HA-Tag (F7) and Myc-Tag (9E10); Abcam for anti-H4 K16Ac, H3 K4Met, ORC1, ORC2, TFIIB and Histone H3; Upstate Biotechnology for anti-H4 tetra-Ac (K5, K8, K12 and K16), H3 di-Ac (K9 and K14), H4 K12Ac, H4 K8Ac, H4 K5Ac and H4 K16Ac; Bethyl Laboratories for MCM proteins and Cdt1; Covance for Pol II; Novus Biological for HBO1; Sigma-Aldrich for Flag-Tag (M2), Actinβ and Cdc6. TBP antibody was obtained from Arnie Berk. Protein A and protein G sepharose beads as well as control IgG-sepharose beads used in co-immunoprecipitation and ChIP assays were purchased from Amersham Biosciences.
Western blot analysis of histone modifications
Histones were acid-extracted from cells, electrophoretically separated, blotted, and assayed for H4, and H3 modifications using the appropriate antibodies.
H4 acetylation assays
In vitro acetylation reactions were performed with minor modifications from a previously described protocol (Ait-Si-Ali et al., 1998). Acetylation of the histone H4 tail peptide (Abcam) was detected with antibodies specific for histone H4 acetylated on K5 or K12. Purified piccolo NuA4 complex was kindly provided by Danesh Moazed, and recombinant Geminin was produced in E. coli. Flag-HBO1 derivatives were immuno-purified from HeLa cells 48 hours following transfection. Cells were lysed in RIPA buffer, and material from 5 X 106 cells was immunoprecipitated overnight with an anti-FLAG sepharose resin. The resulting material was extensively washed in buffer (500 mM KCl, 1% triton X-100, 0.1% SDS), and the amount of FLAG-fusion proteins retained on the beads was monitored by Western blot and normalized for in vitro acetylation activity. When FLAG-fusions were co-expressed with HA-Cdt1 and co-purified on anti-HA beads, the KCl concentration during the washing steps was 240 mM to avoid HBO1 dissociation from Cdt1.
Cell manipulation, binding assays, chromatin fractionation, immunofluorescence, flow cytometry, and chromatin immunoprecipitation
These procedures were performed as previously reported (Miotto and Struhl, 2008).
Supplementary Material
Acknowledgments
We are very grateful to Johannes Walter for insightful comments during the course of this work and on the manuscript and Pierre-Antoine Defossez for critical reading of the manuscript. We thank Anindya Dutta, Danesh Moazed, Herbert Cohen, Donald Chang and Marcel Huber for providing reagents. This work was supported by grants to K.S. from the National Institutes of Health (GM30186).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aggarwal BD, Calvi BR. Chromatin regulates origin activity in Drosophila follicle cells. Nature. 2004;430:372–376. doi: 10.1038/nature02694. [DOI] [PubMed] [Google Scholar]
- Ait-Si-Ali S, Ramirez S, Robin P, Trouche, HB A. A rapid and sensitive assay for histone acetyl-transferase activity. Nucl Acids Res. 1998;26:3869–3870. doi: 10.1093/nar/26.16.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias EE, Walter JC. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 2007;21:497–518. doi: 10.1101/gad.1508907. [DOI] [PubMed] [Google Scholar]
- Ballabeni A, Melixetian M, Zamponi R, Masiero L, Marinoni F, Helin K. Human geminin promotes pre-RC formation and DNA replication by stabilizing CDT1 in mitosis. EMBO J. 2004;23:3122–3132. doi: 10.1038/sj.emboj.7600314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Burke TW, Cook JG, Asano M, Nevins JR. Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HBO1. J Biol Chem. 2001;276:15397–15408. doi: 10.1074/jbc.M011556200. [DOI] [PubMed] [Google Scholar]
