Abstract
The DNA damage response is a global phosphorylation signaling cascade process involved in sensing the damaged DNA condition and coordinating responses to cope with and repair the perturbed cellular state. We utilized a label-free liquid chromatography-mass spectrometry approach to evaluate changes in protein phosphorylation associated with PP5 activity during the DNA damage response. Biological replicate analyses of bleomycin-treated HeLa cells expressing either WT-PP5 or mutant inactive PP5 lead to the identification of six potential target proteins of PP5 action. Four of these putative targets are known to be involved in DNA damage responses. Using phospho-site specific antibodies, we confirmed that phosphorylation of one target, ribosomal protein S6, was selectively decreased in cells overexpressing catalytically inactive PP5. Our findings also suggest that PP5 may play a role in controlling translation and in regulating substrates for proline-directed kinases, such as MAP kinases and cyclin-dependent protein kinases that are involved in response to DNA damage.
Keywords: Label-free quantitation, DNA damage, Comparative phosphoproteomics, Immobilized metal ion affinity chromatography (IMAC), Mass spectrometry (MS), nano reverse phase HPLC, Protein phosphatase 5, PP5, Ser/Thr protein phosphatase, Bleomycin
INTRODUCTION
Protein phosphatases and kinases work together to control cellular processes and signaling pathways. 1, 2 Although much more is known about the protein kinases and their relevant substrates compared to protein phosphatases, 3–8 the importance of studying protein phosphatase enzymes and their targets has been demonstrated for disease states attributed in part to malfunctioning protein phosphatase enzymes. 9–11 Protein phosphate 5 (PP5), a ubiquitously expressed Ser/Thr protein phosphatase involved in cellular responses to DNA damage, regulates three related key Ser/Thr kinases in response to DNA damage: DNA-PKcs (DNA-dependent protein kinase catalytic subunit), ATM (ataxia telangiectasia mutated), and ATR (ATM and Rad3-related). Complexes containing DNA-PKcs mediate repair of double-stranded DNA breaks often caused by certain chemotherapeutic drugs or ionizing radiation, by non-homologous end joining. PP5 appears to mediate dephosphorylation of one or more key residues on DNA-PKcs that control the DNA repair activity. 12 While ATM also is activated by double-strand breaks in DNA, ATR is usually activated by DNA replication-associated problems brought about by ultraviolet light, hypoxia, or hydroxyurea treatment, 13, 14 which leads to cell cycle arrest and DNA repair or to apoptosis if DNA damage cannot be repaired. The specific role of PP5 in activating these kinases has not been defined, although experiments have shown that when PP5 is absent or over-expressed in a catalytically inactive state, ATM and ATR are not activated. 15, 16
These findings imply that PP5 is activated by multiple forms of DNA damage and may have multiple regulatory targets during the DNA damage response. In the study reported herein, we applied a label-free comparative phosphoproteomics approach17 based on immobilized metal ion affinity chromatography and liquid chromatography-tandem mass spectrometry (LC-MS/MS) to identify these targets. Analysis of phosphoproteins in cells expressing wild type (WT) PP5 and a catalytically inactive mutant form of PP5 in the presence of DNA damage resulted in the identification of 6 potential target proteins of PP5 action, some of which were confirmed using Western blot analysis. Our results suggest a role for PP5 in regulating substrates for proline-directed kinases, as well as for proteins involved in translation control.
EXPERIMENTAL PROCEDURES
Cell Culture and expression of PP5 variants
Stable HeLa Tet-ON cell lines (BD Biosciences Clontech, Palo Alto, CA) that express either WT-PP5 or catalytically inactive PP5(H304Q) under inducible conditions were generated following the manufacturer’s guidelines. Constructs used for generating these cell lines include pCI-Flag WT-PP5 18 and pCI-Flag PP5(H304Q). 19
Cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) / high glucose (Invitrogen, Carlsbad, CA) supplemented with 10% Tet System approved fetal bovine serum (FBS) (Clontech, Mountain View, CA), penicillin (100 units/mL) / streptomycin (100 µg/mL) (Invitrogen), 200 µg/mL geneticin and 200 µg/mL hygromycin B (BD Biosciences, San Jose, CA) at 37°C in 5% CO2. Twenty four hours (24 h) after plating, cells were treated with 2 µg/mL doxycycline (BD Biosciences Clontech) for 48 h to induce overexpression of either WT-PP5 or PP5(H304Q), or left untreated as a control. After induction, cells were treated with 12.5 µg/mL bleomycin sulfate (EMD Biosciences Calbiochem, San Diego, CA) for 1 h at 37°C to induce DNA damage. As a negative control, un-induced cells were also treated with bleomycin and processed in parallel. Two independent sets of four samples (WT and H304Q controls, and over expressed WT-PP5 and PP5(H304Q)) were generated. These sample sets are referred to as biological replicates 1 and 2 throughout this study.
SDS-PAGE and Immunoblot Analysis
Duplicate samples from bleomycin-treated control and induced cells were subjected to SDS-PAGE, using a 10% acrylamide resolving gel to probe separately for phospho-S6 and total S6. Proteins were transferred to a nitrocellulose membrane (Bio-Rad Laboratories, Hercules, CA), and the membrane was incubated with blocking buffer (0.1% Tween-20 in TBS containing 5% BSA for the phospho-S6 blot or 5% milk for all other blots) overnight at 4 °C. The membrane was probed with antibodies that recognize either phospho-Ser 235/236 S6 (Cell Signaling Technology, Danvers, MA, 1:1000), total S6 (Cell Signaling Technology, 1:1000), or cadherin (Abcam Inc., Cambridge, MA, 1/1000). Blots were then washed and probed with secondary antibodies. To detect total S6, blots were incubated with anti-mouse IgG secondary antibody conjugated to horseradish peroxidase (Cell Signaling Technology, 1:5000) and the bound antibody was visualized using enhanced chemiluminescence (GE Healthcare UK Limited, Little Chalfont, Buckinghamshire). To detect phospho-S6 and cadherin, blots were incubated with secondary antibody conjugated with IR-680 dye (1:1000, Molecular Probes, Eugene, OR), and then analyzed using the Odyssey infrared imaging system (Li-Cor Biosciences, Lincoln, NE). Fluorescent signals for phospho-S6 were normalized to cadherin. Total S6 chemiluminescence signals were quantitated using ImageJ software (NIH) and normalized to cadherin. The ratio of normalized phospho-S6 to normalized total S6 was calculated to define the relative level of phospho-S6 per sample. The Western blot for YB-1 was processed in the same manner, except that a 15% acrylamide resolving gel was used and chemilumenescence signals for YB-1 were normalized relative to signals of Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) determined from the same blot.
