Abstract
Adaptation to endoplasmic reticulum (ER) stress depends on the activation of an integrated signal transduction pathway known as the unfolded protein response (UPR). Bax inhibitor-1 (BI-1) is an evolutionarily conserved ER-resident protein that suppresses cell death. Here we have investigated the role of BI-1 in the UPR. BI-1 expression suppressed IRE1α activity in fly and mouse models of ER stress. BI-1 deficient cells displayed hyperactivation of the ER stress sensor IRE1α, leading to increased levels of its downstream target X-Box binding protein-1 (XBP-1) and upregulation of UPR target genes. This phenotype was associated with the formation of a stable protein complex between BI-1 and IRE1α, decreasing its ribonuclease activity. Finally, BI-1 deficiency increased the secretory activity of primary B cells, a phenomenon regulated by XBP-1. Our results suggest a new role for BI-1 in early adaptive responses against ER stress which contrasts with its known downstream function in apoptosis.
Introduction
A number of conditions interfere with oxidative protein folding processes in the endoplasmic reticulum (ER) lumen (Ron and Walter, 2007), leading to a cellular condition referred to as “ER stress”. Adaptation to ER stress is mediated by engagement of the unfolded protein response (UPR), an integrated signal transduction pathway that transmits information about protein folding status in the ER lumen to the cytosol and nucleus to increase protein folding capacity. Conversely, cells undergo apoptosis if these mechanisms of adaptation and survival are insufficient to handle the unfolded protein load.
Expression of the UPR transcription factor X-Box binding protein-1 (XBP-1) is essential for the proper function of plasma B cells (Reimold et al., 2001; Iwakoshi et al., 2003), exocrine cells of pancreas, and salivary glands (Lee et al., 2005) and for liver lipogenesis (Lee et al., 2008). Active XBP-1 is generated by the direct processing of its mRNA by the ER stress sensor IRE1α, an ER resident Ser/Thr protein kinase and endoribonuclease (Calfon et al., 2002; Lee et al., 2002). This unconventional splicing event leads to a shift in the codon reading frame, resulting in the expression of an active transcription factor termed XBP-1s that control genes related to protein quality control, ER translocation, glycosylation, and ER/Golgi biogenesis (Shaffer et al., 2004; Lee et al., 2003; costa-Alvear et al., 2007). In addition, IRE1α operates by the formation of a complex signaling platform at the ER membrane through the binding of adaptor proteins, controlling the activation the c-Jun N-terminal kinase (JNK), ERK and NF-κB pathways (reviewed in Hetz and Glimcher, 2008b).
IRE1α activity is specifically regulated by different factors including the phosphatase PTP-1B (Gu et al., 2004), ASK1-interacting protein 1 (AIP1) (Luo et al., 2008), and some members of the BCL-2 protein family (Hetz et al., 2006). The BCL-2 family is a group of evolutionarily conserved regulators of cell death composed of both anti- and pro-apoptotic members, that operate at the mitochondrial membrane to control caspase activation (Danial and Korsmeyer, 2004). We recently described a new function for the pro-apoptotic BCL-2 family members BAX and BAK at the ER where they regulate the amplitude of IRE1α signaling by modulating its activation possibly by a physical interaction (Hetz et al., 2006). These findings suggested a novel role for BCL-2 family members as accessory factors for the instigation of certain UPR signaling events. It is unknown whether or not other apoptosis-related components regulate the UPR.
A recent study suggested that the IRE1α pathway may be modulated by additional proteins such as BAX inhibitor-1 (BI-1) (Bailly-Maitre et al., 2006). Under ischemic conditions, BI-1 deficient mice displayed increased expression of XBP-1s in the liver and kidney (Bailly-Maitre et al., 2006). However, the mechanism underlying this phenotype was not investigated. BI-1 is a six transmembrane containing protein functionally related to the BCL-2 family of proteins and is primarily located in the ER membrane (Xu and Reed, 1998). BI-1 has no obvious homology with BCL-2-related proteins, yet it physically interacts with different members of this family such as BCL-2 and BCL-XL (Xu and Reed, 1998; Chae et al., 2004). In mammalian cells BI-1 is an anti-apoptotic protein that protect cells against many different intrinsic death stimuli (Xu and Reed, 1998), including ER stress among others (Chae et al., 2004). Further studies revealed that BI-1 is well conserved in yeast, plants, viruses and many other organisms (Chae et al., 2003; Huckelhoven, 2004) where its function remains poorly explored. Here we investigated the possible role of BI-1 in the UPR. Overall our results reveal a new function for BI-1 where it negatively modulates the IRE1α/XBP-1 pathway. Our findings suggest a model wherein the expression of anti- and pro-apoptotic proteins at the ER membrane determines the amplitude of UPR responses.
Results
BI-1 Deficiency Increases XBP-1 mRNA Splicing
Although IRE1α is the most evolutionarily conserved pathway of the UPR, little is known about its regulation. To define the possible regulation of IRE1α by BI-1, we determined the levels of xbp-1 mRNA splicing using two different methods in BI-1 knockout (BI-1 KO) murine embryonic fibroblasts (MEFs). We titrated down the dose of the experimental ER stressor tunicamycin (Tm) to a point where wild-type (WT) MEFs displayed only minimal processing of XBP1 mRNA (Figure 1A). Notably, under these conditions, BI-1 KO MEFs displayed pronounced splicing of the XBP1 mRNA. The inhibitory effects of BI-1 on XBP-1 mRNA splicing were minor at very high concentrations of Tm (>1.6 μg/ml, Figure 1B), indicating that BI-1 is a modulator of IRE1α activity. Consistent with our previous findings (Hetz et al., 2006), low doses of Tm treatment revealed positive modulation of IRE1α activity by the pro-apoptotic molecule BAK (Figure 1C). In addition, we were able to validate these results by using other ER-stress inducing agents, such as brefeldin A (inhibits ER to Golgi trafficking) and thapsigargin (blocks the ER-calcium pump SERCA) (Supplementary Information Figure S1).
Figure 1. BI-1 negatively regulates IRE1α signaling.
(A)BI-1 KO and control MEFs were treated with indicated concentrations of Tm for 2.5 h and levels of XBP-1 mRNA splicing were determined in total cDNA by RT-PCR. Spliced and unspliced PCR fragments are indicated. In addition, total levels of XBP-1, spliced XBP-1 and actin mRNA were determined using RT-PCR.
