Abstract
We are reporting on the synthesis of fluorescent nucleoside analogs with modified sugar moieties (e.g., sugars other than ribose and 2’-deoxyribose). Four novel derivatives of the fluorescent thymidine analog 6-methyl-3-(β-D-2’-deoxyribofuranosyl) furano-[2,3-d]pyrimidin-2-one were synthesized via Sonogashira reaction and subsequent copper-catalyzed cycloaddition. These compounds represent promising tools for studying nucleoside metabolism inside living cells, as well as for screening directed evolution libraries of 2’-deoxyribonucleoside kinases with new and improved activity for the corresponding nucleoside analogs.
The ability to detect small-molecule metabolites with high sensitivity in complex mixtures such as a cell’s cytoplasm greatly benefits studies of their cellular uptake and metabolism.1,2 The application of fluorophors as reporters has proven a particularly versatile strategy and hence was recently adapted by our group to study the phosphorylation of nucleosides and nucleoside analogs by nucleoside kinases (dNKs).3 The dNKs are part of the nucleoside salvage pathway and also play a critical role in activating nucleoside analog prodrugs used for antiviral and cancer therapy.
Although nucleosides possess intrinsic fluorescence properties through their pyrimidine and purine moieties, low quantum yields and overlapping absorption maxima with aromatic amino acids in proteins and small-molecule metabolites such as flavines and NADH make their selective detection in vivo impractical. To overcome cellular autofluorescence and improve signal-to-noise ratios, fluorescent substrates with absorption maxima of >300 nm are necessary.4–6
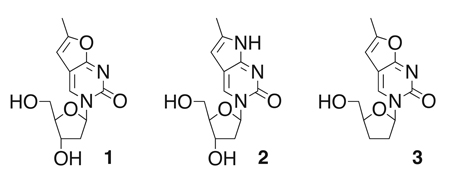
Fortunately, relatively small synthetic modifications of the nucleobase moieties can augment the spectral properties of nucleosides, red-shifting their absorption spectra and increasing quantum yields. In recent work, Berry and coworkers reported the decoration of pyrimidines with furano and pyrrolo moieties, creating fluorescent analogs of thymidine (1) and 2’-deoxycytidine (2) to study DNA structure.7 The modification of the nucleobase shifted the excitation wavelength to 331 – 335 nm while the emission maximum reached 413 – 415 nm.
Our kinetic experiments with 1 showed that the expanded heterocycle does not have a major effect on the performance of dNK from Drosophila melanogaster (DmdNK), a reference enzyme for the phosphorylation of 2’-deoxynucleosides and nucleoside analogs.3 More qualitatively, activation of the fluorescent nucleoside by the fruitfly dNK resulted in accumulation of the phosphorylated analog which could be monitored via fluorescence microscopy (Fig. 1). In the same study, we also prepared the fluorescent version of the nucleoside analog prodrug 3’-deoxy-thymidine (fddT, 3). Fluorescent ddT was successfully used to screen directed evolution libraries of DmdNK for variants with changed substrate specificity and enhanced activity for the prodrug. Host cells that express kinase variants with the highest activity for 3 were identified and sorted by flow cytometry and yielded enzymes with up to 10,000-fold change in substrate specificity. 3
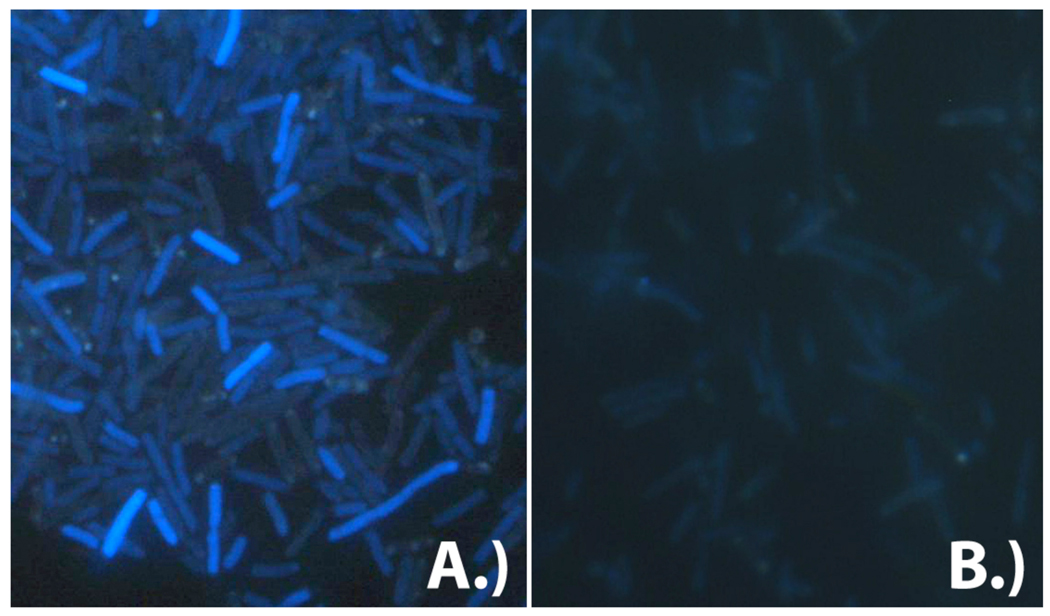
Figure 1.