Protein Extraction
Nearly confluent 100 mm plates of HeLa cells were used for PP5(H304Q) and WT-PP5 samples (7 plates each for the control samples and 8 plates each for samples with overexpressed forms of PP5). Proteins were extracted from the cells using the Roche Complete Lysis-M, EDTA-free kit (Roche Applied Science, Mannheim, Germany) according to the manufacturer’s suggested guidelines. Urea was then added to a final concentration of 8 M, and samples were centrifuged (100,000 × g, 1 h, 4 °C) to remove nucleic acids prior to tryptic digestion. 20 Proteins were reduced with dithiothreitol, and free – SH groups were alkylated with iodoacetamide prior to digestion using modified trypsin at a 1:50 ratio for 4 h at 37 °C, followed by a second trypsin treatment overnight at 37 °C. The digestions were stopped by adding glacial acetic acid to a final pH in the range of 3.5 – 4. STRATA C18-T columns were employed to desalt the tryptic digests, after which tryptic peptides were converted to peptide methyl esters to minimize the non-specific binding of free carboxyl groups during immobilized metal affinity chromatography (IMAC). The procedure of White et al. 21 was followed, except that a second methylation step was added to ensure complete methylation.16 Samples were reconstituted in IMAC loading solution (1:1:1 methanol/acetonitrile/0.01% acetic acid) at a ratio of 100 µL solution to 100 – 200 µg peptides.
Phosphopeptide Enrichment by IMAC
A custom packed IMAC Macrotrap cartridge (3 mm i.d. × 8 mm length) (Michrom BioResources, Inc., Auburn, CA) was employed for phosphopeptide enrichment. 22–24 Our IMAC methodology included most of the advances. 25 Briefly, the column was stripped with 500 µL of 50 mM EDTA (adjusted to pH 9–10 with ammonium hydroxide) at a flow rate of 50 µL/min, washed with 1 mL nanopure water at 100 µL/min, and activated with 375 µL of 100 mM FeCl3 at 25 µL/min. Excess metal ions were removed with 400 µL of 0.1% acetic acid solution at 50 µL/min. The sample was loaded onto the column at 4 µL/min, washed with 400 µL of wash buffer (100 mM NaCl, 1% acetic acid, and 25% acetonitrile) at 25 µL/min, and re-equilibrated with 300 µL of 0.01% acetic acid. Phosphopeptides were eluted with 250 µL of 50 mM Na2HPO4 (pH ~8.5), and the eluant was immediately acidified with glacial acetic acid to a pH of ~4.
Reversed Phase/Nano-HPLC Separation
Peptide mixtures were separated using an automated dual-column phosphoproteome nano-HPLC platform assembled in-house,26 which includes two pairs of SPE and analytical columns. With the exception of the autosampler syringe, all portions of the separation system that come in contact with the peptide mixtures are made of non-metal materials to minimize phosphopeptide losses. HPLC mobile phases consisted of 0.1 M acetic acid in nano-pure water (A) and 70% acetonitrile/0.1 M acetic acid in nano-pure water (B). An exponential HPLC gradient of ~180 min (from 0 – 70% B) was used for each analysis.
LTQ-FT MS/MS
A linear ion trap/Fourier transform (LTQ-FT) hybrid mass spectrometer (Thermo Electron Corp., Bremen, Germany) was used for quantitative measurement and identification of phosphopeptides. Full scan FT-MS spectra (m/z 400 – 2000) were acquired with a resolution of 100,000 for label-free quantitative measurements, which included six technical replicates each of the induced WT-PP5 and PP5(H304Q) samples, and three and four technical replicates of the WT-PP5 and PP5(H304Q) control samples, respectively. MS/MS scans were collected for the 10 most abundant species in each high resolution full MS scan. Datasets were also collected with high mass accuracy precursor FT scans (100,000 resolution), data-dependent MS/MS of the top 5 peptides, followed by MS3 of the neutral loss peak in the MS/MS scan that was correlated with a precursor peak loss associated with phosphorylation, i.e., a neutral loss of 32.7 Da (+3), 49.0 (+2), or 98.0 (+1). This approach was also applied for shorter, predefined full MS scan ranges such as m/z 300 – 850, and m/z 750 – 1575 (so called gas phase fractionation 27) to improve identification of the phosphorylated peptides.
Protein Identification and False Discovery Rate Determination
All results collected from LC-MS/MS analyses were searched by SEQUEST as fully tryptic with static methylation on D and E residues, and the C-terminus of peptides in conjunction with dynamic phosphorylation of S, T, and Y residues. The following filtering criteria were applied to achieve a false discovery rate (FDR) of ≤ 5%: DelCn2 ≥ 0.13; +1 charge state (CS), XCorr ≥ 1.4; +2 CS, XCorr ≥ 2.4; +3 CS, XCorr ≥ 3.3; and +4 CS XCorr ≥ 3.3. The identified phosphorylated peptides were also constrained to a precursor mass error within +/− 6.5 ppm. The Human International Protein Index (IPI) database (Version 3.20 containing 61225 protein entries, www.ebi.ac.uk/IPI) was used to identify proteins. The IPI database also was searched using a decoy database where the reversed human IPI was appended to the forward database and included in the SEQUEST search to determine the FDR. The error rate was estimated from the forward and reverse (decoy) filtered matches, and the FDR was calculated as the percentage of false positive identifications relative to the total number of identified phosphorylated peptides. 28 Tandem MS spectra for all phosphopeptides along with corresponding SEQUEST identification information are included in the SpectrumLook Software Package (see Supplementary Materials) in compliance with recent standards for identifying phosphorylation sites. 29
Quantitative Comparison of Differences in Phosphopeptide Levels
Several programs developed in-house, i.e., Decon-2LS”, 30 “MultiAlign”, 31 and “DAnTE”, 32 (available at http://ncrr.pnl.gov/software) were employed to process mass spectral data and generate matrices for quantitative comparisons of the phosphoproteomes from bleomycin-treated cells expressing either WT-PP5 or catalytically inactive PP5(H304Q). Peptide mass and retention time “features” were extracted from de-isotoped datasets using Decon-2LS, and phosphopeptides with FDR ≤ 5% were used to construct an accurate mass and time tag (AMT) database. 33 Abundances of the same phosphorylated peptide measured in multiple LC-MS analyses were then compared using MultiAlign program. Briefly, features from the LC-MS analyses were collapsed into mass, elution time, and abundance values using the Decon2LS and MultiAlign software packages, and abundance profiles of common features (referred to as clusters) were collated into a master list. Clusters were identified by aligning and matching them based on mass and elution time tolerances to peptides in the AMT tag reference database.
Phosphorylated peptide abundances were transformed (log base 2) and normalized using DAnTE (http://omics.pnl.gov/software/) 32 to remove any systematic variation. First, datasets within each biological replicate group were adjusted to maximize the linear regression correlation against a reference, i.e., the dataset with the fewest missing values in the replicate group. A scaling adjustment based on the median absolute deviation (MAD) was applied to all datasets to adjust variance differences. Finally, the datasets were mean centered. The normalized phosphopeptides for each biological replicate underwent an analysis of variance (ANOVA) to identify statistically significant differences (filtered for a probability factor p ≤ 0.05) in phosphopeptide levels between the four conditions used in the study.
