Abstract
Cell penetrating peptides (CPPs) have attracted recent interest as drug delivery tools, although the mechanisms by which CPPs are internalized by cells are not well defined. Here we report a new experimental approach for the detection and secondary structure determination of CPPs in live cells using Raman microscopy with heavy isotope labeling of the peptide. As a first demonstration of principle Penetratin, a sixteen-residue CPP derived from the Antennapedia homeodomain protein of Drosophila, was measured in single, living melanoma cells. Carbon-13 labeling of the Phe residue of penetratin was used to shift the intense aromatic ring-breathing vibrational mode from 1003 cm−1 to 967 cm−1, thereby enabling the peptide to be traced in cells. Difference spectroscopy and principal components analysis (PCA) were used independently to resolve the Raman spectrum of the peptide from the background cellular Raman signals. Based on the position of the amide I vibrational band in the Raman spectra, the secondary structure of the peptide was found to be mainly random coil and β-strand in the cytoplasm, and possibly assembling as β-sheets in the nucleus. The rapid entry and almost uniform cellular distribution of the peptide, as well as the lack of correlation between peptide and lipid Raman signatures, indicated that the mechanism of internalization under the conditions of study was probably non-endocytotic. This experimental approach can be used to study a wide variety of CPPs as well as other classes of peptides in living cells.
Introduction
Cell penetrating peptides (CPPs) have attracted widespread interest due to their high potential for use as vectors for the intracellular delivery of a range of biologically active compounds. CPPs are rapidly internalized by cells, even when conjugated to cargo up to one hundred times the size of the peptide. A variety of cargo have been successfully conjugated to CPPs and delivered into cells, including small molecules, liposomal drug carriers, peptides and proteins, DNA and RNA oligonucleotides, and metal nanoparticles.1–4
Despite their well-demonstrated potential, the issue of the mechanism(s) by which CPPs and their cargo are internalized by cells is not yet resolved and has generated some controversy.1–3,5–8 Early studies posited a non-endocytotic mechanism that is temperature-, energy (ATP)-, transporter-, and receptor-independent.9–15 This led to the proposal of several models of membrane translocation such as the inverted micelle model,10 the electroporation model,16 and the carpet and pore formation models.2 However, a subsequent study showed that cell fixation, a procedure commonly employed in earlier fluorescence microscopy work, can lead to the artifactual uptake of CPPs.7 More recent studies of CPP uptake in live cells have generally supported an endocytosis mechanism, especially when the CPP is conjugated to high molecular weight cargo, as well as non-endocytotic cell uptake mechanisms depending on the identity of the peptide, its concentration, and the cell type.3,5,6,8,17,18
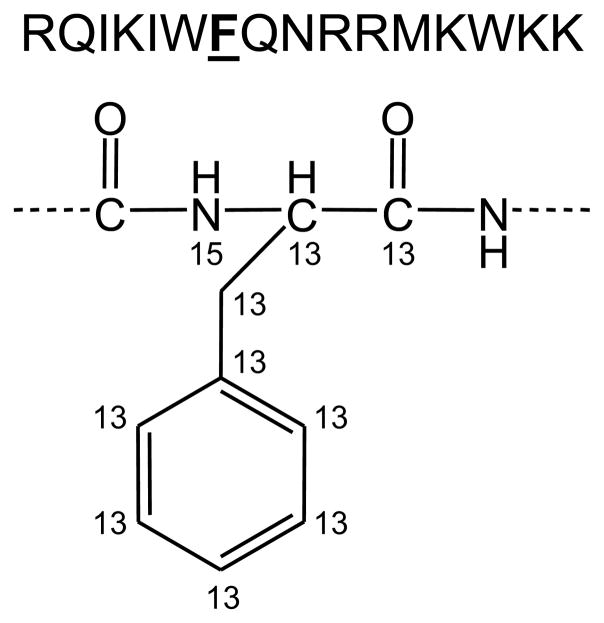
Penetratin is a sixteen-residue CPP derived from the Antennapedia homeodomain protein of Drosophila that was the first CPP to be discovered and is one of the most studied to date.10 The sequence of Penetratin is rich in arginine and lysine residues (Figure 1), which is a structural feature common to many CPPs. Arginine residues in particular are thought to play a key role in facilitating cellular uptake, probably due to the ability of the guanidinium head-group of arginine to form stable ion pairs with negatively charged plasma membrane constituents.1,17,19
Figure 1.
Sequence of penetratin and heavy isotope (13C, 15N) labeling scheme of the Phe residue.
Another structural aspect of CPPs that is thought to be important for cellular uptake is their secondary structure. Penetratin is derived from the third helix of the Antennapedia homeodomain protein and as such has a strong propensity for α-helix formation in lipid environments.20–26 Despite this, experimental studies have shown that α-helical structure is not necessary and possibly even inhibitory for membrane translocation.9,20,21,23 A recent computational study on the molecular structure and dynamics of penetratin interacting with lipid bilayers indicated a high degree of structural polymorphism for the peptide.27 Experimental results support this as in biomimetic phospholipid membranes penetratin was reported to adopt both α-helical and β-type (β-strand and β-turn) conformations depending on the phospholipid composition and peptide to lipid ratio.22,25,26,28,29 The ability of penetratin to change conformations when bound to the cell membrane may be an important aspect of its cell penetrating ability.22
In light of this previous work we hypothesized that confocal Raman microscopy would be a very useful experimental technique for directly measuring the microenvironment and secondary structure of CPPs in live cells to gain new insights on the cell penetrating mechanism. Raman spectroscopy is a vibrational spectroscopic technique that has already been used extensively to characterize the secondary structure of isolated proteins and peptides.30–36 The use of a confocal microscope to perform Raman spectroscopic measurements enables Raman spectra to be measured from ≥1 μm3 volumes within the confines of single, living cells.37–39 In the present study confocal Raman microscopy is employed for the first time to measure the Raman spectrum of a CPP, penetratin, in single, living cells.
Experimental Section
Peptide synthesis and purification
The uniformly labeled penetratin peptide with sequence RQIKIWF[13C9, 15N]QNRRMKWKK-NH2 (“penetratin-F-heavy” = penetratin-Fh) was assembled on a Protein Technology PS3 Peptide Synthesizer by using Rink Amide MBHA resin with an initial load of 0.72 mmol/g. Isotopically labeled Phe, Fmoc-[U-13C9, 15N]-Phe-OH was purchased from Cambridge Isotope Laboratories (Andover, MA). Standard Fmoc-chemistry was used throughout with a 4-fold molar excess of the acylating amino acids, and HCTU and HOBt as coupling reagents.40 The isotopically labeled Fmoc-Phe was coupled manually in 1.5-fold molar excess to reduce consumption of this amino acid. Peptide was cleaved from the resin using thioanisole-water-trifluoroacetic acid (5:5:90) for 2 h. Cleavage solution was extracted with methyl tert-butyl ether prior to purification. Reversed-phase high-performance liquid chromatography (RP-HPLC) purification was performed on a Rainin AutoPrep System with a Vydac 218TP152022 C18 column (15–20 μm particle size, 300 Å pore size, 250 × 22 mm) at a flow rate of 10.0 mL/min. Eluants were 0.1% TFA in water (A) and 0.1% TFA in acetonitrile (B). The elution gradient was 0% B for the first 5 min followed by 0–90% B in 80 min. Detection was at λ = 220 nm. Fractions were analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) and by analytical RP-HPLC. Analytical RP-HPLC was performed on a Hewlett-Packard 1100 liquid chromatograph equipped with a Vydac 218TP5415 C18 RP column (5 μm particle size, 300 Å pore size, 150 × 4.6 mm). Eluants were 0.1% TFA in water (A) and 0.1% TFA in acetonitrile (B). The elution gradient was 0–100% B in 20 min with a flow rate of 1.0 mL/min. Detection was at λ = 220 nm. MALDI-TOF MS was performed on an Applied Biosystems Voyager MALDI-TOF mass spectrometer using α-cyano-4-hydroxycinnamic acid matrix. The determined peptide mass value was [M + H]+ = 2257 Da (theoretical, 2255.8 Da). Peptide stocks were made up as 20 mM solutions in water (pH 7) and kept at 4°C for no longer than 10 days. Stock concentrations were determined by the absorbance at 280 nm (ε280 = 11,000 M−1.cm−1).