(B) BI-1 KO and control MEFs were treated with indicated concentrations of Tm for 2.5h and levels of XBP-1 mRNA splicing determined. Data presented are representative of at least ten independent experiments
(C) BAX and BAK DKO MEFs were reconstituted with a BAK retroviral expression vector or empty vector (mock). After 48 h the levels of XBP-1 splicing were determined after treatment with indicated concentrations of Tm. Data is representative of 3 independent experiments.
(D) BI-1 WT and KO cells were treated with 100 ng/ml Tm for indicated time points, and the levels of XBP-1s were determined in nuclear extracts by Western blot analysis. The levels of SP-1 were used as internal control.
(E) In parallel, BI-1 WT and KO cells were treated with indicated concentrations of Tm for 4h and then the expression levels of XBP-1s, ATF4 and SP1 were determined in nuclear extracts. In addition the levels of phospho-eIF2α, and eIF2α were determined by Western blot in total cell extracts.
(F) BI-1 KO MEFs were reconstituted with a retroviral expression vector encoding a BI-1WT-EGFP fusion protein (BI-1) and then XBP-1 mRNA splicing measured by RT-PCR after treatment with different doses of Tm for 2.5 h. C: Control BI-1 KO cells. Left panel: BI-1-EGFP and HSP90 expression were determined by Western blot.
(G) WT MEFs were transduced with lentiviral vectors expressing shRNA against the bi-1 (shBI-1) or luciferase (shLuc) mRNA and levels of XBP-1 mRNA splicing determined by RT-PCR in cells treated with 100 ng/ml Tm for 2.5 h. As control, total BI-1 mRNA levels were determined by real time PCR. Mean and standard deviation are presented. p value was calculated using student’s t-Test.
(H) In parallel, BAX and BAK DKO cells were transduced with shRNA against BI-1 mRNA or luciferase (Luc) and analyzed as described in (G).
In agreement with the increased XBP-1 mRNA splicing observed above, enhanced expression of XBP-1s protein was observed in BI-1 cells undergoing ER stress when compared with control BI-1 WT cells (Figure 1D and E). BI-1 deficiency did not significantly affect the expression of IRE1-independent events such as ATF4 and eIF2α phosphorylation (Figure 1E), suggesting that BI-1 specifically affects UPR events initiated by IRE1α and not by the stress sensor PERK. We confirmed our results by reconstituting BI-1 KO cells with a human BI-1 which drastically decreased the levels of XBP-1 mRNA splicing (Fig. 1F).
In order to rule out possible compensatory effects associated with BI-1 deficiency, we targeted BI-1 mRNA with small hairpin RNA (shRNA) and lentiviral vectors. This strategy led to a ~75% decrease in BI-1 mRNA levels (Figure 1G). BI-1 knockdown cells displayed increased levels of XBP-1 mRNA splicing when compared with control cells (Figure 1G). Interestingly, knockdown of BI-1 in BAX/BAK DKO cells did not restore the normal levels of XBP-1 mRNA splicing suggesting that BI-1 operates upstream of BAX and BAK in the control of the IRE1α/XBP-1 pathway (Figure 1H). To complement these experiments, we analyzed the expression levels of BI-1 mRNA, the stability of ectopically expressed BI-1, and its subcellular distribution under ER stress conditions. No alteration in the levels of BI-1 expression or its distribution pattern was observed under these conditions (Supplementary Information Figure S2).
Increased Upregulation of XBP-1s Target Genes in BI-1 KO Cells
Previous work has demonstrated that XBP-1s regulates the expression of ER stress-induced genes that promote folding, degradation of misfolded ER proteins through the ER-associated degradation (ERAD) pathway and genes involved in the translocation of proteins into the ER. XBP-1s target genes were previously defined in MEFs by our laboratory using cDNA microarray analysis (Lee et al., 2003), and include chaperones (i.e. ERdj4), ERAD-related genes (i.e. edem and herp), genes involved in protein translocation into the ER (i.e. Sec61), and many others (Lee et al., 2003; Shaffer et al., 2004).. To define the impact of BI-1 on UPR adaptive responses, we determined the levels of XBP-1s target genes in BI-1 deficient cells by real-time PCR. Dose-response experiments demonstrated an increased upregulation of the mRNAs encoding Sec61 and EDEM in BI-1 KO cells when compared with control cells (Figure 2A). Analysis of a broad panel of XBP-1s target genes in cells treated with 100 ng/ml Tm revealed a marked activation of the UPR in BI-1 KO MEFs as evidenced by increases in XBP-1s target genes (Figure 2B and Supplementary Information Figure S1C). As control, we knocked-down XBP-1 with shRNA in BI-1 KO cells and then assessed the mRNA levels of edem, observing a decreased upregulation when compared with control cells (Figure 2C and 4A), similar to the phenotype of XBP-1 KO MEFs (Supplementary Information Figure S3A). Detailed time-course experiments indicated a more rapid and more pronounced upregulation of XBP-1s target genes in BI-1 KO cells (Figure 2D).
Figure 2. Increased UPR responses in BI-1 deficient cells.
(A) BI-1 WT and KO MEFs were treated with indicated concentrations of Tm for 8h and the mRNA levels of the XBP-1 target genes Sec61 and EDEM were determined by real-time PCR.
(B) A panel of UPR target genes was analyzed by real time PCR in BI-1 WT and KO cells treated with 100 ng/ml of Tm for 8h. p values were calculated with t-student test comparing BI-1 WT and KO cells treated with Tm (*: p = 0.05, **: p = 0.01, ***: p <0.001).
(C) As control, the mRNA levels of EDEM were determined in BI-1 KO cells expressing shRNA against XBP-1 or control shRNA (luciferase).
(D) The mRNA levels of EDEM, Sec61, and HERP were determined at indicated time points in cells treated with 100 ng/ml Tm. In A and B p values were calculated by two-way anova to compare the effects of BI-1 ablation on UPR target gene upregulation. In experiments A-D data represents average and standard deviation representative of three experiments.
Figure 4. BI-1 regulates pro-survival responses dependent on IRE1α/XBP-1.
(A) BI-1 KO MEFs were stably transduced with lentiviral vectors expressing shRNA against the xbp-1, ireα (shXBP-1 and shIRE1α) or luciferase (shLuc) mRNA and levels of XBP-1s were determined by Western blot in total nuclear extracts. As controls, ATF4 and SP-1 levels were determined.
(B) BI-1 WT and KO MEFs were stably transduced with lentiviral vectors expressing shRNA against the xbp-1 (shXBP-1) or luciferase (shLuc) mRNA and then treated with 0.1 or 1 μg/ml of Tm for 24h, and cell death was determined by PI staining and FACS analysis.