RESULTS AND DISCUSSION
Identification of Phosphopeptides and Potential Target for Regulation by PP5
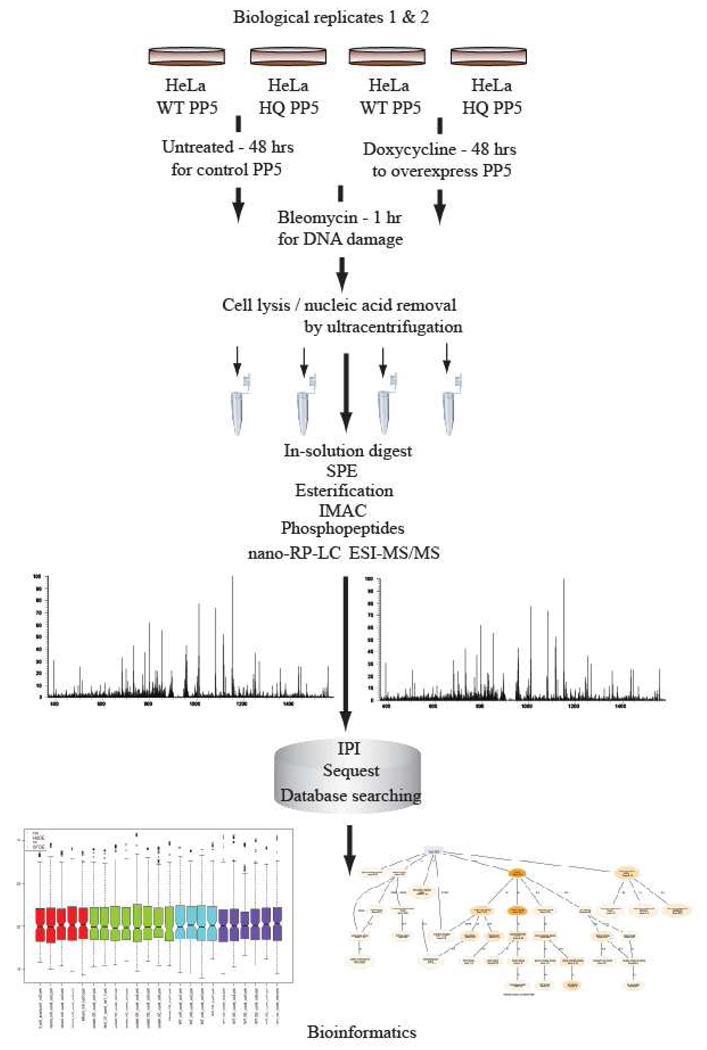
Following the analytical approach illustrated in Figure 1, a total of 122 and 227 unique phosphorylated peptides were identified (FDR ≤ 5%) from LC-MS analyses of biological replicates 1 and 2 respectively. The two biological replicate analyses were performed at different times. The overall differences due to the instrument platform and conditions are reflected in the overall phosphopeptide identifications.
Figure 1.
Major steps involved in the search for targets of PP5 including the cellular treatment with doxycycline for PP5 overexpression, DNA damage induction by bleomycin treatment, nucleic acid removal by ultracentrifugation, label-free differential quantification by MS, followed by bioinformatic discovery treatment of the data.
Based on the overall limited coverage of the analysis, to increase the confidence in those differentially regulated phosphorylation sites, we performed ANOVA significance tests to consider only those phosphopeptides seen as differentially regulated across both biological replicates. Overall analysis resulted in the identification of 6 unique phosphoproteins/15 phosphopeptides that exhibited significant quantitative trends in both biological replicates, which are divided into two sets, shown in Table 1 and 2 respectively.
Table 1.
Overlap of differentially expressed phosphoproteins between biological replicates (BR) 1 and 2, with identical phosphopeptides
| Reference (IPI) |
Peptide | BR 1 Cntl1 |
BR 1 OE1 |
BR 2 Cntl1 |
BR2 OE1 |
Protein | Residue (previously identified) |
Involved in DDR |
Regulation |
|---|---|---|---|---|---|---|---|---|---|
| 00031812 | NYQQNY*QNSESGEKNEGSES*AP EGQAQQR |
0.37 | −2.19 | 0.83 | −1.61 | Y box-binding protein 1 (YB-1) |
Y162 54–56, S176 2 | Yes33–35 | Down |
| 00021405 | SGAQASSTPLS*PTR | 0.82 | 1.05 | −2.39 | −2.07 | Lamin A/C | S22 5, 57–61 | Yes 37 | Up |
| 00007334 | S*KSPS*PPRLTEDR | 0.12 | −1.11 | −0.46 | −1.98 | Apoptotic chromatin condensation inducer in the nucleus (Acinus) |
S384 59, S388 5, 59, 62 | Yes38–40 | Down |
| 00099730 | AQT*PPGPSLSGSKS*PCPQEK | 0.52 | −1.42 | 0.61 | 0.02 | Splicing Coactivator | T1003 5, 57, 59, S1014 57 | Yes 44 | Down |
| VKAQT*PPGPSLSGSKS*PCPQEK | −0.36 | −2.09 | SRm300 |
The values in each of the following columns- BR1 Cntl, BR1 OE, BR2 Cntl, BR2 OE, represent the difference of the (log base 2) average response from PP5(H304Q) and (WT-PP5) (i.e. average PP5(H304Q) response – average (WT-PP5) response). If the average difference (within each biological replicate) going from the control (CnTl) to the overexpressed (OE) condition increases then the regulation is up, if the average difference from the control to the OE condition decreases then the regulation is down. DDR, DNA damage response
Table 2.
Overlap of differentially expressed phosphoproteins between biological replicates (BR) 1 and 2, with similar but not identical peptides
| Reference (IPI) |
Peptide | BR 1 Cntl1 |
BR 1 OE1 |
BR 2 Cntl1 |
BR2 OE1 |
Protein | Residue (previously identified) |
Protein involved in DDR |
Regulation |
|---|---|---|---|---|---|---|---|---|---|
| 00099730 |
2[SGT]PPRQGSIT*SPQANEQ[ SVT]PQRR |
1.24 | −1.16 | Splicing Coactivator SRm300 |
S846 5, T848 5, T856 (No), T866 5, 60 |
Yes 44 | Down | ||
| [SGT]PPRQGSITSPQANEQ[S VT]PQRR |
1.25 | 1.10 | Down | ||||||
| 00021840 | L[SS]LRAS*TSKSESSQK | 2.35 | 0.52 | 40S ribosomal protein S6 | S235 2, 59, 62–64, | Yes [41–43] | Down | ||
| RL[SS]LRAS*TSKSESSQK | 1.33 | −0.91 | S236 2, 59, 62–64, S240 43, 59, | Down | |||||
| L[SS]LRAS*TSKS*ESSQK | 1.36 | 0.77 | 62, 64–66, S244 43, 59, 64–66 | Down | |||||
| RLS*S*LRAS*TSKSESSQK | −2.06 | −5.19 | Down | ||||||
| RLS*S*LRASTSK | −1.35 | −3.20 | Down | ||||||
| RLS*S*LRASTSKSESSQK | −0.53 | −2.87 | Down | ||||||
| 00008274 | SGPKPFS*APKPQTSPS*PK | −1.61 | −2.28 | Adenylyl cyclase- | S310 (No) | No | Down | ||
| SGPKPFSAPKPQT*SPSPK | −0.97 | 0.95 | associated protein 1 | T307 (No) | Up |
The values in each of the following columns- BR1 Cntl, BR1 OE, BR2 Cntl, BR2 OE, represent the difference of the (log base 2) average response from PP5(H304Q) and (WT-PP5) (i.e. average PP5(H304Q) response – average (WT-PP5) response). If the average difference (within each biological replicate) going from the control (Cntl) to the overexpressed (OE) condition increases then the regulation is up, if the average difference from the control to the OE condition decreases then the regulation is down. DDR, DNA damage response
Residues placed within brackets represent ambiguity in phosphorylation site assignment.