Cell culture
Human metastatic melanoma cells (Sk-Mel-2) were purchased from American Type Culture Collection (ATCC) (Manassas, VA). Trypan Blue and DMEM were purchased from Fisher Scientific and CellGro (Herndon, VA), respectively. Opti-MEM and all other reagents required for cell culture were purchased from Invitrogen (Carlsbad, CA). Cells were maintained in Opti-MEM medium supplemented with 4% fetal bovine serum (FBS), 50 units/mL penicillin, and 0.05 mg/mL streptomycin at 37 °C in a humidified atmosphere of 5% CO2 in air. Cells for experiments were harvested from sub-confluent (< 80%) cultures using a trypsin-EDTA solution and then re-suspended in fresh medium.
Raman spectroscopy instrumentation
All Raman spectroscopy measurements were performed with a 0.8 m dispersive spectrograph (Horiba-JY, HR800) with 600 groove/mm grating, 100–125 μm entrance slit width, and liquid-N2-cooled 1024 × 256 CCD detector. Confocal Raman microscopy measurements were performed with an Olympus IX-71 inverted microscope equipped with a Marzhauser automated X-Y scanning stage. Laser beams (514 nm and 647 nm) were generated by a Spectra Physics Beamlok 2060 mixed argon/krypton ion laser. Specimens under the microscope were visualized and targeted using phase contrast or bright field imaging with a digital video camera. Spectral calibrations were performed against the lines of silicon, polystyrene and room light.
Drop deposition Raman spectroscopy of peptides
For Raman drop deposition measurements,41 2 μL of 1 mM aqueous solutions of penetratin and penetratin-Fh were deposited adjacent to each other on a #1.5 glass coverslip (Fisher Scientific) and allowed to dry at room temperature. The coverslip was then immediately placed under the microscope and the Raman spectrum of each deposited peptide was recorded using the same measurement conditions. Raman excitation was performed with a 514 nm laser beam focused by a 40× microscope objective to provide 5 mW of power at the sample. Spectral acquisition time was 2 min. For data analysis the spectra were x-shifted according to the calibration standards and manually baseline corrected. Difference spectra were first calculated using the raw Raman spectra prior to baseline correction. The results presented are representative of three independent measurements.
Raman spectroscopy of aqueous/TFE peptide solutions
Macroscopic Raman spectroscopy of water/TFE peptide solutions was performed through a port of the spectrograph separate from the microscope. Solutions of the peptide (10 μL, 20 mM) were placed in 0.6 mm square I.D. glass capillary tubes (VitroCom, NJ) and sealed at both ends. A 5 cm focal length convex lens was used to focus a 250 mW, 514 nm laser beam through the sample and the Raman scattered light was collected 90° to the beam. Spectral acquisition times were 5 min. Solvent spectra were run under the same conditions for subsequent background correction from the raw peptide spectra. Solvent-subtracted spectra were then manually baseline corrected. The results presented are representative of two independent sets of measurements.
Confocal Raman microscopy of live cells
Cells were grown to ~50% confluency (~10,000 cells/cm2) in 35 mm diameter, collagen coated, #1.5 glass coverslip bottom dishes (MatTek Corp.). Before placement on the microscope stage the adhered cells were washed 1–2 times with PBS and then covered in serum- and phenol red-free DMEM (“DMEM”). In a typical experiment, Raman spectra were first recorded from multiple points within a localized region of a chosen cell prior to the addition of penetratin-Fh. These spectra served as the background (control) spectra. Penetratin-Fh was then added to the dish (without disturbing the dish) to the specified final concentration (25–50 μM) and left to incubate for the specified time before Raman spectra were again recorded from the same region of the cell. This procedure ensured that the best possible spectral difference between peptide-containing and peptide-free cells could be determined. For some experiments (e.g., nuclei measurements) the control measurements were performed immediately after penetratin-Fh addition, before the peptide had reached the region of the cell being probed. All Raman microscopy measurements were performed at room temperature (RT) under normal atmosphere with a 647 nm, 6 mW excitation laser beam focused with a 100× phase contrast, oil immersion microscope objective. A 200 μm confocal hole-size was used and spectral collection times were 10–30 s per point. Automated X-Y area scanning was performed with a step-size of 0.5 μm.
In addition to measurements within individual cells, control measurements were performed for each dish where the laser was focused in unoccupied areas of the dish, between cells. No peptide Raman signals were detected outside of cells. Also for each dish, at the end of the Raman microscopy measurements the dish was kept on the microscope stage and washed of excess peptide with PBS. Trypan Blue solution was then added to the dish to test the viability of cells. Uptake of Trypan Blue by cells either directly irradiated by the laser or not irradiated by the laser was visually assessed with bright-field microscopy. Cell viability was found to be independent of direct exposure to the laser beam. Overall cell viability was ≥80% for cells treated with 50 μM of peptide after 2 h at room temperature on the microscope stage. Although the cells remained viable, we did observe some phase contrast changes to the cells that correlated with penetratin-Fh uptake (but not laser exposure). Typically, penetratin-Fh uptake by cells induced brightening of the cell nucleus and slight rounding under phase contrast. The significance of these peptide-induced morphological changes to the cells was not investigated further.
To check that uptake of penetratin-Fh by cells was not induced by the Raman measurements, experiments were performed where cells were incubated with penetratin-Fh for 2 h in DMEM at 37°C, washed with PBS, and then covered again with DMEM not containing penetratin-Fh. Raman microscopy measurements detected penetratin-Fh in these cells at comparable levels to cells in dishes with DMEM containing penetratin-Fh, thus demonstrating that penetratin-Fh uptake by cells was not laser-induced.
Intracellular peptide stability
Intracellular peptide stability was investigated as described previously with some modifications.5 A confluent cell layer of SK-Mel-2 cells in a 9.6 cm2 dish (the same type of dish used for Raman measurements) was washed once with DMEM and incubated with 1.5 mL of DMEM containing 50 μM penetratin-Fh for 2 h at RT covered from light. The cells were then washed twice with PBS before being detached by trypsinization for 10 min at 37°C. Cells from a total of three 9.6 cm2 dishes were treated according to this procedure and combined in a fresh tube. The combined cells were washed three times with PBS and then lysed in 200 μL of 0.1% HCl for 10 min at RT. The cell lysate was then centrifuged for 15 min at RT and 14,000 rpm (7,000 × g). The peptide-containing supernatant was immediately desalted using a PD Minitrap G-10 column (GE Healthcare) and then concentrated to a volume of ~10 μL using a SpeedVac concentrator (Thermo Scientific). MALDI-TOF MS analysis of the sample, and a control sample of cells not treated with penetratin-Fh, was performed on an Applied Biosystems Voyager MALDI-TOF mass spectrometer using α-cyano-4-hydroxycinnamic acid matrix. The results presented are representative of two independent experiments.
Circular Dichroism spectroscopy
CD spectra were recorded on a Jasco-810 spectropolarimeter (Jasco, Easton, MD) using a quartz cell of 1 mm optical path length. Spectra were measured over a wavelength range of 180–250 nm with an instrument scanning speed of 100 nm/min and a response time of 1 s. The peptide concentration was 40 μM and the CD spectra are the result of eight averaged scans taken at 25 °C. All CD spectra are background-corrected for signal contributions due to the water and/or TFE. CD spectra were modeled using the CDPro suite of programs employing the SP37A protein data set.42 The reported percentage values are the average results computed by the CONTINLL, SELCON3 and CDSSTR programs.