Fluorescence microscopy images of E. coli KY895 expressing A.) an exogenous 2’-deoxyribonucleoside kinase from Drosophila melanogaster or B.) glycinamide ribonucleotide formyltransferase from E. coli (negative control) upon incubation with compound 1 (λex = 331 nm, λem = 413 nm). In the presence of a kinase, intracellular accumulation of 1 is observed as reflected in higher fluorescence intensity.
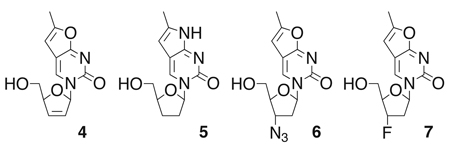
To explore the use of fluorescent nucleoside analogs more broadly, we herein report the successful preparation of four additional fluorescent nucleoside analogs, namely 6-methyl-3-(β-D-2’,3’-dideoxy-2’,3’-didehydro-ribofuranosyl)furano-[2,3-d]pyrimidin-2-one ( 4 ; fd4T) , 6 -methyl-3-(β-D-2’,3’-dideoxy-ribofuranosyl) pyrrolo-[2,3-d]pyrimidin-2-one (5; fddC), 6 - methyl-3-(β-D-3’ azido - 2 ’ , 3 ’ - dideoxy-ribofuranosyl)furano-[2,3-d]pyrimidin-2-one (6; fAZT), and 6-methyl-3-(β-D-3’-fluoro-2’,3’-dideoxy-ribofuranosyl) furano-[2,3-d]pyrimidin-2-one (7; fFT).
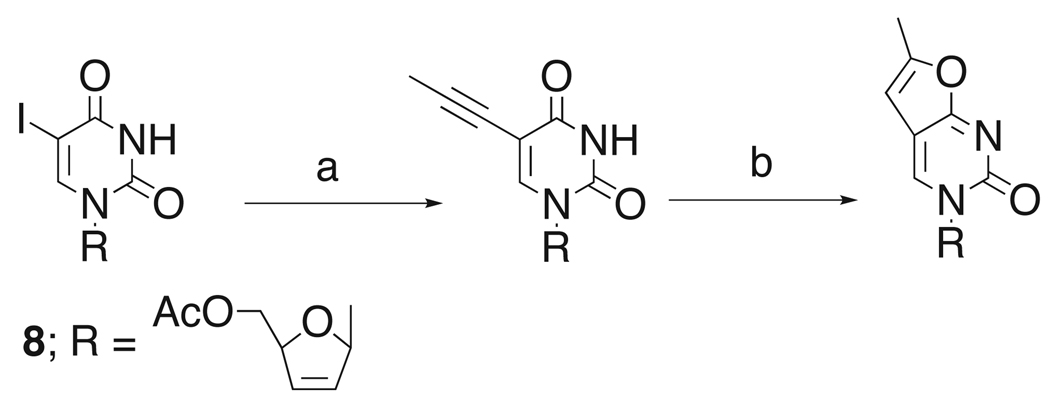
The synthesis of 4 and 5 is relatively straight forward and follows the preparation previously described for 3.3 For 4, we started from the known 5-iodo intermediate 8, assembling the furano pyrimidine via the established Sonogashira route using propyne in the presence of Pd/Cu8,9 and subsequent Cu-catalyzed cycloaddition to afford the fluorescent moiety (Scheme 1).10 The pyrrolo analog 5 can be prepared directly from 3 by treatment of the furano pyrimidine with ammonic methanol to yield the cytidine variant. 11,12
Scheme 1.
