Abstract
A virally encoded superfamily-2 (SF2) helicase (NS3h) is essential for the replication of hepatitis C virus, a leading cause of liver disease worldwide. Efforts to elucidate the function of NS3h and to develop inhibitors against it, however, have been hampered by limited understanding of its molecular mechanism. Here we show x-ray crystal structures for a set of NS3h complexes, including ground-state and transition-state ternary complexes captured with ATP mimics (ADP·BeF3 and 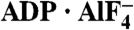 ). These structures provide, for the first time, three conformational snapshots demonstrating the molecular basis of action for a SF2 helicase. Upon nucleotide binding, overall domain rotation along with structural transitions in motif V and the bound DNA leads to the release of one base from the substrate base-stacking row and the loss of several interactions between NS3h and the 3′ DNA segment. As nucleotide hydrolysis proceeds into the transition state, stretching of a “spring” helix and another overall conformational change couples rearrangement of the (d)NTPase active site to additional hydrogen-bonding between NS3h and DNA. Together with biochemistry, these results demonstrate a “ratchet” mechanism involved in the unidirectional translocation and define the step size of NS3h as one base per nucleotide hydrolysis cycle. These findings suggest feasible strategies for developing specific inhibitors to block the action of this attractive, yet largely unexplored drug target.
). These structures provide, for the first time, three conformational snapshots demonstrating the molecular basis of action for a SF2 helicase. Upon nucleotide binding, overall domain rotation along with structural transitions in motif V and the bound DNA leads to the release of one base from the substrate base-stacking row and the loss of several interactions between NS3h and the 3′ DNA segment. As nucleotide hydrolysis proceeds into the transition state, stretching of a “spring” helix and another overall conformational change couples rearrangement of the (d)NTPase active site to additional hydrogen-bonding between NS3h and DNA. Together with biochemistry, these results demonstrate a “ratchet” mechanism involved in the unidirectional translocation and define the step size of NS3h as one base per nucleotide hydrolysis cycle. These findings suggest feasible strategies for developing specific inhibitors to block the action of this attractive, yet largely unexplored drug target.
Keywords: antivirals, motor protein, step size, superfamily 2, transition state
Hepatitis C virus (HCV) is a 9.6 kb positive-sense, single-stranded RNA (ssRNA) virus of the Flaviviridae family. It is a major cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma worldwide (1). A protective vaccine is not available and the existing treatment is frequently ineffective. Like other positive-strand RNA viruses, HCV replicates in close association with modified intracellular membranes (2), where the viral replicase complex catalyzes accumulation of progeny RNA genomes through a negative-strand intermediate. The helicase domain of viral nonstructural protein 3 (NS3h) is an enzymatic component of this replication apparatus and is essential for HCV propagation; the precise role of the helicase, however, remains obscure [reviewed in (2, 3)].
NS3h is classified as a superfamily-2 (SF2) DExH helicase. It has polynucleotide-stimulated (d)NTPase activity (4) and can unwind both RNA and DNA in a 3′-5′ direction (5, 6). Structures of NS3h in complex with single-stranded DNA (ssDNA) have shown that DNA binds in a groove between domain 3 and the RecA-like domains, with the bases stacked in a row between two “bookend” residues (V432 in the domain 2 β-hairpin and W501 in domain 3) (7, 8). The structure of the substrate DNA remains ambiguous, however, as the pucker conformations assigned to the sugar groups differ between reports. Whereas ATP binding has been suggested between NS3h domains 1 and 2 (7), the atomic details of nucleotide binding and hydrolysis have not been described. Kinetic studies of HCV NS3h have begun to reveal its duplex unwinding features detecting periodic pauses of this helicase along the unwinding process. The minimum step size per nucleotide hydrolysis cycle, however, remains to be defined (9–12).
As the largest helicase family, SF2 includes many other medically important enzymes (13–15). Although crystal structures of SF2 helicases have been reported, no enzyme in complex with its substrate has been visualized in different conformational states and the structural details pertaining to the actions of these helicases are lacking (15, 16). Despite the structural snapshots obtained for the SF1 helicases (16–18), the motif distinctions between SF1 and SF2 members suggest divergent mechanisms and there is no consensus on how the SF2 enzymes translocate unidirectionally along ssDNA/ssRNA and destabilize duplexes for unwinding (3, 15). This deficiency has hampered efforts to understand the functional details of SF2 helicases and slowed the search for potent inhibitors targeting medically important candidates, such as the HCV helicase (3, 19).
The difficulty in structurally characterizing nucleotide-bound NS3h is the lack of ATP analogs that can inhibit and cocrystallize with the helicase. Here, we report a set of structures of NS3h complexes, including ground-state and transition-state ternary complexes captured with ATP mimics (ADP·BeF3 and 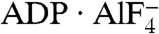 ), to recapitulate the actions of the helicase activity along the reaction pathway of ATP binding and hydrolysis. Our study reveals a series of structural transitions that lead to a ratchet-like action driving unidirectional translocation, define the minimum step as one base translocation per ATP hydrolyzed, and illuminate divergent features in the actions of SF1 and SF2 helicases. The structural details of ATP coordination and DNA substrate binding, as well as nucleotide-dependent structural transitions of motif V and a spring helix, suggest several sites for rational drug design.
), to recapitulate the actions of the helicase activity along the reaction pathway of ATP binding and hydrolysis. Our study reveals a series of structural transitions that lead to a ratchet-like action driving unidirectional translocation, define the minimum step as one base translocation per ATP hydrolyzed, and illuminate divergent features in the actions of SF1 and SF2 helicases. The structural details of ATP coordination and DNA substrate binding, as well as nucleotide-dependent structural transitions of motif V and a spring helix, suggest several sites for rational drug design.