Results and Discussion
Heavy-isotope labeling of penetratin
Raman spectra recorded from cells are dominated by the vibrational bands associated with endogenous cellular proteins.38,43 These vibrational bands include the backbone amide modes as well as bands associated with the amino acid side chains. Phenylalanine generates an intense vibrational band at 1003 cm−1 that is associated with its aromatic ring-breathing mode.44 The ubiquitous appearance of this and other protein-associated Raman bands throughout cells presents a significant challenge for the detection of exogenous peptides in cells because the endogenous protein bands generally overlap with and mask the Raman spectrum of the exogenous peptide. Therefore, in order to trace penetratin in cells in the present study we labeled the phenylalanine residue of the peptide with 13C atoms (Figure 1), which was reported to shift the wavenumber position of the ring-breathing mode from 1003 cm−1 to 967 cm−1.45 The 967 cm−1 band represents a strong, distinguishable Raman signal that allows penetratin to be clearly detected in cells (vide infra).
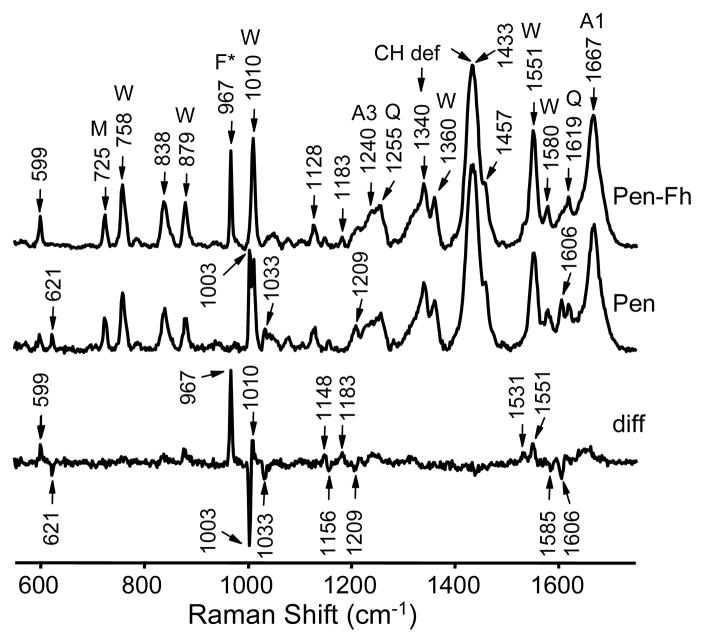
To confirm the isotopic shift the Raman spectrum of penetratin and of heavy-labeled penetratin (penetratin-Fh) was measured by drop deposition (Figure 2). The top spectrum corresponding to penetratin-Fh indeed shows a strong band at 967 cm−1. In comparison, the spectrum of unlabeled penetratin (center) displays the phenylalanine band in the normal position of 1003 cm−1, overlapping strongly with the nearby tryptophan ring-breathing mode at 1010 cm−1.34 The penetratin-Fh – penetratin difference spectrum (bottom) highlights the 1003/967 cm−1 isotopic shift. The difference spectrum also reveals other, much weaker phenylalanine-associated vibrational bands appearing at (12C/13C position) 621/599, 1033/1010, 1156/1148, 1209/1183, 1585/1531, and 1606/1551 cm−1 in accordance with previous vibrational band assignments.34,46,47 Additional tryptophan bands include those with peaks at 758, 879, 1360, 1551 and 1580 cm−1.34,44,46,47 Other noteworthy bands include the amide I band at 1667 cm−1,30,31 the aliphatic –CH2 and –CH3 bending bands at 1433/1457 cm−1,33,44 and the amide III band at 1240 cm−1.30,31 A probable assignment for the nearby 1255 cm−1 band is a glutamine side chain vibration.35
Figure 2.
Raman drop deposition spectra of heavy-labeled penetratin (Pen-Fh), unlabeled penetratin (Pen), and Pen-Fh – Pen difference spectrum (diff) showing the 967 cm−1 13C/15N-Phe (F*) mode and other Phe-associated modes. Other peak assignments: Trp (W),34,44,46,47 Met C-S stretch (M),61 Gln (Q),35,44 –CH2 and –CH3 deformations (CH def),33,44 amide I (A1) and amide III (A3) modes.30,31
Characterization of penetratin-Fh secondary structure in water/TFE solutions
The main aim of the present study was to show that penetratin can be detected in living cells using Raman microscopy and that the Raman spectrum of the peptide can be resolved and potentially interpreted to reveal its secondary structural features. A necessary precursor to these cell experiments was therefore the measurement and interpretation of Raman spectra of the peptide in solution, aided by circular dichroism (CD) spectroscopic measurements of the same peptide solutions. Previous CD studies of penetratin reported that it is mainly a random coil in aqueous solution and that it becomes partly α-helical in TFE solution.21,23,24,26 1H-NMR of penetratin in 90:10 TFE:water solution showed that the peptide forms a bent, irregular helical structure between residues 4–12, which begins and ends with a β-turn.48
The CD spectrum of penetratin-Fh was measured under increasingly hydrophobic solvent conditions by varying the volumetric ratio of water with TFE (Figure S1). Modeling of the data indicated that in 100% water penetratin-Fh possesses mainly random coil character (54%) with also a significant proportion of β-strand structure (21%). The proportion of α-helix structure was very low (3%). In 50:50 TFE:water and 100% TFE solvent the proportion of α-helix increased to 11% and 25% respectively (see Table 1 in Supporting Information). The CD data are therefore in good overall agreement with previous findings.
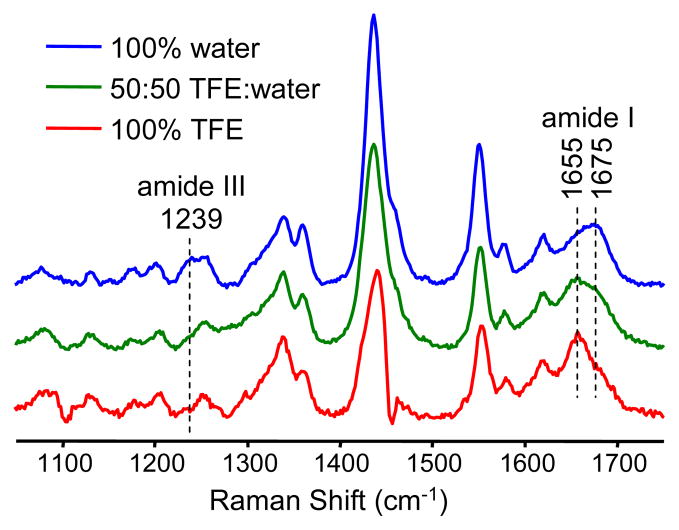
The Raman spectrum of penetratin-Fh was measured under the same three solvent conditions to test whether Raman spectroscopy could detect the secondary structural changes observed by CD spectroscopy. As expected, major changes to the spectrum were observed in the amide I and III band regions (Figure 3). The amide I and III vibrational modes are known to be very sensitive to the secondary structure and as such are frequently used to determine the secondary structure composition of proteins and peptides by both IR and Raman spectroscopy.30,36,49 The amide I mode possesses mainly C=O stretching character, whereas the amide III mode involves mainly C-N stretching and N-H in-plane bending.31,35,44 In Raman spectra the amide I mode is easier to interpret because it is much more intense and has a less convoluted normal mode composition than the amide III mode. We therefore focus our attention on the amide I mode in this study.
Figure 3.