Introduction of furano moiety. (a) propyne, Pd(PPh3)4, CuI, NEt3, DMF, rt, 22 h, 60%; (b) CuI, NEt3, MeOH, reflux, 28–45%.
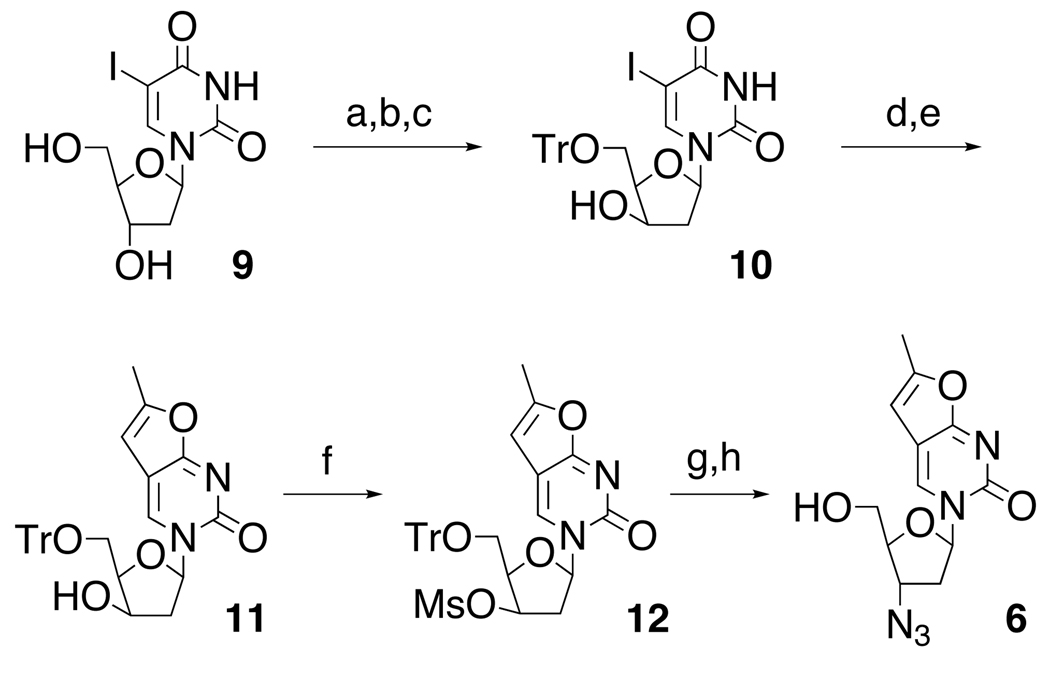
A similar strategy to prepare 6 was complicated by the azido group’s interference with the Pd chemistry, forcing a change in the synthetic approach as outlined in Scheme 2. Following selective protection of the 5’-OH group in 9 with trityl chloride and treatment of the intermediate with mesyl chloride, the configuration of the 3’-group was readily inverted upon refluxing in ethanolic sodium hydroxide to give the xylo compound 10.13 Successive treatment of 10 with propyne in the presence of Pd/Cu and Cu-catalyzed cyclization produced 11.8,9 Next, reaction with MsCl yielded the mesylate 12 and subsequent SN2 substitution with LiN3 and deprotection with acetic acid afforded the azido substituted fluorescent nucleoside analog 6.14, 15
Scheme 2.