In the first set of differentially regulated phosphopeptides, four unique phosphopeptides representing four unique potential target proteins of PP5 overlapped between the two biological replicates (Table 1). For the proteins listed in Table 2, the same differentially regulated peptides were observed in both biological replicates; however these peptides contained additional or alternative phosphorylation sites. In the case of S/R repetitive matrix 1, also known as SRm160, multiple differentially regulated phosphopeptides were identified in both biological replicates, but none of the peptides were shared between replicates (data not shown). The presence of some non-overlapping phosphorylation sites in shared peptides (Table 2) is not surprising, since each phosphoprotein in a cell is likely to be phosphorylated at different sites and with different levels of stoichiometry in any given moment.
Four of these putative targets are known to be involved in DNA damage responses. Y box-binding protein 1 (YB-1) is a multifunctional protein that regulates transcription, translation and mRNA splicing. 34–36 The intermediate filament protein Lamin A/C is a major component of the nuclear matrix that regulates DNA transcription, replication and chromatin organization. 37 Apoptotic chromatin condensation inducer in the nucleus (Acinus) is involved in both pre-mRNA processing and in apoptosis, which can be triggered by irreversible DNA damage and other catastrophic cell cycle disturbances. 38–40 Ribosomal protein S6, a component of the 40S ribosomal subunit is a key target for regulatory phosphorylation by incoming signals that control protein translation and cell size. 41–43 Although a specific role for the splicing factor SRm300 in DNA damage response is not established, other phosphorylation sites within SRm300 have recently been reported to change during DNA damage. Matsuoka and colleagues used immunoaffinity enrichment to identify differentially phosphorylated peptides containing consensus sites for ATM and ATR, phospho-SQ or TQ in SILAC-labeled cells subjected to irradiation. 44 Both consensus and non-consensus phosphorylation sites were identified in immunopurified SRm300-derived peptides in their study. Together with SRm300, SRm160 is a component of multiprotein pre- and post-mRNA splicing complexes. The identification of two spliceosome subunits suggests that this complex is a target for PP5 regulation. Modification of spliceosome subunits during bleomycin treatment is not surprising, since mRNA splicing is altered by genotoxic stress. 45 The final putative target for PP5, Adenylate cyclase-associated protein (CAP-1), is a multifunctional protein required for Ras regulation of adenylate cyclase and cytoskeletal dynamics. Mutations in yeast CAP-1 lead to increased susceptibility to a wide variety of stress inducers. 46
Validation of Target Phosphorylation Sites
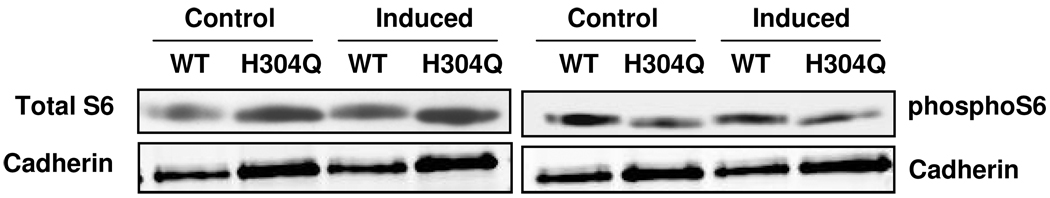
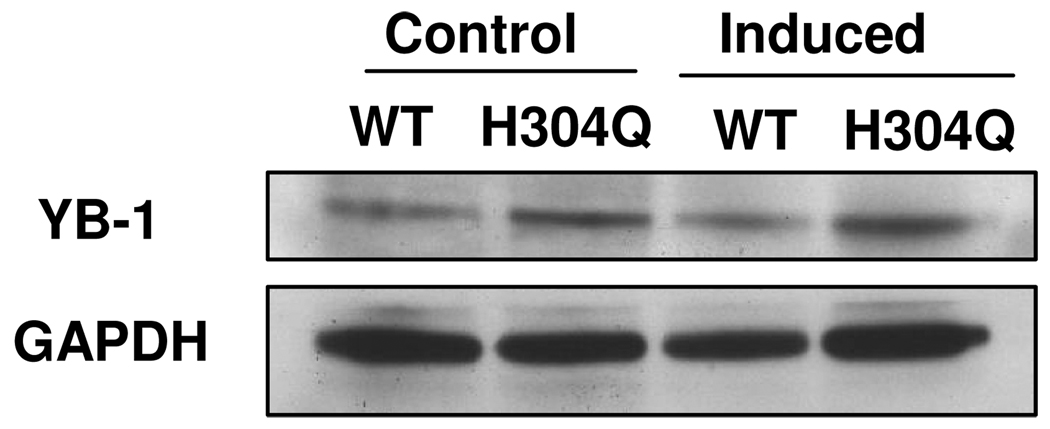
In cells overexpressing PP5 (H304Q), the phosphorylation of PP5 substrates is predicted to be upregulated. In addition, the phosphorylation of targets indirectly influenced by PP5 activity should also be observed; in these cases target sites may increase or decrease. Because WT and inactive PP5 were overexpressed for 48 h, it is also possible that the expression, rather than phosphorylation of the candidate targets identified here was altered. This scenario would also constitute indirect regulation, since PP5 influences several pathways that modify protein expression. 47 In the case of ribosomal S6, we confirmed the LC-MS finding that phosphorylation of S235 and/or S236 was decreased in cells overexpressing PP5 (H304Q) using quantitative Western blot analysis with a phospho-site specific antibody together with antibody recognizing total S6. (Fig 2 and Table 2). Phospho-specific antibodies are not available for analyzing the other target proteins. Nevertheless, for two other putative PP5 targets, we documented that changes in protein expression do not account for the observed changes in phosphopeptide levels. Quantitative Western blot analysis shows that levels of YB-1, a nuclear protein involved in translational regulation, remains unchanged in the conditions represented by the four different samples (Fig 3). In the case of SRm300, we also identified phosphopeptides whose levels were not changed compared to cells overexpressing WT-PP5 and PP5 (H304Q), which is consistent with the conclusion that phosporylation of a subset of sites, rather than protein expression, was selective altered by PP5 (H304Q). For all overlapping peptides found to be differentially regulated in both biological replicates, the trend in regulation was consistent.