Raman spectra of penetratin-Fh in three different water/TFE solvent conditions. TFE solvent peaks were removed by subtraction. The shape of the intense ~1440 cm−1 band in the 100% TFE spectrum was affected by partial overlap with a strong solvent peak.
At the simplest level of interpretation, the position of the amide I mode can be interpreted in terms of a predominant or “average” secondary structure. Obviously this approach is most effective for proteins/peptides that possess relatively uniform secondary structures and generally proves most useful for distinguishing between α-helix, β-strand, and random coil conformations. For example, Raman and IR spectroscopy and theoretical calculations of model polypeptides with uniform secondary structures established that α-helices can be characterized by the appearance of amide I bands in the range ~1650–1655 cm−1, which can be clearly distinguished from β-sheets/strands that produce amide I bands in the range 1665–1675 cm−1.30,31,44 Random coil conformations on the other hand are characterized by a high degree of structural heterogeneity with respect to the peptide backbone dihedral angles and therefore give rise to broadened amide I bands, typically peaking around ~1665 cm−1.31 β-turns are also associated with a high dispersion of amide I frequencies, which can occur in the range ~1645–1690 cm−1.30,44
In 100% aqueous solution the Raman spectrum of penetratin-Fh displays a broad, asymmetric amide I band with a peak at 1675 cm−1 (Figure 3). The relatively high peak wavenumber value of 1675 cm−1 and the asymmetry of the band is indicative of a significant β-strand contribution. The broadness of the band is also indicative of a high degree of structural heterogeneity, which is consistent with a high proportion of random coil character, as was indicated by the CD data. It should be noted that β-strand and PII conformations cannot be distinguished by unpolarized Raman spectroscopy, as employed here, although they can be distinguished by way of polarized Raman measurements (the same can also be said of α-versus 310-helix).50 We did not perform polarized Raman measurements in this study, although based on the CD data we assume that mainly the β-strand conformation contributes to the secondary structure of penetratin-Fh in aqueous solution.
With increasing TFE solvent, a shift in the amide I peak position from 1675 cm−1 to 1655 cm−1 occurred (Figure 3). This change is consistent with a transition of the peptide toward higher α-helical content, as was observed by CD spectroscopy. The band remains relatively broad however, and retains a weak shoulder on its high wavenumber edge that together is indicative of the retention of some β-strand and random coil character. Overall, the Raman and CD spectroscopic data for penetratin-Fh in water/TFE solutions are in very good agreement. Also, despite the relatively simple, qualitative approach to the interpretation of the amide I band of penetratin-Fh, the overall trends observed for this band in the Raman spectrum are in good agreement with the established “fingerprints” from model proteins, especially the distinction between α-helix and β-strand.
Detection of penetratin-Fh in cell nuclei by Raman difference spectroscopy
Next we treated live melanoma (SK-Mel-2) cells with penetratin-Fh and performed confocal Raman microscopy on individual cells. Cells were treated at RT with 50 μM penetratin-Fh and Raman measurements were performed immediately after treatment using 647 nm laser excitation. Although Penetratin-Fh was detected throughout most regions of the cell (vide infra) it was observed to accumulate in the cell nuclei (especially the nucleoli) over time, first detectable on average ~15–20 min after addition and sometimes as early as 5 min after addition of the peptide. Nucleolar accumulation was reported previously for penetratin and other CPPs.3,5,6 The observed rate of accumulation was only weakly dependent on concentration in the range 25–300 μM (data not shown). MALDI-TOF MS analysis of lysates of SK-Mel-2 cells previously treated with 50 μM penetratin-Fh for 2 h (see Experimental Procedures) confirmed that penetratin-Fh remained fully intact and unmodified inside the cells (Figure S2), as was reported for other studies of penetratin with different cell lines.5
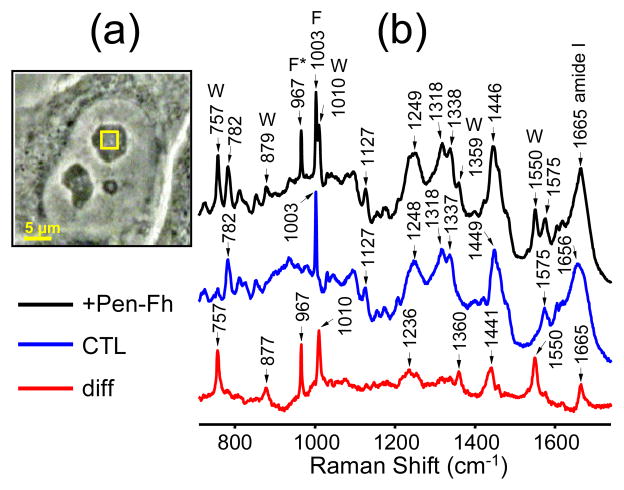
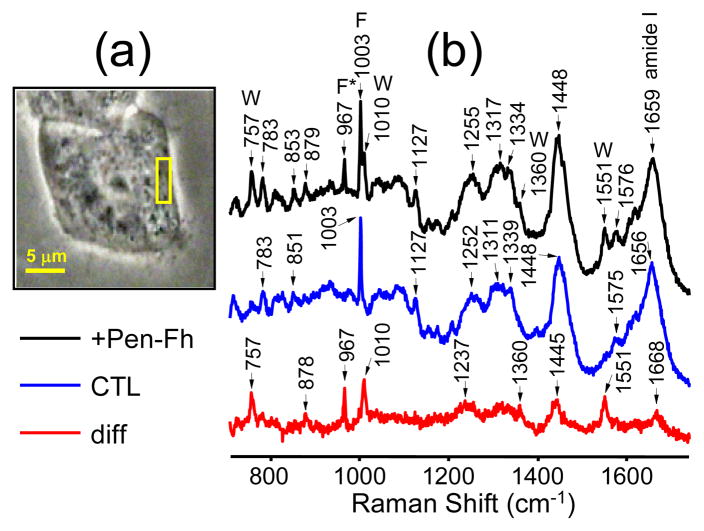
Figure 4a presents a phase contrast image of a cell. The yellow square represents a 3.5 × 3.5 μm scan region within a nucleolus of the cell from which Raman spectra were recorded. Prior to peptide addition the laser was scanned with 0.5 μm step-size to collect a total of 49 spectra within this region, which were then averaged (Figure 4b, center spectrum). The same procedure was then performed 2 h after 50 μM peptide treatment (Figure 4b, top spectrum). The 967 cm−1 vibrational band due to penetratin-Fh can be clearly seen in the top spectrum. The bottom spectrum in Figure 4b is the difference between the treatment and control spectra. The difference spectrum shows the 967 cm−1 band and five other bands peaking at 757, 877, 1010, 1360 and 1550 cm−1, all of which can be assigned to the vibrations of Trp in penetratin-Fh.34,46,47 Trp is the least naturally abundant of all the essential amino acids and therefore is not normally detectable in the Raman spectra of the endogenous proteins of cells. The difference spectrum also shows two prominent bands peaking at 1441 and 1665 cm−1 that can be assigned to the C-H deformations and amide I vibrations of penetratin-Fh, respectively. It is not likely that these bands are associated with the endogenous proteins of the cell because all other regions of the difference spectrum are devoid of endogenous protein signatures, most notably the ubiquitous 1003 cm−1 phenylalanine band. The amide bands of penetratin-Fh should appear in the difference spectrum alongside its Phe- and Trp-related bands.
Figure 4.
(a) Phase contrast image of a melanoma (SK-Mel-2) cell captured with 100× magnification. (b) Averaged Raman spectra derived from the area indicated by the yellow box within the nucleolus of the cell shown in (a). Spectra were measured either ~120 min after treatment with 50 μM penetratin-Fh (+Pen-Fh) or before treatment (control, CTL). Pen-Fh – CTL difference spectrum (diff) is also shown. Bands due to Trp (W), Phe (F) and 13C/15N-labeled Phe (F*) are indicated above the spectra. Scan parameters: 3.5 × 3.5 μm, 0.5 μm step-size, 2 × 10 s dwell-time.