Preparation of fAZT. (a) TrCl, Py, reflux; (b) MsCl, CH2Cl2, 67%; (c) NaOH, EtOH, reflux, 65%; (d) propyne, Pd(PPh3)4, CuI, NEt3, DMF, rt, 22 h, 62%; (e) CuI, NEt3, MeOH, reflux, 70%;(f) MsCl, Et3N, CH2Cl2, rt, 1h, 87%; (g) LiN3, DMF, 110°C, 2.5h, 80%; (h) AcOH, 90°C, 15 min, 60%.
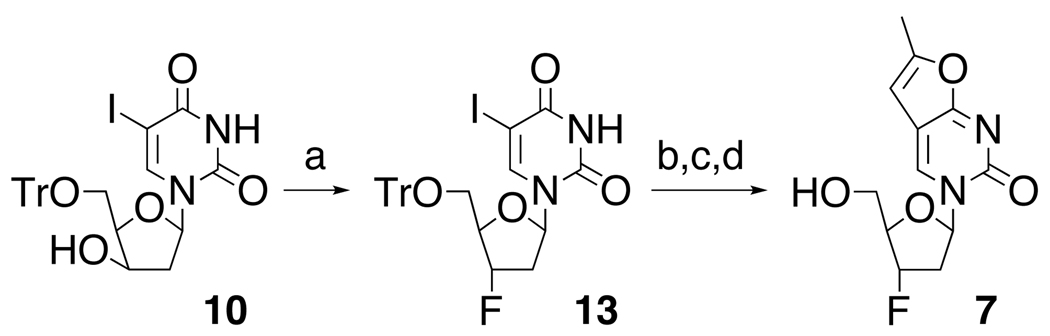
Alternatively, intermediate 10 could readily be converted to the 3’-fluoro analog 13 with DAST (Scheme 3). Subsequent substitution of the 5-iodo group with propyne, followed by cyclization again yielded the corresponding furano-pyrimidine 7.16
Scheme 3.
Fluorination of xylo-intermediate 10. (a) DAST, CH2Cl2, rt, 1h, 52%; (b) propyne, Pd(PPh3)4, CuI, NEt3, DMF, rt, 22 h, 65%; (c) CuI, NEt3, MeOH, reflux, 38%; (d) AcOH, 90°C, 15 min, 60%.
The fluorescence properties of all four compounds were determined and found to be very similar to the previously reported 3, having excitation wavelengths of 333 ± 2 nm with a molar absorptivity of 3460 M−1 cm−1 and emission maxima ranging from 413 – 415 nm. The quantum yield is approximately 0.3. Interestingly, the fluorescence intensity of 4 is approximately 2-fold higher than the other nucleosides and nucleoside analogs, presumably a reflection of the heterocycle’s electronic interactions with the alkene bond in the sugar moiety.
Separately, we also tested the compounds’ substrate properties with wild type DmdNK (Table 1). Experiments were limited to specific activity measurements at 100 µM substrate concentration as the determination of accurate Michaelis-Menten parameters was complicated by limited solubility of fluorescent analogs above 1 mM and KM values of >250 µM for some substrates.
Table 1.
Substrate specificities of wild type DmdNK
| substrate (100 µM) | specific activity (µM min−1 mg−1) |
|---|---|
| THY * | 60,000 ± 1010 |
| fTHY (1) * | 25,511 ± 455 (43%) |
| dC * | 43,734 ± 660 |
| fdC (2) | 56,034 ± 1050 (128%) |
| ddT | 1310 ± 87 |
| fddT (3) | 397 ± 36 (30%) |
| ddC | 2005 ± 148 |
| fddC (5) | 52 ± 9 (2.6%) |
| FT | 1189 ± 17 |
| fFT (7) | 144 ± 27 (12%) |
| AZT | 1054 ± 14 |
| fAZT (6) | 128 ± 7 (12%) |
| d4T | 70 ± 15 |
| fd4T (4) | 232 ± 20 (330%) |
Specific activities were measured by spectrophotometric coupled-enzyme assay with 100 µM substrate and 1 mM ATP except for substrates marked with an asterisk (measured at 10 µM substrate concentration under vmax conditions). Experiments were performed in triplicates. Percentage in parentheses is relative activity of fluorescent analog over non-fluorescent analog.