Figure 2.
Phosphorylation of S6 Ser 235/236 in bleomycin-treated HeLa cells overexpressing PP5 (H304Q) or WT-PP5. HeLa cells expressing control or induced WT-PP5 or PP5(H304Q) were treated with bleomycin, then lysates prepared and subjected to western blot analysis for phospho-Ser 235/236 S6 or total S6. Cadherin was monitored as the loading control. Blots were quantified as described in Material and Methods. The log base 2 value of the difference of control PP5(H304Q) response – WT-PP5 response was 1.842 and for overexpressed PP5(H304Q) – WT-PP5 was 0.580 in the blots shown. Western blot analyses performed for 2 independent biological replicates yielded similar results.
Figure 3.
YB-1 levels in bleomycin-treated cells overexpressing PP5 (H304Q) relative to cells overexpressing WT-PP5. HeLa cells expressing control or induced WT-PP5 or PP5(H304Q) were treated with bleomycin, then lysates prepared and subjected to western blot analysis for YB-1. GAPDH was monitored as the loading control. Western blot analyses were performed twice, with similar results.
Preferential Changes in Proline-directed Phosphorylation as a Function of PP5 Activity During DNA Damage
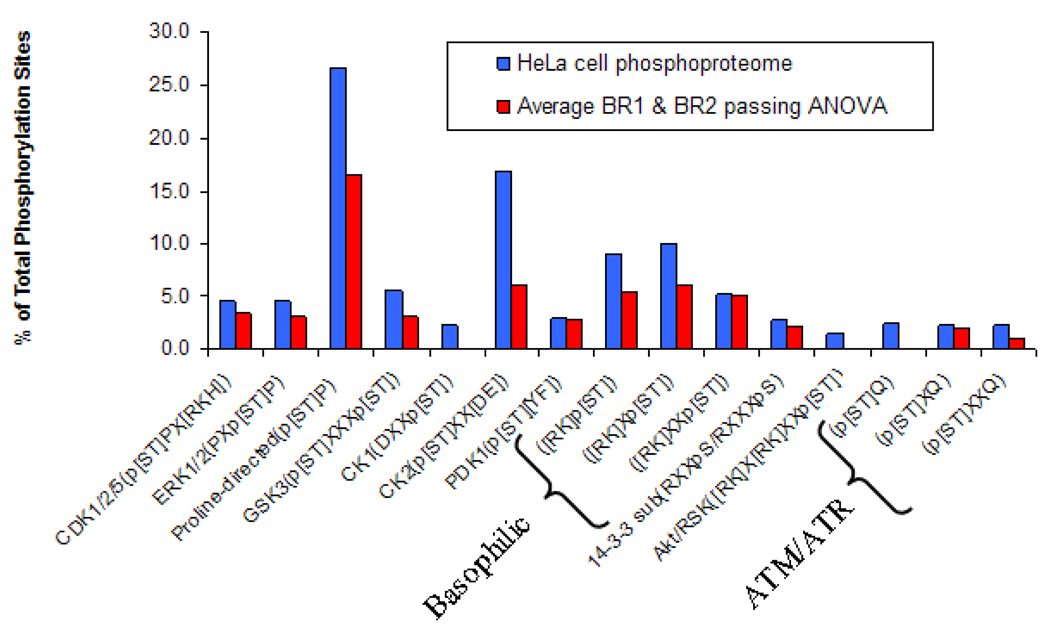
We also performed analyses to explore whether PP5 preferentially dephosphorylates certain kinase consensus sequences. Figure 4 compares phosphorylation sites identified in HeLa cells that express only native PP5 26 with phosphorylated peptides that pass the ANOVA test across the four conditions (the average of biological replicates 1 and 2) for 11 distinct kinase phosphorylation motifs. The percentage of the phosphorylation sites appears similar for each of the motifs with the exception of proline-directed (p[ST]P) and casein kinase II (p[ST]XX[DE]) signature sites. Among the differentially abundant phosphopeptides representing the six PP5 candidates, over 50% of the S/T phosphorylation sites were proline directed compared to 25% in an untreated extract prepared in a similar manner from HeLa cells expressing only native PP5. 26 This finding suggests that targets for proline-directed S/T kinases such as cyclin-dependent protein kinases, MAP kinases, and GSK3 kinase were selectively altered by changing PP5 activity, which is consistent with the established involvement of these pathways and the associated kinases in DNA damage responses, 48, 49 and with reports showing that PP5 can inhibit or block MAP kinase pathways. 50, 51 Although PP5 is required for activation of ATM and ATR, no SQ or TQ-directed phosphorylation sites, which represent optimal sites for ATM and related DNA damage activated-kinases, were found in our study. Peptides containing these sites may be in low abundance and require selective enrichment to be effectively detected. 44
Figure 4.
A comparison of 11 distinct kinase phosphorylation motifs, correlating phosphorylation sites identified in HeLa cells expressing only native PP525 and in phosphorylated peptides passing the ANOVA test across all 4 PP5 conditions (the average of biological replicates 1 and 2).