(C) BI-1 KO shIRE1, shXBP-1 and shLuc cells were treated with indicated concentrations of Tm for 24h, and cell death was determined by PI staining and FACS analysis.
(D) BI-1 KO cells transduced with indicated shRNA constructs were exposed to 0.1 μg/ml Tm, 40 μM etoposide, 50 ng/ml TNF-α together with 1 μg/ml actinomycin D or 10 μM taxol for 24 h and cell viability was analyzed by the MTS assay. Inset: The levels of XBP-1 splicing were assessed in WT MEFS after similar treatments for 3h. Average and standard deviations represent three determinations. * indicates p < 0.001 using student’s t-Test.
(E) The levels of XBP-1 splicing (right panels) and cell death (left panels) were assessed in BCL-2 WT and KO MEFs, or MCL-1 WT and KO MEFs treated with indicated concentrations of Tm for 2.5 h (splicing) or 24 h (cell viability, PI staining and FACS analysis).
BI-1 Expression Regulates the Inactivation of IRE1α/XBP-1 Signaling
We have recently reported that XBP-1 mRNA splicing levels decline after prolonged ER stress (Lin et al., 2007). Here we corroborated these observations in BI-1 WT cells, observing a decrease in the levels of splicing around 18h of Tm treatment (Figure 3A). Surprisingly, we observed a sustained maintenance of XBP-1 mRNA splicing in BI-1 deficient cells, even after 24h of treatment, suggesting that BI-1 may be involved in the inactivation of IRE1α signaling (Figure 3A). These effects correlated well with the prolonged upregulation of EDEM and Sec61 mRNA up to 24h after Tm treatment in BI-1 deficient cells (Figure 3B).
Figure 3. Delayed inactivation of XBP-1 mRNA splicing in BI-1 deficient cells.
(A) Lower panel: XBP-1 mRNA splicing was monitored over time in BI-1 WT and KO cells treated with 100 ng/ml Tm. Upper panel: Quantification of the percentage of XBP-1 mRNA splicing was calculated after the densitometric analysis.
(B) XBP-1 target genes edem and sec61 were evaluated in BI-1 WT and KO MEFs after 18h and 24h of treatment with 100 ng/ml Tm using real time PCR. p values were calculated by two-way anova to compare the effects of BI-1 ablation on UPR target gene upregulation.
(C) BI-1 WT and KO cells were treated for 2h with 1 μg/ml of Tm and washed three times with PBS. Then, mRNA splicing was evaluated by RT-PCR during the recovery period at indicated time points.
(D) BI-1 WT and KO cells were treated with 10 μg/ml of Tm for 3h to trigger complete XBP-1 mRNA splicing. Then cells were treated with 3 μg/ml actinomycin D to block transcription and the decay of XBP-1 mRNA was followed over time by real time PCR of total cDNA and normalized with the XBP-1 mRNA levels of control cultures not treated with actinomycin D.
To further evaluate the possible participation of BI-1 in the inactivation of IRE1α, we treated BI-1 WT and KO cells for only 2h with high doses of Tm to trigger almost full XBP-1 mRNA splicing in both cell types. Tm-containing media was then washed out, and XBP-1 mRNA splicing monitored during the recovery period. Under these experimental conditions, XBP-1 mRNA levels decreased by half in BI-1 WT cells by 24h post-treatment, whereas complete retention of XBP-1 splicing was still observed in BI-1 KO cells (Figure 3C). Taken together, these results suggest that BI-1 regulates the amplitude of IRE1α signaling possibly by down-regulating its activity. In control experiments, we monitored XBP-1 mRNA stability in BI-1 WT and KO cells undergoing ER stress. No significant differences in the decay of XBP-1 mRNA were observed in either cell type (Fig. 3D).
Dual Role of BI-1 in the Regulation of UPR Signaling and Downstream Apoptosis
Activation of the IRE1α/XBP-1 pathway confers cellular protection in adaptation to ER stress (Lin et al., 2007). To determine the consequences of BI-1-regulated XBP-1 mRNA splicing on survival and adaptation to ER stress, we introduced IRE1α and XBP-1 shRNAs into BI-1 KO and control cells (Figure 4A) and then assessed the effects on cell survival. As shown in Fig. 4b, inhibition of XBP-1s expression in BI-1 KO cells further enhanced their susceptibility to ER stress, as evidenced by increased cell death. Interestingly, when experiments were performed with 100 ng/ml of Tm, the effects of knocking down XBP-1 on cell viability were only evident in BI-1 deficient cells and not control cells (Fig. 4B), consistent with the dramatic differences observed in the levels of XBP-1 mRNA splicing under these conditions (Fig. 1A). At high doses of Tm, the protective effects of XBP-1 expression were also observed in BI-1 WT cells (Fig. 4B). Similar results were observed when the levels of IRE1α were reduced with shRNA in BI-1 KO cells (Fig. 4C). Thus, the previously described increased susceptibility of BI-1 deficient cells to ER stress-induced apoptosis (Supplementary Information Fig. S3B and C) is a combination of the balance between its downstream control of the apoptosis machinery and the early regulation of pro-survival signals mediated by IRE1α/XBP-1 activation.
No effects were observed on the viability of XBP-1 or IRE1α knockdown cells when they were exposed to non-ER stress related challenges, including TNFα (death receptors), paclitaxel (Taxol ™, destabilize cytoskeleton), or etoposide (DNA damage) (Figure 4D). Consistent with this observation, these drugs did not induce XBP-1 splicing (Figure 4D, inset). As additional controls we analyzed levels of XBP-1 mRNA splicing in MEFs deficient in the anti-apoptotic genes MCL-1 or BCL-2. We did not observe any significant increase in XBP-1 splicing in these cells, despite a clear increase in susceptibility to ER stress-dependent cell death (Figure 4E). Hence an augmented susceptibility to cell death does not by itself increase XBP-1 mRNA splicing activity.