Figure S3 provides a detailed view of the amide I band region for Raman spectra recorded from nucleoli before and after 50 μM peptide treatment. The spectral overlay clearly highlights the shift in the amide I band due to the appearance of penetratin-Fh in the nucleoli, and the difference spectrum reveals the amide I band for penetratin-Fh peaking at 1665 cm−1. It is interesting that this peak position does not match either of the peak positions observed for the peptide in water or TFE solution (see Figure 3). The band is also significantly narrower and more symmetrical at its center in the spectrum from the nucleolus than in the solution spectra (c.f. Figure 3 and 4). In fact the amide I band in the nucleolus peptide spectrum more closely resembles that shown in the spectrum of the peptide measured by drop deposition (Figure 2), where the peptide was dropped and dried on a glass slide. The high intensity, sharpness and symmetry of the amide I band in the spectrum of the drop deposition sample suggest a uniformity of structure, probably β-sheet based on the 1667 cm−1 peak position.51 Therefore, by implication due to the spectral similarity, it appears that penetratin-Fh may also be similarly assembled as β-sheets within the condensed volume of the nucleolus.
Another conceivable contribution to the amide I band appearing in the difference spectrum from cells could be a change in the secondary structure of the endogenous proteins (e.g., nuclear histone proteins) induced by the presence of a high local concentration of the exogenous peptide. However, we consider it unlikely that this is a major contributing factor in the present case for the following reasons. Firstly, such a change in the secondary structure would probably require a major rearrangement of the overall chromatin structure, which would likely lead to the appearance of other spectral features in the difference spectrum as well (e.g., from DNA). Also, if such a shift of the amide I band did occur it should appear in the difference spectrum as a classical difference feature with a positive peak and a negative peak, with the latter appearing at ~1655 cm−1 (the usual peak position for endogenous cellular proteins). Instead, all of the observed features in the difference spectrum are positive, which indicates that the background Raman signals remained relatively constant and that the difference spectrum reflects a case of (a + b) − a = b, where “b” is the exogenous peptide and “a” is the cell background.
Detection of penetratin-Fh in cell nuclei with PCA
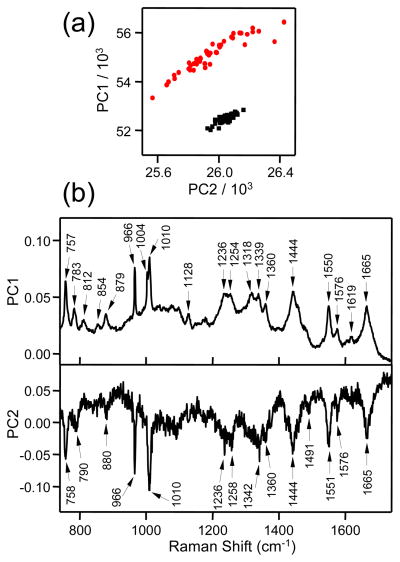
Difference spectroscopy is perhaps the simplest method that can be employed to extract the spectrum of the exogenous peptide from the background cellular Raman signals. While useful, a potentially more powerful method of analysis is that of principal components analysis (PCA). PCA is a multivariate statistical technique that linearly transforms an original set of variables (in the present case wavenumber points) into a substantially smaller set of orthogonal variables (principal components or PCs) that encapsulate most of the information (i.e., correlation or covariance) in the original set of variables.52 PCA is a highly sensitive method of data analysis that can detect very small peak shifts and subtle intensity changes of bands in large sets of Raman spectroscopic data. It has been successfully applied in the analysis of Raman microscopy data to classify different cell types and morphologies.52–56 Here we use PCA to distinguish between cells treated and not treated with penetratin-Fh.
As a first example, PCA was applied to the same nucleolus data set analyzed above by difference spectroscopy in Figure 4. The PCA calculations were performed with Minitab® 15 by decomposing the covariance matrix. We preferred to input raw, uncorrected Raman spectra into the calculations because the experimental conditions for the control and treatment measurements were carefully matched and we wanted to avoid introducing any possible artifacts in the results through data manipulation. Each individual Raman spectrum gets a specific score for each PC. The scores represent the distribution of the spectra for a given PC. Figure 5a plots the PC1 scores versus the PC2 scores for all of the spectra measured from the cell nucleolus before peptide treatment (control, black squares) and following peptide treatment (red circles). This scatter plot displays a clear separation of treatment and control spectra along both PC1 and PC2. I.e., PCA clearly detects the presence of the peptide in the treatment group. In addition to the PC scores, PCA calculates for each variable (i.e., wavenumber point) a load for each PC, which represents the importance of a given variable to a given PC. Figure 5b plots the PC1 and PC2 loads for the nucleolus data. The spectral features in these plots represent the main chemical differences between the treatment and control groups. The plot of the PC1 loads closely resembles the raw, averaged spectrum observed for the nucleolus containing penetratin-Fh (c.f., Figure 4, top spectrum). In other words, the largest variation in the data is the overall spectral intensity in the presence of the peptide. The plot of the PC2 loads displays the full spectrum of penetratin-Fh, free of virtually all other background cellular Raman bands. Of particular note is the absence of the 1003 cm−1 phenylalanine band (which would signify any endogenous protein contribution), the appearance of the amide I band at 1665 cm−1, and the almost perfect overall agreement between this PCA-derived spectrum and the difference spectrum shown in Figure 4. PCA however seems to be more effective at resolving the amide I band. In the PCA-derived spectrum the amide I band appears very sharp and symmetrical, which is again supportive of a β-sheet structure.
Figure 5.
(a) PC1 versus PC2 scores calculated from PCA of Raman spectra measured within the area of the cell marked by the yellow box in Figure 4a. Data corresponding to spectra recorded before penetratin-Fh treatment are shown as black squares, and those spectra recorded 2 h after 50 μM penetratin-Fh treatment are indicated by the red circles. (b) Plots of the PC1 and PC2 loads versus relative wavenumber (cm−1).
Detection of penetratin-Fh in the cytoplasm of cells
Penetratin-Fh was also detected throughout the cytoplasm of cells using Raman microscopy. Measurements performed within the cytoplasm have the potential to provide very useful information on the mechanisms of CPP uptake by cells, especially when performed at early time-points in the vicinity of the cell membrane. However, measurements in the cytoplasm are more difficult to perform than measurements within the nucleus because the local concentrations of exogenous peptide are generally lower in the cytoplasm. Additionally, the cytoplasm is inherently a more heterogeneous and dynamic environment than the nucleus, which causes the background cellular Raman signals to vary considerably both spatially and temporally. This makes the extraction and interpretation of the exogenous peptide spectrum from the cytoplasm more complicated, but not intractable.
The resolution of the exogenous peptide spectrum requires that Raman spectra be recorded from a region of the cell before the peptide is added, and then measured from the same region of the cell after the peptide is added. If the cellular background remains relatively constant between the two measurements the exogenous peptide spectrum can be resolved quite effectively by spectral subtraction (Figure 6). Overlaid on the phase contrast image of a cell (Figure 6a) is the scan area from which Raman measurements were performed before and after peptide treatment. Just 2–3 min after treatment with 25 μM penetratin-Fh, the peptide was detected throughout the indicated scan region. Figure 6b shows the treatment, control and difference spectra. The difference spectrum is comparable to those obtained for penetratin-Fh in the nucleus, except the amide I band position in this case appears slightly higher at 1668 cm−1.
Figure 6.