Comparing Gene Ontology Differences in Cells with DNA Damage Overexpressing WT or Inactive PP5
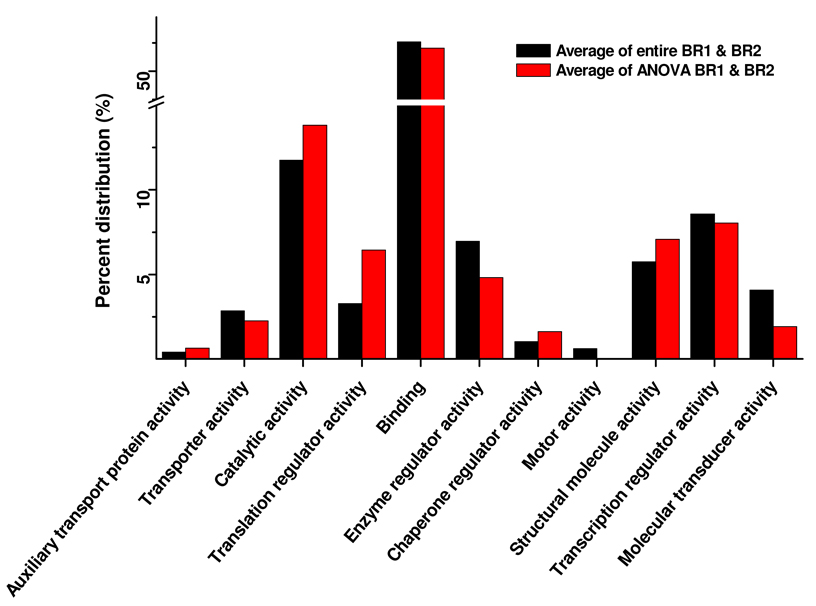
We also explored the molecular function of the differentially phosphorylated proteins identified. To do so, GO terms were determined for the entire complement of identified phosphorylated proteins using the Blast2GO 52 suite of bioinformatic programs. The terms were then compared to the statistically significant regulated phosphorylated proteins for each of the biological replicates. Figure 5 compares the functional categories and percent distributions for the entire complement of phosphorylated proteins identified (average of the two biological replicates) with the phosphorylated proteins passing the ANOVA. The proportion of proteins identified in the two major functional categories, i.e., binding proteins and proteins with catalytic activity was the same in both cases. Proteins involved in translation regulator activity, (e.g., ribosomal protein S6), doubled, increasing from 3% (total phosphoproteins identified) to 6% (phosphoproteins elevated or decreased in bleomycin-treated cells expressing PP5(H304Q), compared to phosphoproteins from cells expressing WT PP5). This result suggests that PP5 influences the phosphorylation status and/or expression levels of proteins controlling translation, a process that is dramatically affected by DNA damage. 53 Because the level of statistically significant differentially expressed phosphorylated protein overlap between biological replicates can be influenced by the phosphorylation action of multiple components of a signaling pathways,44 classes or categories of proteins as opposed to specific key individual proteins may undergo differential regulation under study conditions. This observation was made in the analysis of ATM and ATR substrates during DNA damage, where kinases activated by DNA damage influenced the phosphorylation of multiple components of particular pathways rather than key individual proteins within the process. The increase in phosphoproteins controlling translation activity in cells overexpressing inactive PP5 and subjected to DNA damage may represent a similar pattern.
Figure 5.
Functional categories and the percent distributions (the average of biological replicates 1 and 2) for the entire complement of phosphorylated proteins identified versus the phosphorylated proteins passing the ANOVA test.
CONCLUSIONS
We have identified several potential substrates for PP5 regulation in DNA damaged-cells. Several of these phosphoproteins are known to function in the DNA damage pathway, whereas several candidate targets have not previously been associated with DNA damage. The sites identified in these phosphoproteins exhibit a bias for proline-directed kinases. Analysis of functional categories of all differentially regulated phosphoproteins also suggests that PP5 could potentially play a role in translational regulation.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs. Jon Jacobs and Weijun Qian for helpful discussions and input. This research was supported by NIH grants NS031221 (SR) and RR018522 (RDS). Work was performed in the Proteomics NCRR Biomedical Technology Resource Center, located in the Environmental Molecular Sciences Laboratory (EMSL), a U.S. Department of Energy (DOE) Office of Biological and Environmental Science national scientific user facility on the Pacific Northwest National Laboratory (PNNL) campus in Richland, Washington. PNNL is a multi-program national laboratory operated by Battelle for the DOE under Contract DE-AC05-76RL01830.
REFERENCES
- 1.Chalmers MJ, Kolch W, Emmett MR, Marshall AG, Mischak H. Identification and analysis of phosphopeptides. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;803(1):111–120. doi: 10.1016/j.jchromb.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127(3):635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 3.Amanchy R, Kalume DE, Iwahori A, Zhong J, Pandey A. Phosphoproteome analysis of HeLa cells using stable isotope labeling with amino acids in cell culture (SILAC) J Proteome Res. 2005;4(5):1661–1671. doi: 10.1021/pr050134h. [DOI] [PubMed] [Google Scholar]
- 4.Ballif BA, Roux PP, Gerber SA, MacKeigan JP, Blenis J, Gygi SP. Quantitative phosphorylation profiling of the ERK/p90 ribosomal S6 kinase-signaling cassette and its targets, the tuberous sclerosis tumor suppressors. Proc Natl Acad Sci U S A. 2005;102(3):667–672. doi: 10.1073/pnas.0409143102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A. 2004;101(33):12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffert JD, Pisitkun T, Wang G, Shen RF, Knepper MA. Quantitative phosphoproteomics of vasopressin-sensitive renal cells: regulation of aquaporin-2 phosphorylation at two sites. Proc Natl Acad Sci U S A. 2006;103(18):7159–7164. doi: 10.1073/pnas.0600895103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang F, Stenoien DL, Strittmatter EF, Wang J, Ding L, Lipton MS, Monroe ME, Nicora CD, Gristenko MA, Tang K, Fang R, Adkins JN, Camp DG, 2nd, Chen DJ, Smith RD. Phosphoproteome profiling of human skin fibroblast cells in response to low- and high-dose irradiation. J Proteome Res. 2006;5(5):1252–1260. doi: 10.1021/pr060028v. [DOI] [PubMed] [Google Scholar]
- 8.Zhou H, Watts JD, Aebersold R. A systematic approach to the analysis of protein phosphorylation. Nat Biotechnol. 2001;19(4):375–378. doi: 10.1038/86777. [DOI] [PubMed] [Google Scholar]
- 9.Arroyo JD, Hahn WC. Involvement of PP2A in viral and cellular transformation. Oncogene. 2005;24(52):7746–7755. doi: 10.1038/sj.onc.1209038. [DOI] [PubMed] [Google Scholar]
- 10.Brady MJ, Saltiel AR. The role of protein phosphatase-1 in insulin action. Recent Prog Horm Res. 2001;56:157–173. doi: 10.1210/rp.56.1.157. [DOI] [PubMed] [Google Scholar]
- 11.Oliver CJ, Shenolikar S. Physiologic importance of protein phosphatase inhibitors. Front Biosci. 1998;3:D961–D972. doi: 10.2741/a336. [DOI] [PubMed] [Google Scholar]
- 12.Wechsler T, Chen BP, Harper R, Morotomi-Yano K, Huang BC, Meek K, Cleaver JE, Chen DJ, Wabl M. DNA-PKcs function regulated specifically by protein phosphatase 5. Proc Natl Acad Sci U S A. 2004;101(5):1247–1252. doi: 10.1073/pnas.0307765100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammond EM, Giaccia AJ. The role of ATM and ATR in the cellular response to hypoxia and re-oxygenation. DNA Repair (Amst) 2004;3(8–9):1117–1122. doi: 10.1016/j.dnarep.2004.03.035. [DOI] [PubMed] [Google Scholar]