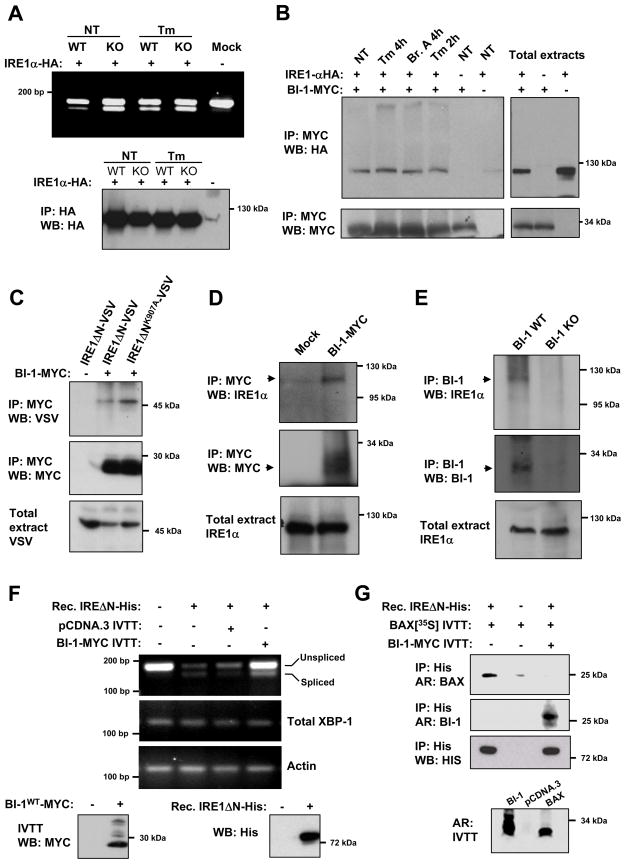
BI-1 Forms a Protein Complex with IRE1α
The activation of IRE1α requires dimerization which then triggers its auto-phosphorylation and RNAse activity. We predicted that the immunoprecipitation of IRE1α would promote its dimerization and activation in the absence of an ER stressor. We developed an assay to monitor the RNAse activity of IRE1α in vitro purified from MEFs by the immunoprecipitation (IP) of an N-terminus HA-tagged form of IRE1α (IRE1α-HA) followed by the incubation of the extracted protein complexes with a total mRNA mixture in the presence of ATP to trigger XBP-1 mRNA splicing. IRE1α-containing IP protein complexes from BI-1 KO cells were more active than IRE1α extracted from control cells (Figure 5A). These data suggest that the expression of BI-1 negatively modulates the RNAse activity of IRE1α in our cell free assay. We also analyzed the rate of IRE1α phosphorylation in BI-1 KO cells undergoing ER stress. BI-1 KO cells stimulated with 100 ng/ml of Tm for 30 min showed a characteristic phosphorylation shift of IRE1α that was absent in control cells (Supplementary Information Figure S4A).
Figure 5. BI-1 forms a protein complex with IRE1α and regulates its endoribonuclease activity.
(A) BI-1 WT and KO MEFs expressing HA-tag IRE1α (IRE1α-HA) were treated with 100 ng/ml Tm or left untreated. IRE1α-HA was immunoprecipitated (IP) and then incubated with total brain mRNA (substrate). After 30 min, mRNA was re-extracted and the levels of XBP-1 mRNA splicing were determined by RT-PCR. Bottom panel: the levels of IRE1α-HA expression were determined by Western blot of the immunoprecipitates.
(B) 293T cells were co-transfected with expression vectors for BI-1-MYC and IRE1α-HA. After 48h cells were treated with 0.5 μg/ml Tm or 20 μM brefeldin A (Bref. A) for indicated time points and then the co-precipitation of MYC-BI-1 with IRE1α-HA was evaluated by immunoprecipitation and Western blot.
(C) 293T cells were transfected with BI-1-MYC or an IRE1α inactive mutant (K907A) in the presence of VSV-tagged IRE1α lacking its ER luminal domain (IRE1ΔN-VSV) and then immunoprecipitation and Western blot analysis performed as in (B).
(D) HEK cells were transiently transfected with a BI-1-MYC expression vector or empty pCDNA.3 vector. After 48h, BI-1-MYC was immunoprecipitated and its association with endogenous IRE1α was assessed by Western blot.
(E) Endogenous BI-1 was immunoprecipitated from MEFs cells and its association with endogenous IRE1α was determined by Western blot analysis. As a control experiment, immunoprecipitation was performed from BI-1 KO cells.
(F) The endoribonuclease activity of recombinant (rec.) IRE1ΔN-HIS was monitored in vitro using the conditions described in materials and methods. IVTT BI-1-MYC or control IVTT from empty vector (pCDNA.3) were pre-incubated with IRE1ΔN-HIS for 1h at 30°C and then total mRNA was added to the reaction and incubated for 1h. Then, the ribonuclease activity of IRE1α was analyzed by RT-PCR using regular XBP-1 mRNA splicing primers evidenced as decreased PCR product of the non-spliced fragment. Total XBP-1 mRNA and actin were monitored as control. Lower panel: Western blot analysis of IVTT BI-1-MYC and IRE1ΔN-HIS is shown.
(G) IVTT BI-1 was incubated with recombinant IRE1ΔN-HIS in the presence or absence of IVTT BAX Met-35S labeled (upper panel). Then IRE1ΔN-HIS was pulled-down and its association with radiolabel BAX was determined by electrophoresis and autoradiograph (AR). As control, IRE1ΔN-HIS levels were determined by Western blot. To address the binding of BI-1 to IRE1, IVTT BI-1 Met-35S labeled was used with non-labeled BAX in the same experimental conditions. Bottom panel: Met-35S labeled BI-1, BAX or mock (pCDNA.3) were analyzed by electrophoresis and autoradiograph.
Based on the results of our in vitro splicing assay, we searched for a physical interaction between BI-1 and IRE1α. Co-IP experiments using lysates from cells co-transfected with IRE1α-HA and MYC-tagged BI-1 showed an association between both proteins (Figure 5B). BI-1 interaction required the cytosolic C-terminal region of IRE1α, which encodes the kinase and endoribonuclease domains (Figure 5C and 6B). Finally, the interaction of BI-1 with IRE1α was not altered in cells undergoing ER stress triggered by Tm or brefeldin A treatments (Figure 5B), indicating constitutive binding of BI-1 and IRE1α under resting conditions. In agreement with this result, BI-1 associated equally well in co-immunoprecipitation experiments with an inactive IRE1α mutant (K907A) and wild type IRE1α (Figure 5C). More importantly, we were able to confirm our experiments by monitoring the interaction between ectopically expressed BI-1-MYC and endogenous IRE1α in human cells (Fig. 5D). We were also successful in detecting a physical association between endogenous BI-1 and endogenous IRE1α (Fig. 5E).
Figure 6. BI-1 regulates IRE1α through its cytosolic C-terminal region.