(a) Phase contrast image of a melanoma (SK-Mel-2) cell captured with 100× magnification. (b) Averaged Raman spectra derived from the area indicated by the yellow rectangle within the cytoplasm of the cell shown in (a). Spectra were recorded either ~2–10 min after treatment with 25 μM penetratin-Fh (+Pen-Fh) or before treatment (control, CTL). Pen-Fh – CTL difference spectrum (diff) is also shown. Bands due to Trp (W), Phe (F) and 13C/15N-labeled Phe (F*) are indicated above the spectra. Scan parameters: 2 × 6 μm, 0.5 μm step-size, 2 × 5 s dwell-time.
More commonly in the cytoplasm the cell background of a chosen scan area changed with time, rendering difference spectroscopy infeasible and necessitating PCA. The most common change that was observed in the cells (both spatially and temporally) was that of lipid Raman signals. Lipid vesicles are scattered throughout the cytoplasm of cells and can be detected by Raman microscopy.57 We frequently detected strong, highly localized lipid signals in the cells that also changed position with time. This is likely reflective of the natural movement of transport vesicles within the living cell. The appearance or disappearance of lipid vesicles into or from the scan region between the control and treatment scans manifests as positive or negative lipid-associated Raman bands in the difference spectrum. This poses a problem because lipid generates a strong C=C stretching Raman band at ~1655 cm−1 that overlaps with the peptide amide I band furnishing the secondary structure information of the peptide.57 However, the multi-dimensional nature of PCA enables the exogenous peptide spectrum to be resolved from the Raman signals of lipids and other endogenous cellular constituents.
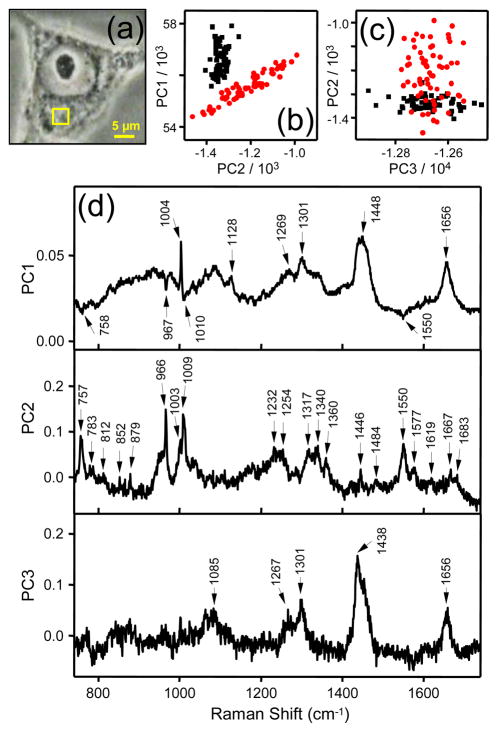
Figure 7 presents PCA results for measurements performed within the cytoplasm of a cell. In this experiment 50 μM of penetratin-Fh was added and the laser was scanned within the indicated area (Figure 7a) before and 20 min after peptide addition. Figures 7b and 7c present the PC1 v PC2 and PC2 v PC3 scatter plots, respectively. A clear separation between treatment and control spectral groups is shown along PC1 and PC2 in the scatter plots. Figure 7d presents a plot of the loads for the first three PCs. PC1 displays endogenous protein signatures (e.g., 1003 cm−1 Phe) and some moderately strong lipid-associated Raman signatures (e.g., the prominent 1301 cm−1 peak).57 Also evident in the plot of the PC1 loads are very small, negative peaks associated with penetratin-Fh, at 758, 967, 1010 and 1550 cm−1. Thus, the PC1 loads plot indicates a slight negative correlation between penetratin-Fh and lipid. I.e., lipid signatures were slightly more prominent in the control scan, prior to the appearance of the peptide. This was a common occurrence during the course of our experiments and we will discuss the implications of this for the mechanism of penetratin-Fh uptake later in the discussion. The next two PCs clearly separate the penetratin-Fh and lipid Raman signatures. The PC3 loads plot is dominated by lipid Raman signatures, while the plot of the PC2 loads is dominated by Raman signatures of penetratin-Fh (Figure 7d). Of note, the amide I band appears with a somewhat broad and asymmetrical profile with a peak at 1667 cm−1 and shoulder at ~1683 cm−1. Generally we observed an amide I band for penetratin-Fh in the cytoplasm that was broader and higher in peak position than in the nucleolus. While the spectra of penetratin-Fh recorded from the nucleoli appeared more like the drop deposition spectrum of the peptide, the spectra recorded from the cytoplasm more closely resembled the aqueous solution spectrum of the peptide. Thus, in the cytoplasm the structure of penetratin-Fh is probably best described as monomeric, with mainly random coil and β-strand characteristics. Similar results are presented in Supporting Information for another experiment performed within the cytoplasm of a cell using similar experimental conditions (Figure S4).
Figure 7.
(a) Phase contrast image of a melanoma (SK-Mel-2) cell captured with 100× magnification. (b) PC1 versus PC2 scores and (c) PC2 versus PC3 scores calculated from PCA of Raman spectra measured within the area of the cell marked by the yellow box in (a). Data corresponding to spectra recorded before penetratin-Fh treatment are shown as black squares, and those spectra recorded ~20–40 min after 50 μM penetratin-Fh treatment are indicated by the red circles. (d) Plots of the PC1, PC2 and PC3 loads versus relative wavenumber (cm−1). Scan parameters: 4 × 4 μm, 0.5 μm step-size, 2 × 10 s dwell-time.
Implications for the mechanism of cell internalization
The great potential of the Raman microscopy method for the study of CPPs in live cells stems from the fact that it can provide both structural information on the peptide as well as detailed chemical information on the microenvironment of the peptide within cells. For example, if penetratin-Fh entered cells via a classical endocytosis mechanism then the peptide should be encapsulated within endosomes and therefore transported into cells with lipid, which should be evident in the Raman spectra. Lipid should be consistently co-localized with the exogenous peptide, and therefore a strong, positive correlation between penetratin-Fh and lipid should be seen by PCA. Although lipid and penetratin-Fh occasionally did appear together at certain points within cells, these occurrences were probably coincidental. As shown above, and in general, either no change or a decrease in the overall amount of lipid was detected within a scan region upon arrival of the exogenous peptide. Interestingly, we observed no instances of an increase in lipid. This is somewhat surprising because if lipid vesicles were randomly migrating in and out of the laser scan regions independent of the exogenous peptide then one might expect an equal frequency of occurrences of positive and negative peptide-lipid correlations with PCA. The significance of this apparent bias towards a negative correlation is not presently clear. It may be reflective of a mutual repulsion between the exogenous peptide and lipid in the cytoplasm of the cells.
The above observations seem to support a non-endocytotic cell internalization mechanism for penetratin-Fh. Additionally, we detected penetratin-Fh inside cells near the plasma membrane as early as ~2–3 min after peptide treatment at RT, and the Raman signals of the peptide were usually quite homogenous across the laser scan region (Figure S5). This rapid entry and non-punctate distribution of penetratin-Fh in cells is again supportive of a non-endocytotic internalization mechanism. Recent confocal fluorescence microscopy studies of penetratin uptake in live cells supported endocytosis.5,17,18 However, these studies employed peptide in the concentration range of 1–10 μM and another recent study showed that non-endocytotic uptake can occur if the concentration of the peptide is above a threshold level of about 10 μM.3 The concentrations of peptide employed in the present work (25–50 μM) were above this threshold and therefore under the conditions of our experiments a non-endocytotic uptake mechanism is consistent with previous findings. Interestingly, we did not observe peptide uptake at 4°C (data not shown), which is an observation frequently used to support endocytosis. However at 4°C the fluidity of the plasma membrane is likely to be drastically reduced, which could also conceivably inhibit other types of non-endocytotic mechanisms of membrane translocation as well. We therefore do not consider the lack of uptake at 4°C to be necessarily indicative of endocytosis.