- 14.Roy K, Wang L, Makrigiorgos GM, Price BD. Methylation of the ATM promoter in glioma cells alters ionizing radiation sensitivity. Biochem Biophys Res Commun. 2006;344(3):821–826. doi: 10.1016/j.bbrc.2006.03.222. [DOI] [PubMed] [Google Scholar]
- 15.Ali A, Zhang J, Bao S, Liu I, Otterness D, Dean NM, Abraham RT, Wang XF. Requirement of protein phosphatase 5 in DNA-damage-induced ATM activation. Genes Dev. 2004;18(3):249–254. doi: 10.1101/gad.1176004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Bao S, Furumai R, Kucera KS, Ali A, Dean NM, Wang XF. Protein phosphatase 5 is required for ATR-mediated checkpoint activation. Mol Cell Biol. 2005;25(22):9910–9919. doi: 10.1128/MCB.25.22.9910-9919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang F, Jaitly N, Jayachandran H, Luo Q, Monroe ME, Du X, Gritsenko MA, Zhang R, Anderson DJ, Purvine SO, Adkins JN, Moore RJ, Mottaz HM, Ding SJ, Lipton MS, Camp DG, 2nd, Udseth HR, Smith RD, Rossie S. Applying a targeted label-free approach using LC-MS AMT tags to evaluate changes in protein phosphorylation following phosphatase inhibition. J Proteome Res. 2007;6(11):4489–4497. doi: 10.1021/pr070068e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Messner D, Romeo C, Boynton A, Rossie S. Inhibition of PP2A, but not PP5, mediates p53 activation by low levels of okadaic acid in rat liver epithelial cells. J Cell Biochem. 2005;99(1):241–255. doi: 10.1002/jcb.20919. [DOI] [PubMed] [Google Scholar]
- 19.Sanchez-Ortiz E, Hahm BK, Armstrong DA, Rossie S. Protein phosphatase 5 protects neurons against amyloid β toxicity. J Neurochem. 2009 doi: 10.1111/j.1471-4159.2009.06337.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giavalisco P, Nordhoff E, Lehrach H, Gobom J, Klose J. Extraction of proteins from plant tissues for two-dimensional electrophoresis analysis. Electrophoresis. 2003;24(1–2):207–216. doi: 10.1002/elps.200390016. [DOI] [PubMed] [Google Scholar]
- 21.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20(3):301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 22.Cao P, Stults JT. Mapping the phosphorylation sites of proteins using on-line immobilized metal affinity chromatography/capillary electrophoresis/electrospray ionization multiple stage tandem mass spectrometry. Rapid Commun Mass Spectrom. 2000;14(17):1600–1606. doi: 10.1002/1097-0231(20000915)14:17<1600::AID-RCM68>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 23.Kocher T, Allmaier G, Wilm M. Nanoelectrospray-based detection and sequencing of substoichiometric amounts of phosphopeptides in complex mixtures. J Mass Spectrom. 2003;38(2):131–137. doi: 10.1002/jms.422. [DOI] [PubMed] [Google Scholar]
- 24.Posewitz MC, Tempst P. Immobilized gallium(III) affinity chromatography of phosphopeptides. Anal Chem. 1999;71(14):2883–2892. doi: 10.1021/ac981409y. [DOI] [PubMed] [Google Scholar]
- 25.Ndassa YM, Orsi C, Marto JA, Chen S, Ross MM. Improved immobilized metal affinity chromatography for large-scale phosphoproteomics applications. J Proteome Res. 2006;5(10):2789–2799. doi: 10.1021/pr0602803. [DOI] [PubMed] [Google Scholar]
- 26.Ham BM, Yang F, Jayachandran H, Jaitly N, Monroe ME, Gritsenko MA, Livesay EA, Zhao R, Purvine SO, Orton D, Adkins JN, Camp DG, 2nd, Rossie S, Smith RD. The influence of sample preparation and replicate analyses on HeLa Cell phosphoproteome coverage. J Proteome Res. 2008;7(6):2215–2221. doi: 10.1021/pr700575m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yi EC, Marelli M, Lee H, Purvine SO, Aebersold R, Aitchison JD, Goodlett DR. Approaching complete peroxisome characterization by gas-phase fractionation. Electrophoresis. 2002;23(18):3205–3216. doi: 10.1002/1522-2683(200209)23:18<3205::AID-ELPS3205>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 28.Li X, Gerber SA, Rudner AD, Beausoleil SA, Haas W, Villen J, Elias JE, Gygi SP. Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J Proteome Res. 2007;6(3):1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- 29.Bradshaw RA, Burlingame AL, Carr S, Aebersold R. Reporting protein identification data: the next generation of guidelines. Mol Cell Proteomics. 2006;5(5):787–788. doi: 10.1074/mcp.E600005-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Mayampurath AM, Jaitly N, Purvine SO, Monroe ME, Auberry KJ, Adkins JN, Smith RD. DeconMSn: a software tool for accurate parent ion monoisotopic mass determination for tandem mass spectra. Bioinformatics. 2008;24(7):1021–1023. doi: 10.1093/bioinformatics/btn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaitly N, Monroe ME, Petyuk VA, Clauss TR, Adkins JN, Smith RD. Robust algorithm for alignment of liquid chromatography-mass spectrometry analyses in an accurate mass and time tag data analysis pipeline. Anal Chem. 2006;78(21):7397–7409. doi: 10.1021/ac052197p. [DOI] [PubMed] [Google Scholar]
- 32.Polpitiya AD, Qian WJ, Jaitly N, Petyuk VA, Adkins JN, Camp DG, 2nd, Anderson GA, Smith RD. DAnTE: a statistical tool for quantitative analysis of - omics data. Bioinformatics. 2008;24(13):1556–1558. doi: 10.1093/bioinformatics/btn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith RD, Anderson GA, Lipton MS, Masselon C, Pasa-Tolic L, Udseth H, Belov M, Shen Y, Veenstra TD. High-performance separations and mass spectrometric methods for high-throughput proteomics using accurate mass tags. Adv Protein Chem. 2003;65:85–131. doi: 10.1016/s0065-3233(03)01017-9. [DOI] [PubMed] [Google Scholar]
- 34.Gaudreault I, Guay D, Lebel M. YB-1 promotes strand separation in vitro of duplex DNA containing either mispaired bases or cisplatin modifications, exhibits endonucleolytic activities and binds several DNA repair proteins. Nucleic Acids Res. 2004;32(1):316–327. doi: 10.1093/nar/gkh170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guay D, Gaudreault I, Massip L, Lebel M. Formation of a nuclear complex containing the p53 tumor suppressor, YB-1, and the Werner syndrome gene product in cells treated with UV light. Int J Biochem Cell Biol. 2006;38(8):1300–1313. doi: 10.1016/j.biocel.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 36.Kohno K, Izumi H, Uchiumi T, Ashizuka M, Kuwano M. The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays. 2003;25(7):691–698. doi: 10.1002/bies.10300. [DOI] [PubMed] [Google Scholar]