(A) Amino acid sequence comparison of the C-terminal region of BI-1 from different species including BI-1 from human (hBI-1), mouse (mBI-1), Arabidopsis thaliana (aBI-1), Drosophila melanogaster (dBI-1) and the putative yeast homologue YN1305c. Predicted C-terminal cytosolic domain is highlighted with a gray square.
(B) 293T cells were co-transfected with expression vectors for BI-1WT-MYC or BI-1C9A-MYC with IRE1α-HA (WT) or IRE1α deletion mutant of the cytosolic (ΔC), ER luminal domain (ΔN) or empty vector (m). After 48h cell extracts were prepared and HA-IRE1α was immunoprecipitated and interactions with BI-1 determined by Western blot.
(C) BI-1 KO MEFs were reconstituted with expression vectors for EGFP fusion proteins of BI-1WT or BI-1ΔC and then XBP-1s mRNA levels measured by RT-PCR after treatment with different doses of Tm for 2.5 h. Right panel: BI-1 expression levels were analyzed by monitoring EGFP fluorescence by FACS.
(D) IVTT BI-1WT-MYC and BI-1C9A-MYC labeled with Met-35S were incubated for 3h with recombinant IRE1ΔN-HIS. Then, IRE1ΔN-HIS was pulled-down and its association with radiolabel BI-1 was determined by electrophoresis and autoradiograph (AR). Bottom panel: Levels of radiolabel BI-1WT-MYC, BI-1C9A-MYC or mock (pCDNA.3) were compared by autoradiograph.
(E) 293T cells were co-transfected with expression vectors for BI-1-MYC, and the VSV-cytosolic domain of IRE1α and after 48h BI-1-MYC was immunoprecipitated. Isolated protein complexes were incubated with increasing concentrations (10, 50 and 150 μM) of a synthetic peptide representing the C-terminal ten amino acids of BI-1 for 30 min, and IRE1α association with BI-1 measured by Western blot.
(F) BI-1 WT and KO cells were treated with 10 μM BI-1C-ter peptide or control scrambled peptide for 2h, cells treated with 100 ng/ml Tm for indicated time points and levels of XBP-1 mRNA splicing were determined in total cDNA by RT-PCR.
(G) WT MEFs were pre-treated with 10 μM BI-1C-ter peptide or scrambled peptide for 2h and then treated with 200 ng/ml tunicamycin or 0.2 μM brefeldin A for 2h or with 1 μg/ml tunicamycin as positive control. Then, the phosphorylation shift (P-IRE1) of endogenous IRE1α was monitored by Western blot. HSP90 levels were analyzed as loading control.
We next tested the possible effects of BI-1 on the activity of IRE1α. We established an in vitro assay to monitor the endoribonuclase activity of purified IRE1α. The cytosolic His-tag version of human IRE1α (IRE1ΔNα-HIS) was expressed and purified from insect cells since they express a BAX and BI-1 homologue and because this mutant of IRE1α adopts an active dimeric state (not shown). Then, purified IRE1ΔNα-HIS was incubated with a mixture of total mRNA in the presence or absence of in vitro transcribed/translated (IVTT) BI-1. After 1h of incubation, mRNA was re-extracted and the cleavage of XBP-1 mRNA in the splicing site was monitored by RT-PCR. As shown in Fig. 5F, the activity of IRE1ΔN–HIS was almost completely blocked by the presence of BI-1 in the reaction. These results indicate that the effects of BI-1 on IRE1α activity can be reconstituted in vitro, suggesting a direct regulation.
We tested for possible effects of BI-1 on the binding of BAK to IRE1α. We first performed transient transfection of different combinations of IRE1α-HA, BAK and BI-1-MYC. Co-expression of BAK and BI-1 reduced the interaction of BAK with IRE1α as compared with control (Supplementary Information Figure S4B). Similar results were observed when the binding of BAX to the complex was tested in the same experimental system (not shown). We have previously described that the physical association between IRE1α and BAX are recapitulated with recombinant proteins, indicating a direct interaction (Hetz et al., 2006). To monitor the binding of BI-1 to IRE1α, we first performed pull-down assays with recombinant IRE1ΔN-HIS and IVTT BI-1WT. We were able to detect the formation of a protein complex between IRE1α and BI-1WT in vitro (Fig. 6D and 5G). Interestingly, in the same experimental system the presence of BI-1 drastically reduced the binding of IVTT BAX to IRE1ΔN-HIS (Fig. 5G), suggesting that BAX and BI-1 regulate IRE1α through related mechanisms and may compete for a common binding site. Taken together with the results shown in Fig. 1H, these findings suggest that BI-1 operates upstream of BAX and BAK in the control of IRE1α inactivation. In agreement with previous findings (Xu and Reed, 1998), we did not observe a significant interaction between BI-1 and BAX or BAK, but it associated with BCL-2 or BCL-XL (Supplementary Information Figure S4C–E).
BI-1 Regulates IRE1α Through its C-terminal Region
Bioinformatics analysis of the BI-1 sequence failed to identify known possible protein-protein interaction or catalytic domains present in other proteins. The cytosolic C-terminus of mammalian BI-1 is composed of only 10 amino acids, and it is conserved in multicellular organisms (Fig. 6a), and has been shown to be essential for the regulation of apoptosis (Chae et al., 2003). We expressed a BI-1 mutant in which the last nine amino acids of the protein were replaced by alanines (BI-1C9A) and tested its interaction with IRE1α. As shown in Figure 6B, BI-1C9A did not significantly interact with IRE1α but it still located at the ER (Supplementary Information Figure S5). Hence the lack of physical association between BI-1C9A and IRE1α is not due to a change in the subcellular localization of the mutant protein.
To test the role of the C-terminal region of BI-1 on IRE1α signaling we performed reconstitution experiments in BI-1 KO cells. Ectopic expression of hBI-1 reduced the levels of XBP-1 mRNA splicing in BI-1 KO cells (Figure 6C), an effect that was not observed in cells expressing mutant BI-1 where full XBP-1 mRNA splicing was still observed. To monitor the effects of the C-terminal region of BI-1 on the interaction with IRE1α, we performed pull-down assays with recombinant IRE1ΔN-HIS and IVTT BI-1WT or BI-1C9A. Mutation on the C-terminal region of BI-1 completely abrogated its association with IRE1ΔN-HIS (Fig. 6D).