Our data indicate a relatively minor impact on the substrate properties of 2’-deoxyribonucleosides 1 and 2 upon introduction of the furano and pyrrolo moieties. Beyond the overall lower activities for nucleoside analogs, the same trend is observed for ddT and its corresponding fluorescent analog 3. Furano-pyrimidines with the 3’-azido (6) and 3’-fluoro (7) groups decline about 10-fold, possibly reflecting changes in substrate binding due to steric constraints in the enzyme active site and conformational changes in the substrate. Similar binding effects, albeit this time in favor of the substrate, could explain the 3-fold gain in activity for 4, the fluorescent version of d4T. Unexpectedly, pyrrolo analog 5 shows a 40-fold drop in activity relative to the non-fluorescent ddC, a trend opposite to the slight gain observed with its 2’-deoxyribosoyl analog 2. Conformational differences in the sugar moiety are unlikely to account for the differences as 3 possesses the same 2’,3’-dideoxyribosyl portion, yet shows 10-fold higher activity. Future studies exploring the conformational preferences of furano and pyrrolopyrimidines in regard to their syn-anti orientation might assist in rationalizing their difference in performance.
In summary, the application of fluorescent nucleoside analogs as molecular probes provides a new tool for studying the uptake and metabolism of antiviral prodrugs as demonstrated in our laboratory and by other research groups.3,17,18 Herein, we have shown that nucleoside analogs with furano and pyrrolo modification are synthetically readily accessible and can serve as substrates for the type-I 2’-deoxyribo-nucleoside kinase from Drosophila melanogaster. In bacterial and mammalian cell cultures, these fluorescent analogs have shown no unusual cytotoxicity, hence making them suitable reporters to study cellular uptake and phosphorylation, as well as to evaluate large combinatorial libraries of kinases for variants with substrate specificity for the corresponding prodrugs by fluorescence activated cell sorting (FACS).
Acknowledgement
This work was supported in part by the National Institutes of Health (GM69958), as well as by a grant to the Emory Center for AIDS Research (AI050409) from the National Institutes of Health and by institutional funding from the Emory University Health Science Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Hermanson GT. Bioconjugate Techniques. San Diego: Academic; 2008. Chapter 8. [Google Scholar]
- 2.O'Donnell MJ, McLaughlin LW. In: Bioorganic Chemistry; Nucleic Acids. Hecht SM, editor. Oxford: New York: 1996. pp. 216–243. [Google Scholar]
- 3.Liu L, Li Y, Liotta D, Lutz S. Nucleic Acids Res. 2009;37:4472. doi: 10.1093/nar/gkp400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jameson DM, Eccleston JF. In: Methods in Enzymology. Brand L, Johnson ML, editors. Vol. 278. San Diego: Acadenic; 1997. pp. 363–390. [DOI] [PubMed] [Google Scholar]
- 5.Rist MJ, Marino JP. Curr. Org. Chem. 2002;6:775. [Google Scholar]
- 6.Kool ET. Acc. Chem. Res. 2002;35:936. doi: 10.1021/ar000183u. [DOI] [PubMed] [Google Scholar]
- 7.Berry DA, Jung KY, Wise DS, Sercel AD, Pearson WH, Mackie H, Randolph JB, Somers RL. Tetrahedron Lett. 2004;45:2457. [Google Scholar]
- 8.Robins MJ, Barr PJ. Tetrahedron Lett. 1981;22:421. [Google Scholar]
- 9.Robins MJ, Vinayak RS, Wood SG. Tetrahedron Lett. 1990;31:3731. [Google Scholar]
- 10.Compound 4: UV (H2O) λmax 331 nm; IR (cm−1) 3440 (br), 3195, 1670, 1642; 1H NMR (400 MHz, DMSO-d6) δ 2.29 (s, 3H), 3.61 (m, 2H), 4.88 (s, 1H), 5.05 (t, 1H, J = 5.6 Hz), 6.01 (d, 1H, J = 6.0 Hz), 6.38 (m, 2H), 6.95 (s, 1H), 8.53 (s, 1H); 13C NMR (100 MHz, DMSO-d6) δ 14.27, 62.78, 88.88, 92.61, 100.95, 107.50, 127.22, 135.24, 138.19, 154.98, 155.60, 172.18; HRMS (FAB) m/z 271.0684, calcd for C12H12O4N2Na 271.0689 (M+Na).