- 37.Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22(7):832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu Y, Yao J, Liu Z, Liu X, Fu H, Ye K. Akt phosphorylates acinus and inhibits its proteolytic cleavage, preventing chromatin condensation. Embo J. 2005;24(20):3543–3554. doi: 10.1038/sj.emboj.7600823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joselin AP, Schulze-Osthoff K, Schwerk C. Loss of Acinus inhibits oligonucleosomal DNA fragmentation but not chromatin condensation during apoptosis. J Biol Chem. 2006;281(18):12475–12484. doi: 10.1074/jbc.M509859200. [DOI] [PubMed] [Google Scholar]
- 40.Sahara S, Aoto M, Eguchi Y, Imamoto N, Yoneda Y, Tsujimoto Y. Acinus is a caspase-3-activated protein required for apoptotic chromatin condensation. Nature. 1999;401(6749):168–173. doi: 10.1038/43678. [DOI] [PubMed] [Google Scholar]
- 41.Brenneisen P, Wenk J, Wlaschek M, Krieg T, Scharffetter-Kochanek K. Activation of p70 ribosomal protein S6 kinase is an essential step in the DNA damage-dependent signaling pathway responsible for the ultraviolet B-mediated increase in interstitial collagenase (MMP-1) and stromelysin-1 (MMP-3) protein levels in human dermal fibroblasts. J Biol Chem. 2000;275(6):4336–4344. doi: 10.1074/jbc.275.6.4336. [DOI] [PubMed] [Google Scholar]
- 42.Ruvinsky I, Meyuhas O. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci. 2006;31(6):342–348. doi: 10.1016/j.tibs.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 43.Shah OJ, Ghosh S, Hunter T. Mitotic regulation of ribosomal S6 kinase 1 involves Ser/Thr, Pro phosphorylation of consensus and non-consensus sites by Cdc2. J Biol Chem. 2003;278(18):16433–16442. doi: 10.1074/jbc.M300435200. [DOI] [PubMed] [Google Scholar]
- 44.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316(5828):1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 45.Lal A, Gorospe M. Egad, more forms of gene regulation: the gadd45a story. Cell Cycle. 2006;5(13):1422–1425. doi: 10.4161/cc.5.13.2902. [DOI] [PubMed] [Google Scholar]
- 46.Shima F, Okada T, Kido M, Sen H, Tanaka Y, Tamada M, Hu CD, Yamawaki-Kataoka Y, Kariya K, Kataoka T. Association of yeast adenylyl cyclase with cyclase-associated protein CAP forms a second Ras-binding site which mediates its Ras-dependent activation. Mol Cell Biol. 2000;20(1):26–33. doi: 10.1128/mcb.20.1.26-33.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hinds TD, Jr, Sanchez ER. Protein phosphatase 5. Int J Biochem Cell Biol. 2007 doi: 10.1016/j.biocel.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dent P, Yacoub A, Fisher PB, Hagan MP, Grant S. MAPK pathways in radiation responses. Oncogene. 2003;22(37):5885–5896. doi: 10.1038/sj.onc.1206701. [DOI] [PubMed] [Google Scholar]
- 49.Watcharasit P, Bijur GN, Zmijewski JW, Song L, Zmijewska A, Chen X, Johnson GV, Jope RS. Direct, activating interaction between glycogen synthase kinase-3beta and p53 after DNA damage. Proc Natl Acad Sci U S A. 2002;99(12):7951–7955. doi: 10.1073/pnas.122062299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morita K, Saitoh M, Tobiume K, Matsuura H, Enomoto S, Nishitoh H, Ichijo H. Negative feedback regulation of ASK1 by protein phosphatase 5 (PP5) in response to oxidative stress. Embo J. 2001;20(21):6028–6036. doi: 10.1093/emboj/20.21.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.von Kriegsheim A, Pitt A, Grindlay GJ, Kolch W, Dhillon AS. Regulation of the Raf-MEK-ERK pathway by protein phosphatase 5. Nat Cell Biol. 2006;8(9):1011–1016. doi: 10.1038/ncb1465. [DOI] [PubMed] [Google Scholar]
- 52.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 53.Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34(Pt 1):7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 54.Guo A, Villen J, Kornhauser J, Lee KA, Stokes MP, Rikova K, Possemato A, Nardone J, Innocenti G, Wetzel R, Wang Y, MacNeill J, Mitchell J, Gygi SP, Rush J, Polakiewicz RD, Comb MJ. Signaling networks assembled by oncogenic EGFR and c-Met. Proc Natl Acad Sci U S A. 2008;105(2):692–697. doi: 10.1073/pnas.0707270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, Hu Y, Tan Z, Stokes M, Sullivan L, Mitchell J, Wetzel R, Macneill J, Ren JM, Yuan J, Bakalarski CE, Villen J, Kornhauser JM, Smith B, Li D, Zhou X, Gygi SP, Gu TL, Polakiewicz RD, Rush J, Comb MJ. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131(6):1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 56.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23(1):94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 57.Imami K, Sugiyama N, Kyono Y, Tomita M, Ishihama Y. Automated phosphoproteome analysis for cultured cancer cells by two-dimensional nanoLC-MS using a calcined titania/C18 biphasic column. Anal Sci. 2008;24(1):161–166. doi: 10.2116/analsci.24.161. [DOI] [PubMed] [Google Scholar]
- 58.Lowery DM, Clauser KR, Hjerrild M, Lim D, Alexander J, Kishi K, Ong SE, Gammeltoft S, Carr SA, Yaffe MB. Proteomic screen defines the Polo-box domain interactome and identifies Rock2 as a Plk1 substrate. Embo J. 2007;26(9):2262–2273. doi: 10.1038/sj.emboj.7601683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Molina H, Horn DM, Tang N, Mathivanan S, Pandey A. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci U S A. 2007;104(7):2199–2204. doi: 10.1073/pnas.0611217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24(10):1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 61.Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R. Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci U S A. 2006;103(14):5391–5396. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim JE, Tannenbaum SR, White FM. Global phosphoproteome of HT-29 human colon adenocarcinoma cells. J Proteome Res. 2005;4(4):1339–1346. doi: 10.1021/pr050048h. [DOI] [PubMed] [Google Scholar]
- 63.Gu JJ, Santiago L, Mitchell BS. Synergy between imatinib and mycophenolic acid in inducing apoptosis in cell lines expressing Bcr-Abl. Blood. 2005;105(8):3270–3277. doi: 10.1182/blood-2004-10-3864. [DOI] [PubMed] [Google Scholar]
- 64.Lal L, Li Y, Smith J, Sassano A, Uddin S, Parmar S, Tallman MS, Minucci S, Hay N, Platanias LC. Activation of the p70 S6 kinase by all-trans-retinoic acid in acute promyelocytic leukemia cells. Blood. 2005;105(4):1669–1677. doi: 10.1182/blood-2004-06-2078. [DOI] [PubMed] [Google Scholar]
- 65.Corradetti MN, Inoki K, Guan KL. The stress-inducted proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. J Biol Chem. 2005;280(11):9769–9772. doi: 10.1074/jbc.C400557200. [DOI] [PubMed] [Google Scholar]
- 66.Shah OJ, Hunter T. Critical role of T-loop and H-motif phosphorylation in the regulation of S6 kinase 1 by the tuberous sclerosis complex. J Biol Chem. 2004;279(20):20816–20823. doi: 10.1074/jbc.M400957200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.