We characterized in more detail the function of the C-terminus of BI-1 on the UPR. A synthetic peptide containing the last 13 amino acids of BI-1 was fused with a polyarginine tag to enhance cell permeability (BI-1C-ter). To test the effects of the peptide on the interaction between BI-1 and IRE1α, we first immunoprecipitated the BI-1/IRE1α complex and then incubated it with different concentrations of BI-1C-ter for 30 min. At 150 μM the peptide completely displaced IRE1α from BI-1WT confirming the requirement of the C-terminus for its interaction with IRE1α (Figure 6E). We then assessed the activity of BI-1C-ter in cells treated with Tm. Treatment of cells with the C-terminal BI-1 peptide increased XBP-1 spliced mRNA (Fig. 6F) and augmented the levels of herp mRNA when compared with a control scrambled peptide (Supplementary Information Figure S4G). This effect was not observed in BI-1 KO MEFs (Supplementary Information Figure S4F), indicating that the activity of the BI-1C-ter peptide is specific and depends on the expression of endogenous BI-1. In addition, the BI-1C-ter peptide did not increase XBP-1 mRNA splicing in BAX and BAK DKO cells (Supplementary Information Fig. S4H). To complement these experiments, we monitored the effects of the BI-1C-ter peptide on the phosphorylation of IRE1α, an event associated with its activation. BI-1C-ter peptide drastically increased the rate of IRE1α phosphorylation in cells treated with low doses of Tm or brefeldin A (Fig. 6G). Taken together, these results reinforce the observation that BI-1 is a negative regulator of IRE1α and that this regulation occurs through the formation of a protein complex between the two proteins.
BI-1 Regulates IRE1α in vivo in Multicellular Organisms
Homologues of human BI-1 (hBI-1) have been identified in different species including plants such as Arabidopsis thaliana, invertebrate animals such as Drosophila melanogaster (dBI-1), the budding yeast Saccharomyces cerevisiae (Ynl305c) and other species (Chae et al., 2003). However, when we analyzed the C-terminal sequence of BI-1 from different species we noticed that the amino acids critical for interaction with IRE1α were not conserved in Ynl305c, but were present in other species analyzed (Fig. 6a). To assess the function of BI-1 in regulating the UPR in different species in vivo, we first tested the susceptibility of BI-1 KO mice to a stress response. BI-1 mice were treated once with Tm for 6h and levels of XBP-1s and ATF4 then analyzed in liver nuclear extracts (Figure 7A). We observed a marked increase of XBP-1s levels in BI-1 KO mice treated with Tm when compared with control animals. However, no differences in the induction of PERK-dependent transcription factor ATF4 were observed. Similar results were observed when XBP-1s levels were monitored in the kidney (Figure 7B).
Figure 7. BI-1 regulates XBP-1 mRNA splicing in vivo and modulates IgM secretion in primary B cells.
(A) BI-1 WT and BI-1 deficient mice were injected intraperitoneally with 0.2 μg Tm/g weight, and after 6 h of treatment animals were sacrificed and the expression levels of XBP-1s, ATF4 and SP1 (control) were determined by Western blot of nuclear extracts.
(B) In parallel, expression of XBP-1s and HSP90 were determined in kidney total protein extracts from animals presented in (B).
(C) WT or dBI-1 overexpressing D. melanogaster larva were grown in the presence or absence of 50 μg/ml Tm, 50 mM DTT or 10 μM thapsigargin for 20 h, and levels of XBP-1 splicing were determined by RT-PCR. As control, overexpression levels of dBI-1 mRNA and actin were measured by RT-PCR. Data is representative of three independent experiments.
(D) Physiological model of ER stress: Primary B cells were purified from spleens of BI-1 WT and KO mice and then stimulated with the indicated concentration of LPS. After 2 days of culture, the levels of IgM were measured in the cell culture supernatant by ELISA. The values represent the results of the analysis of four different animals. * indicates p < 0.05 using student’s t-Test.
(E) Splenic B cells from BI-1 WT and KO mice were cultured for 2 days in the presence of 0.1 μg/ml LPS, stained with brefeldin A-BODIPY, and then analyzed by FACS to determine relative content of ER and Golgi.
(F) Cell surface levels of IgM and IgD were measured by FACS analysis of freshly isolated splenocytes from BI-1 WT and KO mice. Average percentages of IgD and IgM positive cells are indicated.
The IRE1α/XBP-1 branch is highly conserved in D. melanogaster (Souid et al., 2007; Plongthongkum et al., 2007). We analyzed the levels of XBP-1 mRNA splicing in flies overexpressing dBI-1. As an experimental model we grew fly larvae in media containing 50 μg/ml Tm for 20h and then measured the levels of XBP-1 mRNA splicing in total tissue extracts as previously described (Plongthongkum et al., 2007). As shown in Figure 7C, overexpression of dBI-1 significantly decreased the levels of XBP-1 splicing in larvae treated with Tm, indicating that BI-1 also regulates IRE1α in invertebrates. Similar results were observed when dBI-1 overexpressing flies were exposed to thapsigargin or DTT (Fig. 7C). Finally, we tested the activity of the putative yeast BI-1 homologue by generating an Yn1305c mutant. Consistent with the lack of conservation of the C-terminal IRE1α interacting motif, mutant Yn1305c yeast cells did not show any significant increase in levels of the XBP-1 functional homologue HAC1p when compared with control yeast grown in DTT-containing culture media (data not shown), suggesting that while showing some limited amino-acid sequence homology, this yeast protein may not be a close ortholog of BI-1 (S. Bernales, J. Weissman and P. Walter, unpublished data).
BI-1 Deficiency Increases ER/Golgi Expansion and Immunoglobulin Secretion in Primary B Cells
Secretory cells require a developed ER for proper function. The first insights about the function of XBP-1 in vivo came from studies in the immune system, where the high demand for immunoglobulin synthesis in B cells constitutes an endogenous source of ER stress (Reimold et al., 2001). XBP-1-deficient B cells are markedly defective in antibody secretion in vivo in response to antigenic challenge (Iwakoshi et al., 2003; Zhang et al., 2005). To evaluate the role of BI-1 in the control of XBP-1-dependent processes in a physiologically-relevant system, we determined the rate of IgM secretion in BI-1 KO primary B cells. Increased levels of IgM were observed in the cell culture media of BI-1 deficient B cells after stimulation with LPS (Figure 7D). This phenomenon was associated with a marked increased staining with Brefeldin A-BODIPY in BI-1 KO cells, which is indicative of an expanded ER and Golgi in these cells compared with controls (Figure 7E), a process previously described to be XBP-1 dependent (Sriburi et al., 2004; Shaffer et al., 2004). Freshly isolated splenic B cells from BI-1 KO mice showed no differences in surface IgM and IgD expression when compared with control mice (Figure 7F). Taken together, these data indicate that BI-1 regulates two distinct known processes mediated by XBP-1 in primary B cells, ER/Golgi expansion and immunoglobulin secretion.