A study of the binding and conformation of a series of CPPs including penetratin in the lipid bilayers of large unilamellar vesicles showed a negative correlation between α-helical content and uptake efficiency.20 Other fluorescence, CD, and EPR spectroscopic studies indicated that the adoption of a predominantly β-strand structure by penetratin may be required for membrane disruption.25,26 The present Raman microscopy results could be considered supportive of this as, throughout the cells, the structure of penetratin-Fh was measured to be predominantly random coil/β-strand, especially in the cytoplasm closer to the cell membrane. No evidence of α-helical structure formation by penetratin-Fh was detected in cells, which (as our solution studies showed) would have been clearly indicated by the appearance of the amide I band at ~1655 cm−1. It is of course possible that the peptide forms α-helical or other transient, functionally important conformations as it crosses the cell membrane. Limitations with the signal intensity and the lateral spatial resolution (~0.5 μm) of the Raman microscopy methods employed in the present work precluded measurements from being performed exclusively from cell membranes. Such measurements using improved techniques in future could provide further insights into the mechanism of membrane crossing by penetratin and other CPPs.
To date there have been only a few previous Raman microscopy cellular studies utilizing heavy isotope labeling of an exogenous compound. For example, one recent study examined carbon nanotubes in live cells58 while another detected liposomes in fixed cells.59 The present study is the first example of an exogenous peptide being measured in live cells. Raman microscopy offers key advantages over fluorescence microscopy, such as the extra dimension of chemical structure information it provides, and the avoidance of attachment of a bulky fluorescent chromophore that can potentially affect the cell-penetrating properties of the peptide.
The full potential of Raman microscopy for determining the secondary structure of exogenous peptides in live cells has not yet been fully realized in the present study. For example, isotope-edited Raman difference spectroscopy,32,35 whereby two or more adjacent peptide links are labeled with 13C to resolve the amide I signature and hence the local secondary structures of those bonds, has already proved useful for the study of proteins and peptides in solution and could also be applied similarly to peptides in live cells. Also, the recent development of theoretical models of vibrational coupling in peptides has now made it possible to fit the amide I band profile of even the most structurally distorted peptide, yielding the dihedral angles (φ and ψ) for all the individual peptide groups.36,60 While such a high resolution approach to the interpretation of the amide I band was beyond the scope of the present study, future studies could employ this approach to determine the detailed secondary structure of exogenous peptides in living cells.
Conclusions
Raman microscopy was used to determine the microenvironment and secondary structure of penetratin in different regions of single, living melanoma cells. 13C-labeling of the Phe residue of penetratin was used to shift the intense aromatic ring-breathing mode to 967 cm−1, thereby enabling the peptide to be traced in cells. Difference spectroscopy and PCA were successfully used to resolve the spectrum of the peptide from the background cellular Raman signals. The peptide was found to have mainly random coil and β-strand structural characteristics throughout the cell. The rapid entry and almost uniform cellular distribution of the peptide as well as the lack of consistent co-localization of peptide and lipid Raman signatures indicated that the mechanism of internalization under the conditions of study was probably non-endocytotic. This experimental approach can potentially be used to study a wide variety of CPPs as well as other classes of peptides in living cells.
Supplementary Material
Acknowledgments
Support for this research was provided by grants from the National Institutes of Health (GM073621 to A.C.T and EB000289 to G.B.F) and the Robert A. Welch Foundation (to G.B.F.).
Footnotes
Supporting Information Available: CD spectroscopic data for penetratin-Fh; MALDI-TOF MS data for intracellular penetratin-Fh recovery; cellular Raman spectroscopic data in the amide I band region; Raman spectroscopic and PCA data for a cell cytoplasm measurement; confocal Raman maps from cells. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Brooks H, Lebleu B, Vives E. Adv Drug Deliv Rev. 2005;57:559–77. doi: 10.1016/j.addr.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Lundberg P, Langel U. J Mol Recognit. 2003;16:227–33. doi: 10.1002/jmr.630. [DOI] [PubMed] [Google Scholar]
- 3.Tunnemann G, Martin RM, Haupt S, Patsch C, Edenhofer F, Cardoso MC. FASEB J. 2006;20:1775–84. doi: 10.1096/fj.05-5523com. [DOI] [PubMed] [Google Scholar]
- 4.Wadia JS, Dowdy SF. Curr Opin Biotechnol. 2002;13:52–6. doi: 10.1016/s0958-1669(02)00284-7. [DOI] [PubMed] [Google Scholar]
- 5.Fischer R, Kohler K, Fotin-Mleczek M, Brock R. J Biol Chem. 2004;279:12625–35. doi: 10.1074/jbc.M311461200. [DOI] [PubMed] [Google Scholar]
- 6.Rhee M, Davis P. J Biol Chem. 2006;281:1233–40. doi: 10.1074/jbc.M509813200. [DOI] [PubMed] [Google Scholar]
- 7.Richard JP, Melikov K, Vives E, Ramos C, Verbeure B, Gait MJ, Chernomordik LV, Lebleu B. J Biol Chem. 2003;278:585–90. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- 8.Tunnemann G, Ter-Avetisyan G, Martin RM, Stockl M, Herrmann A, Cardoso MC. J Pept Sci. 2008;14:469–76. doi: 10.1002/psc.968. [DOI] [PubMed] [Google Scholar]
- 9.Derossi D, Calvet S, Trembleau A, Brunissen A, Chassaing G, Prochiantz A. J Biol Chem. 1996;271:18188–93. doi: 10.1074/jbc.271.30.18188. [DOI] [PubMed] [Google Scholar]
- 10.Derossi D, Chassaing G, Prochiantz A. Trends Cell Biol. 1998;8:84–7. [PubMed] [Google Scholar]
- 11.Derossi D, Joliot AH, Chassaing G, Prochiantz A. J Biol Chem. 1994;269:10444–50. [PubMed] [Google Scholar]
- 12.Prochiantz A. Curr Opin Cell Biol. 2000;12:400–6. doi: 10.1016/s0955-0674(00)00108-3. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki T, Futaki S, Niwa M, Tanaka S, Ueda K, Sugiura Y. J Biol Chem. 2002;277:2437–43. doi: 10.1074/jbc.M110017200. [DOI] [PubMed] [Google Scholar]
- 14.Thoren PE, Persson D, Karlsson M, Norden B. FEBS Lett. 2000;482:265–8. doi: 10.1016/s0014-5793(00)02072-x. [DOI] [PubMed] [Google Scholar]