- 11.Woo JS, Meyer RB, Gamper HB. Nucleic Acids Res. 1996;24:2470. doi: 10.1093/nar/24.13.2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Compound 5: UV (H2O) λmax 335 nm; IR (cm−1) 3350 (br), 1670, 1560, 1090; 1H NMR (400 MHz, CDCl3/CD3OD) δ 1.91–1.93 (m, 2H), 2.18 (m, 1H), 2.41(s, 3H), 2.54 (m, 1H) 3.82 (dd, 1H, J = 4.0, 12 Hz), 4.08 (dd, 1H, J = 2.8, 12.4 Hz), 4.26 (m, 1H), 4.80 (d, 1H, J = 1.6 Hz), 6.22 (dd, 1H, J = 2.4, 6.8 Hz), 8.44 (s, 1H); 13C NMR (100 MHz, CDCl3) δ 13.5, 24.2, 33.9, 62.5, 83.2, 88.8, 97.9, 110.6, 134.9, 138.6, 155.5, 158.6; HRMS (FAB) m/z 250.1183, calcd for C12H16O3N3 250.1186 (M+H).
- 13.McGuigan C, Carangio A, Snoeck R, Andrei G, De Clercq E, Balzarini J. Nucleosides Nucleotides Nucleic Acids. 2004;23:1. doi: 10.1081/ncn-120027813. [DOI] [PubMed] [Google Scholar]
- 14.Czernecki S, Valery JM. Synthesis. 1991:239. [Google Scholar]
- 15.Compound 6 was crystallized from ethyl acetate. UV (H2O) λmax 331 nm; IR (cm−1) 3469 (br), 2839, 1667, 1638, 1573, 1482; 1H NMR (400 MHz, CDCl3) δ 2.27 (d, 3H, J = 7.2 Hz), 2.30–2.36 (m, 1H), 2.57–2.64 (m, 1H), 3.29 (br, 1H), 3.71–3.75 (m, 1H), 3.90–3.95 (m, 2H), 4.18 (dd, 1H, J = 6.8, 13.2 Hz), 6.10–6.14 (m, 1H), 8.65 (s, 1H); 13C NMR (100 MHz, CDCl3) δ 14.1, 39.2 58.6, 60.5, 85.7, 88.0, 100.1, 108.4, 136.2, 155.2, 156.2, 171.9; HRMS (FAB) m/z 292.1036, calcd for C12H14O4N5 292.1039 (M+H).
- 16.Compound 7: UV (H2O) λmax 335 nm; IR (cm−1) 3350 (br), 1670, 1560, 1090; 1H NMR (600 MHz, DMSO-d6) δ 2.20 (m, 1H), 2.33 (s, 3H), 2.73 (m, 1H), 3.66 (m, 2H), 4.33 (dt, 1H, J = 25.8 Hz), 5.32 (dd, 1H, J = 56.4 Hz), 6.22 (q, 1H), 6.44 (s, 1H), 8.60 (s, 1H). 13C NMR (150 MHz, DMSO-d6) δ 13.6, 60.6, 86.2, 87.6, 95.4, 100.4, 106.8,136.5, 154.3, 171.56. 19F NMR (376 MHz, DMSO-d6) δ-174.9; HRMS (FAB) m/z 269.0932, calcd for C12H14O4N2F1 269.0932 (M+H).
- 17.Patching SG, Baldwin SA, Baldwin AD, Young JD, Gallagher MP, Henderson PJ, Herbert RB. Org. Biomol. Chem. 2005:462. doi: 10.1039/b414739a. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Sun XJ, Smith KM, Visser F, Carpenter P, Barron G, Peng YS, Robins MJ, Baldwin SA, Young JD. Biochemistry. 2006:1087. doi: 10.1021/bi0520535. [DOI] [PubMed] [Google Scholar]