Discussion
BI-1 is a specialized and evolutionarily conserved regulator of cell death and is present in species even where no BCL-2 family homologues have been described including other eukaryotes, plants, bacteria, and even viruses (Chae et al., 2003; Huckelhoven, 2004). Recent studies indicate that several BCL-2 family members reside in other organelles where they perform novel functions (reviewed in Hetz, 2007). In support of this concept, we recently described that BAX and BAK modulate the stress sensor IRE1α at the ER membrane (Hetz et al., 2006). Here we present evidence indicating that BI-1 negatively regulates the IRE1α/XBP-1 pathway. BI-1 deficient cells showed hyperactivation of IRE1α associated with increased XBP-1 mRNA splicing and upregulation of XBP-1s-dependent responses. Notably, inactivation of IRE1α signaling over time was markedly delayed in BI-1 KO MEFs, indicating an important inhibitory activity of BI-1 on XBP-1 mRNA splicing. This regulation was mediated by the formation of a protein complex between IRE1α and BI-1, and was reconstituted in vitro with purified components. The inhibition of IRE1α by BI-1 was recapitulated in vivo in BI-1 deficient mice and flies overexpressing dBI-1, indicating that this regulation is conserved across species.
Engagement of the IRE1α/XBP-1 pathway confers protection against ER stress (Lin et al., 2007). Our results indicate that BI-1 negatively controls XBP-1s expression, which contrasts with its known general downstream anti-apoptotic activity against intrinsic death stimuli (i.e. growth factor deprivation, oxidative stress and DNA damage) (Xu and Reed, 1998). Interestingly, BI-1’s regulatory effects on the UPR are more evident when moderate to low doses of ER stressors are employed, which resembles in vivo conditions where cells are equipped to cope with injury (adaptive conditions). In agreement with these findings, we observed an increased rate of IgM secretion in LPS-stimulated BI-1 deficient primary B cells. Consistent with this idea, it has been reported that mild ER stress conditions evoke distinct signaling processes, where apoptosis-related events are not observed under mild ER stress conditions (Rutkowski et al., 2006). Our results suggest that the changes in apoptosis observed in BI-1 KO cells may reflect a balance between the inhibition of survival signaling mediated by IRE1α and the general downstream anti-apoptotic activity of the intrinsic death machinery (model in Supplementary Information Fig. S7). Recently, it was reported that BI-1 overexpression may negatively affect ER stress responses through the control of the Heme Oxygenase-1 gene (Lee et al., 2007). However, we did not observe any effects on the transcription of Heme Oxygenase-1 in BI-1 deficient MEFs (Supplementary Information Fig. S6). The regulation of IRE1α by BI-1 may be related to the effects of BAX and BAK on the UPR, where BAX and BAK may compete for a similar binding site on IRE1α. Because BAX and BAK are not present in yeast, the acquisition of UPR modulatory activities may have evolved in higher eukaryotes. In fact, we did not observe a significant effect on the activation of the UPR in Ynl305c deficient yeast
Several bifunctional activities for apoptosis-related proteins have been described over the last few years (reviewed in Hetz and Glimcher, 2008). For example, the pro-apoptotic protein BAD controls glucose metabolism (Danial et al., 2003) and insulin secretion by β-cells (Danial et al., 2008). Similarly, expression of the pro-apoptotic protein BID is important for engagement of survival DNA repair responses (Zinkel et al., 2005; Kamer et al., 2005). BCL-2-related proteins have inhibitory activities on autophagy, a survival pathway against nutrient starvation (Maiuri et al., 2007; Pattingre et al., 2005; Maiuri et al., 2007). BCL-2 and BCL-XL also have alternative roles in pro-inflammatory processes through NALP1 regulation (Bruey et al., 2007). Finally, BAX and BAK were shown to control mitochondrial morphogenesis (Karbowski et al., 2006). Thus, mounting evidence indicates that apoptosis-related proteins have alternative functions and vital roles in normal cellular physiology.
Although PERK and IRE1α share functionally similar luminal sensing domains and are both activated in cells treated with ER stress inducers in vitro, they are selectively activated in vivo by the physiological stress of unfolded proteins. For example, XBP-1 deficiency drastically affects the ability of B lymphocytes to secrete immunoglobulins (Reimold et al., 2001; Iwakoshi et al., 2003; Lee et al., 2005), a defect that is not present in PERK deficient mice (Gass et al., 2007). The differences in terms of tissue-specific regulation of the UPR in vivo may be explained by the formation of distinct regulatory protein complexes through specific binding of adaptor and modulator proteins. Since several proteins selectively modulate IRE1α signaling (reviewed in Hetz and Glimcher, 2008b) we envision a model where IRE1α signaling emerge as a highly regulated process than previously appreciated, and may be controlled by the formation of a complex protein scaffold unto which many other regulatory components assemble (previously referred to as the UPRosome (Hetz and Glimcher, 2008b)). Taken together with the current study, increasing evidence suggests a rheostat model in which a balance between anti- and pro-apoptotic proteins at the ER membrane modulates the amplitude of IRE1α signaling, and hence cellular sensitivity to ER stress conditions.