- 15.Vives E, Brodin P, Lebleu B. J Biol Chem. 1997;272:16010–7. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 16.Binder H, Lindblom G. Biophys J. 2003;85:982–95. doi: 10.1016/S0006-3495(03)74537-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amand HL, Fant K, Norden B, Esbjorner EK. Biochem Biophys Res Commun. 2008;371:621–5. doi: 10.1016/j.bbrc.2008.04.039. [DOI] [PubMed] [Google Scholar]
- 18.Thoren PE, Persson D, Isakson P, Goksor M, Onfelt A, Norden B. Biochem Biophys Res Commun. 2003;307:100–7. doi: 10.1016/s0006-291x(03)01135-5. [DOI] [PubMed] [Google Scholar]
- 19.Wender PA, Mitchell DJ, Pattabiraman K, Pelkey ET, Steinman L, Rothbard JB. Proc Natl Acad Sci USA. 2000;97:13003–8. doi: 10.1073/pnas.97.24.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caesar CE, Esbjorner EK, Lincoln P, Norden B. Biochemistry. 2006;45:7682–92. doi: 10.1021/bi052095t. [DOI] [PubMed] [Google Scholar]
- 21.Christiaens B, Grooten J, Reusens M, Joliot A, Goethals M, Vandekerckhove J, Prochiantz A, Rosseneu M. Eur J Biochem. 2004;271:1187–97. doi: 10.1111/j.1432-1033.2004.04022.x. [DOI] [PubMed] [Google Scholar]
- 22.Clayton AH, Atcliffe BW, Howlett GJ, Sawyer WH. J Pept Sci. 2006;12:233–8. doi: 10.1002/psc.715. [DOI] [PubMed] [Google Scholar]
- 23.Letoha T, Gaal S, Somlai C, Czajlik A, Perczel A, Penke B. J Mol Recognit. 2003;16:272–9. doi: 10.1002/jmr.637. [DOI] [PubMed] [Google Scholar]
- 24.Lindberg M, Biverstahl H, Graslund A, Maler L. Eur J Biochem. 2003;270:3055–63. doi: 10.1046/j.1432-1033.2003.03685.x. [DOI] [PubMed] [Google Scholar]
- 25.Magzoub M, Eriksson LE, Graslund A. Biochim Biophys Acta. 2002;1563:53–63. doi: 10.1016/s0005-2736(02)00373-5. [DOI] [PubMed] [Google Scholar]
- 26.Magzoub M, Eriksson LE, Graslund A. Biophys Chem. 2003;103:271–88. doi: 10.1016/s0301-4622(02)00321-6. [DOI] [PubMed] [Google Scholar]
- 27.Polyansky AA, Volynsky PE, Arseniev AS, Efremov RG. J Phys Chem B. 2009;113:1107–19. doi: 10.1021/jp803640e. [DOI] [PubMed] [Google Scholar]
- 28.Su Y, Doherty T, Waring AJ, Ruchala P, Hong M. Biochemistry. 2009;48:4587–95. doi: 10.1021/bi900080d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su Y, Mani R, Doherty T, Waring AJ, Hong M. J Mol Biol. 2008;381:1133–44. doi: 10.1016/j.jmb.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bandekar J. Biochim Biophys Acta. 1992;1120:123–43. doi: 10.1016/0167-4838(92)90261-b. [DOI] [PubMed] [Google Scholar]
- 31.Chen MC, Lord RC. J Am Chem Soc. 1974;96:4750–2. doi: 10.1021/ja00822a004. [DOI] [PubMed] [Google Scholar]
- 32.Dong J, Wan ZL, Chu YC, Nakagawa SN, Katsoyannis PG, Weiss MA, Carey PR. J Am Chem Soc. 2001;123:7919–20. doi: 10.1021/ja011101f. [DOI] [PubMed] [Google Scholar]
- 33.Laporte L, Stulz J, Thomas GJ. Biochemistry. 1997;36:8053–8059. doi: 10.1021/bi970283g. [DOI] [PubMed] [Google Scholar]
- 34.Lord RC, Yu NT. J Mol Biol. 1970;50:509–24. doi: 10.1016/0022-2836(70)90208-1. [DOI] [PubMed] [Google Scholar]
- 35.Overman SA, Thomas GJ., Jr Biochemistry. 1998;37:5654–65. doi: 10.1021/bi972339c. [DOI] [PubMed] [Google Scholar]
- 36.Schweitzer-Stenner R. Vib Spectrosc. 2006;42:98–117. [Google Scholar]
- 37.Diem M, Romeo M, Boydston-White S, Miljkovic M, Matthaus C. Analyst. 2004;129:880–5. doi: 10.1039/b408952a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puppels GJ, de Mul FF, Otto C, Greve J, Robert-Nicoud M, Arndt-Jovin DJ, Jovin TM. Nature. 1990;347:301–3. doi: 10.1038/347301a0. [DOI] [PubMed] [Google Scholar]
- 39.Puppels GJ, Olminkhof JH, Segers-Nolten GM, Otto C, de Mul FF, Greve J. Exp Cell Res. 1991;195:361–7. doi: 10.1016/0014-4827(91)90385-8. [DOI] [PubMed] [Google Scholar]
- 40.Fields GB, Noble RL. Int J Pept Protein Res. 1990;35:161–214. doi: 10.1111/j.1399-3011.1990.tb00939.x. [DOI] [PubMed] [Google Scholar]
- 41.Ortiz C, Zhang D, Xie Y, Ribbe AE, Ben-Amotz D. Anal Biochem. 2006;353:157–66. doi: 10.1016/j.ab.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 42.Sreerama N, Woody RW. Anal Biochem. 2000;287:252–60. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 43.Uzunbajakava N, Lenferink A, Kraan Y, Willekens B, Vrensen G, Greve J, Otto C. Biopolymers. 2003;72:1–9. doi: 10.1002/bip.10246. [DOI] [PubMed] [Google Scholar]
- 44.Krimm S, Bandekar J. Adv Protein Chem. 1986;38:181–364. doi: 10.1016/s0065-3233(08)60528-8. [DOI] [PubMed] [Google Scholar]
- 45.Huang WE, Stoecker K, Griffiths R, Newbold L, Daims H, Whiteley AS, Wagner M. Environ Microbiol. 2007;9:1878–89. doi: 10.1111/j.1462-2920.2007.01352.x. [DOI] [PubMed] [Google Scholar]
- 46.De Gelder J, De Gussem K, Vandenabeele P, Moens L. J Raman Spectrosc. 2007;38:1133–1147. [Google Scholar]
- 47.Overman SA, Thomas GJ., Jr Biochemistry. 1995;34:5440–51. doi: 10.1021/bi00016a015. [DOI] [PubMed] [Google Scholar]
- 48.Czajlik A, Mesko E, Penke B, Perczel A. J Pept Sci. 2002;8:151–71. doi: 10.1002/psc.380. [DOI] [PubMed] [Google Scholar]
- 49.Williams RW. Methods Enzymol. 1986;130:311–31. doi: 10.1016/0076-6879(86)30016-8. [DOI] [PubMed] [Google Scholar]
- 50.Schweitzer-Stenner R. J Phys Chem B. 2004;108:16965–16975. [Google Scholar]
- 51.Measey TJ, Schweitzer-Stenner R. J Am Chem Soc. 2006;128:13324–5. doi: 10.1021/ja0632411. [DOI] [PubMed] [Google Scholar]
- 52.Notingher L, Jell G, Notingher PL, Bisson I, Tsigkou O, Polak JM, Stevens MM, Hench LL. J Mol Struct. 2005;744:179–185. [Google Scholar]
- 53.Crow P, Barrass B, Kendall C, Hart-Prieto M, Wright M, Persad R, Stone N. Br J Cancer. 2005;92:2166–70. doi: 10.1038/sj.bjc.6602638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Notingher I, Jell G, Lohbauer U, Salih V, Hench LL. J Cell Biochem. 2004;92:1180–92. doi: 10.1002/jcb.20136. [DOI] [PubMed] [Google Scholar]
- 55.Short KW, Carpenter S, Freyer JP, Mourant JR. Biophys J. 2005;88:4274–88. doi: 10.1529/biophysj.103.038604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swain RJ, Jell G, Stevens MM. J Cell Biochem. 2008;104:1427–38. doi: 10.1002/jcb.21720. [DOI] [PubMed] [Google Scholar]
- 57.Takai Y, Masuko T, Takeuchi H. Biochim Biophys Acta. 1997;1335:199–208. doi: 10.1016/s0304-4165(96)00138-9. [DOI] [PubMed] [Google Scholar]
- 58.Liu Z, Li X, Tabakman SM, Jiang K, Fan S, Dai H. J Am Chem Soc. 2008;130:13540–1. doi: 10.1021/ja806242t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matthaus C, Kale A, Chernenko T, Torchilin V, Diem M. Mol Pharm. 2008;5:287–93. doi: 10.1021/mp7001158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schweitzer-Stenner R. Biophys J. 2002;83:523–32. doi: 10.1016/S0006-3495(02)75188-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lord RC, Yu NT. J Mol Biol. 1970;51:203–13. doi: 10.1016/0022-2836(70)90137-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.