- Cadoret JC, Meisch F, Hassan-Zadeh V, Luyten I, Guillet C, Duret L, Quesneville H, Prioleau MN. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc Natl Acad Sci USA. 2008;105:15837–15842. doi: 10.1073/pnas.0805208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contzler R, Regamey A, Favre B, Roger T, Hohl D, Huber M. Histone acetyltransferase HBO1 inhibits NF-kappaB activity by coactivator sequestration. Biochem Biophys Res Commun. 2006;350:208–213. doi: 10.1016/j.bbrc.2006.09.030. [DOI] [PubMed] [Google Scholar]
- Cook JG, Chasse DA, Nevins JR. The regulated association of Cdt1 with minichromosome maintenance proteins and Cdc6 in mammalian cells. J Biol Chem. 2004;279:9625–9633. doi: 10.1074/jbc.M311933200. [DOI] [PubMed] [Google Scholar]
- Crampton A, Chang F, Pappas DL, Jr, Frisch RL, Weinreich M. An ARS element inhibits DNA replication through a SIR2-dependent mechanism. Mol Cell. 2008;30:156–166. doi: 10.1016/j.molcel.2008.02.019. [DOI] [PubMed] [Google Scholar]
- Danis E, Brodolin K, Menut S, Maiorano D, Girard-Reydet C, Mechali M. Specification of a DNA replication origin by a transcription complex. Nat Cell Biol. 2004;6:721–730. doi: 10.1038/ncb1149. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Cayrou C, Ullah M, Landry AJ, Côté V, Selleck W, Lane WS, Tan S, Yang XJ, Côté J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Foy RL, Song IY, Chitalia VC, Cohen HT, Saksouk N, Cayrou C, Vaziri C, Cote J, Panchenko MV. Role of Jade-1 in the histone acetyltransferase (HAT) HBO1 complex. J Biol Chem. 2008;283:28817–28826. doi: 10.1074/jbc.M801407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki R, Kim MS, Sasaki Y, Yoshimura K, Kitagawa H, Kato S. Ligand-induced transrepression by VDR through association of WSTF with acetylated histones. EMBO J. 2005;24:3881–3894. doi: 10.1038/sj.emboj.7600853. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Georgiakaki M, Chabbert-Buffet N, Dasen B, Meduri G, Wenk S, Rajhi L, Amazit L, Chauchereau A, Burger CW, Blok LJ, et al. Ligand-controlled interaction of histone acetyltransferase binding to ORC-1 (HBO1) with the N-terminal transactivating domain of progesterone receptor induces steroid receptor coactivator 1-dependent coactivation of transcription. Mol Endocrinol. 2006;20:2122–2140. doi: 10.1210/me.2005-0149. [DOI] [PubMed] [Google Scholar]
- Ghosh M, Kemp M, Liu G, Ritzi M, Schepers A, Leffak M. Differential binding of replication proteins across the human c-myc replicator. Mol Cell Biol. 2006;26:5270–5283. doi: 10.1128/MCB.02137-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren A, Tabib A, Hecht M, Cedar H. DNA replication timing of the human beta-globin domain is controlled by histone modification at the origin. Genes & Dev. 2008;22:1319–1324. doi: 10.1101/gad.468308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi MA, Bochar DA, Schmiesing JA, Dong Y, Barak OG, Speicher DW, Yokomori K, Shiekhattar R. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature. 2002;418:994–998. doi: 10.1038/nature01024. [DOI] [PubMed] [Google Scholar]
- Hartl T, Boswell C, Orr-Weaver TL, Bosco G. Developmentally regulated histone modifications in Drosophila follicle cells: initiation of gene amplification is associated with histone H3 and H4 hyperacetylation and H1 phosphorylation. Chromosoma. 2007;116:197–214. doi: 10.1007/s00412-006-0092-2. [DOI] [PubMed] [Google Scholar]
- Iizuka M, Matsui T, Takisawa H, Smith MM. Regulation of replication licensing by acetyltransferase Hbo1. Mol Cell Biol. 2006;26:1098–1108. doi: 10.1128/MCB.26.3.1098-1108.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka M, Sarmento OF, Sekiya T, Scrable H, Allis CD, Smith MM. Hbo1 links p53-dependent stress signaling to DNA replication licensing. Mol Cell Biol. 2008;28:140–153. doi: 10.1128/MCB.00662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka M, Stillman B. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J Biol Chem. 1999;274:23027–23034. doi: 10.1074/jbc.274.33.23027. [DOI] [PubMed] [Google Scholar]