Experimental Procedures
See online supplementary material for detailed methods and acknowledgments
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Bailly-Maitre B, Fondevila C, Kaldas F, Droin N, Luciano F, Ricci JE, Croxton R, Krajewska M, Zapata JM, Kupiec-Weglinski JW, Farmer D, Reed JC. Cytoprotective gene bi-1 is required for intrinsic protection from endoplasmic reticulum stress and ischemia-reperfusion injury. Proc Natl Acad Sci U S A. 2006;103:2809–2814. doi: 10.1073/pnas.0506854103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruey JM, Bruey-Sedano N, Luciano F, Zhai D, Balpai R, Xu C, Kress CL, Bailly-Maitre B, Li X, Osterman A, Matsuzawa S, Terskikh AV, Faustin B, Reed JC. Bcl-2 and Bcl-XL regulate proinflammatory caspase-1 activation by interaction with NALP1. Cell. 2007;129:45–56. doi: 10.1016/j.cell.2007.01.045. [DOI] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Chae HJ, Ke N, Kim HR, Chen S, Godzik A, Dickman M, Reed JC. Evolutionarily conserved cytoprotection provided by Bax Inhibitor-1 homologs from animals, plants, and yeast. Gene. 2003;323:101–113. doi: 10.1016/j.gene.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Chae HJ, Kim HR, Xu C, Bailly-Maitre B, Krajewska M, Krajewski S, Banares S, Cui J, Digicaylioglu M, Ke N, Kitada S, Monosov E, Thomas M, Kress CL, Babendure JR, Tsien RY, Lipton SA, Reed JC. BI-1 regulates an apoptosis pathway linked to endoplasmic reticulum stress. Mol Cell. 2004;15:355–366. doi: 10.1016/j.molcel.2004.06.038. [DOI] [PubMed] [Google Scholar]
- costa-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD. XBP1 Controls Diverse Cell Type- and Condition-Specific Transcriptional Regulatory Networks. Mol Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Danial NN, Gramm CF, Scorrano L, Zhang CY, Krauss S, Ranger AM, Datta SR, Greenberg ME, Licklider LJ, Lowell BB, Gygi SP, Korsmeyer SJ. BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature. 2003;424:952–956. doi: 10.1038/nature01825. [DOI] [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- Danial NN, Walensky LD, Zhang CY, Choi CS, Fisher JK, Molina AJ, Datta SR, Pitter KL, Bird GH, Wikstrom JD, Deeney JT, Robertson K, Morash J, Kulkarni A, Neschen S, Kim S, Greenberg ME, Corkey BE, Shirihai OS, Shulman GI, Lowell BB, Korsmeyer SJ. Dual role of proapoptotic BAD in insulin secretion and beta cell survival. Nat Med. 2008;14:144–153. doi: 10.1038/nm1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass JN, Jiang HY, Wek RC, Brewer JW. The unfolded protein response of B-lymphocytes: PERK-independent development of antibody-secreting cells. Mol Immunol. 2007 doi: 10.1016/j.molimm.2007.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu F, Nguyen DT, Stuible M, Dube N, Tremblay ML, Chevet E. Protein-tyrosine phosphatase 1B potentiates IRE1 signaling during endoplasmic reticulum stress. J Biol Chem. 2004;279:49689–49693. doi: 10.1074/jbc.C400261200. [DOI] [PubMed] [Google Scholar]
- Hetz C, Bernasconi P, Fisher J, Lee AH, Bassik MC, Antonsson B, Brandt GS, Iwakoshi NN, Schinzel A, Glimcher LH, Korsmeyer SJ. Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science. 2006;312:572–576. doi: 10.1126/science.1123480. [DOI] [PubMed] [Google Scholar]
- Hetz C, Glimcher L. The daily job of night killers: alternative roles of the BCL-2 family in organelle physiology. Trends Cell Biol. 2008;18:38–44. doi: 10.1016/j.tcb.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Hetz C, Glimcher L. The UPRosome and XBP-1: mastering secretory cell function. Curr Immunol Rev. 2008b;4:1–10. [Google Scholar]
- Hetz CA. ER Stress Signaling and the BCL-2 Family of Proteins: From Adaptation to Irreversible Cellular Damage. Antioxid Redox Signal. 2007;9:2345–2356. doi: 10.1089/ars.2007.1793. [DOI] [PubMed] [Google Scholar]
- Huckelhoven R. BAX Inhibitor-1, an ancient cell death suppressor in animals and plants with prokaryotic relatives. Apoptosis. 2004;9:299–307. doi: 10.1023/b:appt.0000025806.71000.1c. [DOI] [PubMed] [Google Scholar]
- Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4:321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- Kamer I, Sarig R, Zaltsman Y, Niv H, Oberkovitz G, Regev L, Haimovich G, Lerenthal Y, Marcellus RC, Gross A. Proapoptotic BID is an ATM effector in the DNA-damage response. Cell. 2005;122:593–603. doi: 10.1016/j.cell.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Karbowski M, Norris KL, Cleland MM, Jeong SY, Youle RJ. Role of Bax and Bak in mitochondrial morphogenesis. Nature. 2006;443:658–662. doi: 10.1038/nature05111. [DOI] [PubMed] [Google Scholar]
- Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 2005;24:4368–4380. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GH, Kim HK, Chae SW, Kim DS, Ha KC, Mike C, Kress C, Reed JC, Kim HR, Chae HJ. Bax inhibitor-1 regulates endoplasmic reticulum stress-associated reactive oxygen species and heme oxygenase-I expression. J Biol Chem. 2007 doi: 10.1074/jbc.M700053200. [DOI] [PubMed] [Google Scholar]
- Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, He Y, Zhang H, Yu L, Chen H, Xu Z, Tang S, Urano F, Min W. AIP1 is critical in transducing IRE1-mediated endoplasmic reticulum stress response. J Biol Chem. 2008 doi: 10.1074/jbc.M710557200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri MC, Le TG, Criollo A, Rain JC, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, Hickman JA, Geneste O, Kroemer G. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Plongthongkum N, Kullawong N, Panyim S, Tirasophon W. Ire1 regulated XBP1 mRNA splicing is essential for the unfolded protein response (UPR) in Drosophila melanogaster. Biochem Biophys Res Commun. 2007;354:789–794. doi: 10.1016/j.bbrc.2007.01.056. [DOI] [PubMed] [Google Scholar]
- Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Arnold SM, Miller CN, Wu J, Li J, Gunnison KM, Mori K, Sadighi Akha AA, Raden D, Kaufman RJ. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 2006;4:e374. doi: 10.1371/journal.pbio.0040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee AH, Qian SB, Zhao H, Yu X, Yang L, Tan BK, Rosenwald A, Hurt EM, Petroulakis E, Sonenberg N, Yewdell JW, Calame K, Glimcher LH, Staudt LM. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Souid S, Lepesant JA, Yanicostas C. The xbp-1 gene is essential for development in Drosophila. Dev Genes Evol. 2007;217:159–167. doi: 10.1007/s00427-006-0124-1. [DOI] [PubMed] [Google Scholar]
- Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167:35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Reed JC. Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol Cell. 1998;1:337–346. doi: 10.1016/s1097-2765(00)80034-9. [DOI] [PubMed] [Google Scholar]
- Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest. 2005;115:268–281. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkel SS, Hurov KE, Ong C, Abtahi FM, Gross A, Korsmeyer SJ. A role for proapoptotic BID in the DNA-damage response. Cell. 2005;122:579–591. doi: 10.1016/j.cell.2005.06.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.