- Karnani N, Taylor C, Malhotra A, Dutta A. Pan-S replication patterns and chromosomal domains defined by genome-tiling arrays of ENCODE genomic areas. Genome Res. 2007;17:865–876. doi: 10.1101/gr.5427007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp MG, Ghosh M, Liu G, Leffak M. The histone deacetylase inhibitor trichostatin A alters the pattern of DNA replication origin activity in human cells. Nucl Acids Res. 2005;33:325–336. doi: 10.1093/nar/gki177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Hong B, Choi JM, Kim Y, Watanabe S, Ishimi Y, Enomoto T, Tada S, Kim Y, Cho Y. Structural basis for inhibition of the replication licensing factor Cdt1 by geminin. Nature. 2004;430:913–917. doi: 10.1038/nature02813. [DOI] [PubMed] [Google Scholar]
- Lipford JR, Bell SP. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol Cell. 2001;7:21–30. doi: 10.1016/s1097-2765(01)00151-4. [DOI] [PubMed] [Google Scholar]
- Lucas I, Palakodeti A, Jiang Y, Young DJ, Jiang N, Fernald AA, LeBeau MM. High-throughput mapping of origins of replication in human cells. EMBO Rep. 2007;8:770–777. doi: 10.1038/sj.embor.7401026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzmann M, Maiorano D, Mechali M. A Cdt1-geminin complex licenses chromatin for DNA replication and prevents rereplication during S phase in Xenopus. EMBO J. 2006;25:5764–5774. doi: 10.1038/sj.emboj.7601436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto B, Sagnier T, Berenger H, Bohmann D, Pradel J, Graba Y. Chameau HAT and DRpd3 HDAC function as antagonistic cofactors of JNK/AP-1-dependent transcription during Drosophila metamorphosis. Genes Dev. 2006;20:101–112. doi: 10.1101/gad.359506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto B, Struhl K. Differential gene regulation by selective association of transcriptional coactivators and bZIP DNA-binding domains. Mol Cell Biol. 2006;26:5969–5982. doi: 10.1128/MCB.00696-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto B, Struhl K. HBO1 histone acetylase is a co-activator of the replication licensing factor Cdt1. Genes & Dev. 2008;22:2633–2638. doi: 10.1101/gad.1674108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H, Taraviras S, Lygerou Z, Nishimoto T. The human licensing factor for DNA replication Cdt1 accumulates in G1 and is destabilized after initiation of S-phase. J Biol Chem. 2001;276:44905–44911. doi: 10.1074/jbc.M105406200. [DOI] [PubMed] [Google Scholar]
- Pappas DL, Jr, Frisch R, Weinreich M. The NAD(+)-dependent Sir2p histone deacetylase is a negative regulator of chromosomal DNA replication. Genes Dev. 2004;18:769–781. doi: 10.1101/gad.1173204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksouk N, Avvakumov N, Champagne KS, Hung T, Doyon Y, Cayrou C, Paquet E, Ullah M, Landry AJ, Cote V, et al. HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol Cell. 2009;33:257–265. doi: 10.1016/j.molcel.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena S, Dutta A. Geminin-Cdt1 balance is critical for genetic stability. Mutat Res. 2005;569:111–121. doi: 10.1016/j.mrfmmm.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Sugimoto N, Kitabayashi I, Osano S, Tatsumi Y, Yugawa T, Narisawa-Saito M, Matsukage A, Kiyono T, Fujita M. Identification of novel human Cdt1-binding proteins by a proteomics approach: proteolytic Regulation by APC/CCdh1. Mol Biol Cell. 2007;19:1007–1021. doi: 10.1091/mbc.E07-09-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter B, Pogoutse O, Guo X, Krogan N, Lewis P, Greenblatt JF, Rine J, Emili A. Association with the origin recognition complex suggests a novel role for histone acetyltransferase Hat1p/Hat2p. BMC Biol. 2007;5:38. doi: 10.1186/1741-7007-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada S, Li A, Maiorano D, Mechali M, Blow JJ. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat Cell Biol. 2001;3:107–113. doi: 10.1038/35055000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei Y, Assenberg M, Tsujimoto G, Laskey R. The MCM3 acetylase MCM3AP inhibits initiation, but not elongation, of DNA replication via interaction with MCM3. J Biol Chem. 2002;277:43121–43125. doi: 10.1074/jbc.C200442200. [DOI] [PubMed] [Google Scholar]
- Takei Y, Swietlik M, Tanoue A, Tsujimoto G, Kouzarides T, Laskey R. MCM3AP, a novel acetyltransferase that acetylates replication protein MCM3. EMBO Rep. 2001;2:119–123. doi: 10.1093/embo-reports/kve026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Diffley JF. Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2-7 during G1 phase. Nat Cell Biol. 2002;4:198–207. doi: 10.1038/ncb757. [DOI] [PubMed] [Google Scholar]
- Tatsumi Y, Sugimoto N, Yugawa T, Narisawa-Saito M, Kiyono T, Fujita M. Deregulation of Cdt1 induces chromosomal damage without rereplication and leads to chromosomal instability. J Cell Sci. 2006;119:3128–3140. doi: 10.1242/jcs.03031. [DOI] [PubMed] [Google Scholar]
- Teer JK, Dutta A. Human Cdt1 lacking the evolutionarily conserved region that interacts with MCM2-7 is capable of inducing re-replication. J Biol Chem. 2008;283:6817–6825. doi: 10.1074/jbc.M708767200. [DOI] [PubMed] [Google Scholar]
- Thommes P, Blow JJ. The DNA replication licensing system. Cancer Surv. 1997;29:75–90. [PubMed] [Google Scholar]
- Vaziri C, Saxena S, Jeon Y, Lee C, Murata K, Machida Y, Wagle N, Hwang DS, Dutta A. A p53-dependent checkpoint pathway prevents rereplication. Mol Cell. 2003;11:997–1008. doi: 10.1016/s1097-2765(03)00099-6. [DOI] [PubMed] [Google Scholar]
- Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M. Histone acetylation regulates the time of replication origin firing. Mol Cell. 2002;10:1223–1233. doi: 10.1016/s1097-2765(02)00702-5. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel JA, Kutok JL, Weng AP, Dutta A. Expression of geminin as a marker of cell proliferation in normal tissues and malignancies. Am J Pathol. 2002;161:267–273. doi: 10.1016/S0002-9440(10)64178-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZQ, Liu X. Role for Plk1 phosphorylation of Hbo1 in regulation of replication licensing. Proc Natl Acad Sci USA. 2008;105:1919–1924. doi: 10.1073/pnas.0712063105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagi K, Mizuno T, You Z, Hanaoka F. Mouse geminin inhibits not only Cdt1-MCM6 interactions but also a novel intrinsic Cdt1 DNA binding activity. J Biol Chem. 2002;277:40871–40880. doi: 10.1074/jbc.M206202200. [DOI] [PubMed] [Google Scholar]
- Yin Y, Yu VC, Zhu G, Chang DC. SET8 plays a role in controlling G1/S transition by blocking lysine acetylation in histone through binding to H4 N-terminal tail. Cell Cycle. 2008;7:1423–1432. doi: 10.4161/cc.7.10.5867. [DOI] [PubMed] [Google Scholar]
- You Z, Masai H. Cdt1 forms a complex with the minichromosome maintenance protein (MCM) and activates its helicase activity. J Biol Chem. 2008;283:24469–24477. doi: 10.1074/jbc.M803212200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Feng H, Santiago FE, Kipreos ET. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature. 2003;423:885–889. doi: 10.1038/nature01747. [DOI] [PubMed] [Google Scholar]
- Zhou J, Chau C, Deng Z, Stedman W, Lieberman PM. Epigenetic control of replication origins. Cell Cycle. 2005;4:889–892. doi: 10.4161/cc.4.7.1823. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