Results
Structure Determination.
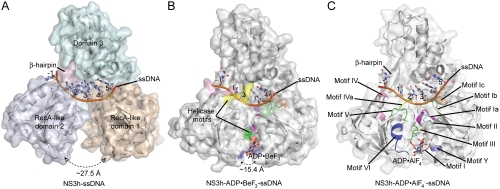
To investigate the mechanism of HCV helicase action, we determined the structures of NS3h bound to ssDNA (dA6 or dT6) alone and in combination with ground-state and transition-state mimics of ATP (Fig. 1 and Table 1). The NS3h structures in NS3h-ssDNA(dA6) and NS3h-ssDNA(dT6) were superimposable to each other, and also to the polypeptide chains from the previously determined NS3h structures in complex with ssDNA. The structure of ssDNA is, however, different from those in previous reports (7, 8). Although several nucleotide analogs, such as AMPPNP/AMPPCP [adenosine 5′-(β,γ-imido/methylene)triphosphate] and ATPγS, have been used to capture helicases in nucleotide-bound states, these commonly used analogs do not inhibit the catalytic activity of the HCV helicase (20) and do not cocrystallize with NS3h. To capture NS3h along the reaction pathway of ATP binding and hydrolysis, we used alternative ATP mimics in cocrystallization attempts. We were initially able to determine structures for NS3h-ADP·BeF3·Mn-ssDNA(dT6) and 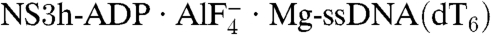 ternary complexes. As the NS3h-ADP·BeF3·Mn-ssDNA(dT6) complex showed binding of NS3h to two short DNA fragments, we further solved the structure of NS3h-ADP·BeF3·Mn-ssDNA(dT12) to reveal the electron density representing one continuous DNA strand (see Experimental Procedures).
ternary complexes. As the NS3h-ADP·BeF3·Mn-ssDNA(dT6) complex showed binding of NS3h to two short DNA fragments, we further solved the structure of NS3h-ADP·BeF3·Mn-ssDNA(dT12) to reveal the electron density representing one continuous DNA strand (see Experimental Procedures).
Fig. 1.
Crystal Structures of NS3h Complexes. (A) NS3h in complex with ssDNA (dA6). (B) NS3h in complex with ADP·BeF3 and ssDNA (dT12, with five deoxynucleosides presented for clarity). (C) NS3h in complex with 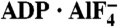 and ssDNA (dT6). The structures are represented by ribbons and transparent surfaces. The DNA bases and deoxyribose groups are shown with sticks and numerically labeled. The DNA phosphodiester backbones are simplified as orange tubes. The DNA atoms are color coded according to elements. The helicase motifs are color coded in the surface and ribbon respectively in (B) and (C). The distances between the Cα atoms of K244 (domain 1) and S403 (domain 2) are noted. ADP·BeF3 and
and ssDNA (dT6). The structures are represented by ribbons and transparent surfaces. The DNA bases and deoxyribose groups are shown with sticks and numerically labeled. The DNA phosphodiester backbones are simplified as orange tubes. The DNA atoms are color coded according to elements. The helicase motifs are color coded in the surface and ribbon respectively in (B) and (C). The distances between the Cα atoms of K244 (domain 1) and S403 (domain 2) are noted. ADP·BeF3 and 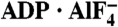 are shown with sticks and color coded.
are shown with sticks and color coded.
Table 1.
Crystallographic Data and Refinement Statistics.
| Complex | 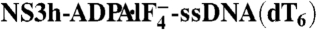 |
NS3h-ADP·BeF3-ssDNA(dT6) | NS3h-ADP·BeF3-ssDNA(dT12) | NS3h-ssDNA(dA6) | NS3h-ssDNA(dT6) |
| PDB ID | 3KQL | 3KQN | 3KQU | 3KQH | 3KQK |
| Data collection | |||||
| Source | NSLS X29 | NSLS X29 | NSLS X29 | NSLS X29 | NSLS X29 |
| Wavelength (Å) | 1.0809 | 1.0809 | 1.0809 | 1.0809 | 1.0809 |
| Resolution (Å) | 50–2.5 (2.59–2.5) | 50–2.05 (2.12–2.05) | 50–2.4 (2.49–2.4) | 50–2.4 (2.49–2.4) | 50–2.7 (2.8–2.7) |
| Space group | P2 | P61 | P1 | P212121 | P212121 |
| Unit cell: a, b, c (Å) | 43.5, 75.4, 144.2 | 116.8, 116.8, 71.2 | 116.4, 116.5, 71.1 | 45.2, 115.4, 197.3 | 45.5, 115.4, 198.7 |
| α, β, γ (°) | 90, 90.2, 90 | 90, 90, 120 | 89.9, 89.9, 119.9 | 90, 90, 90 | 90, 90, 90 |
| Observations | 159870 | 385808 | 296250 | 277054 | 169476 |
| Unique reflections | 31686 | 34176 | 121925 | 40398 | 28955 |
| Redundancy | 5.0 (2.8) | 11.3 (4.3) | 2.6 (1.9) | 6.9 (2.9) | 5.9 (3.1) |
| Completeness (%) | 97.6 (88.3) | 97.7 (92.2) | 96.8 (89.0) | 96.3 (82.3) | 97.0 (79.3) |
| Mean I/σI | 9.6 | 11.6 | 12.1 | 12.8 | 10.5 |
| R-merge on I* | 0.10 (0.28) | 0.08 (0.28) | 0.07 (0.32) | 0.06 (0.30) | 0.08 (0.30) |
| Cutoff criteria l/σl | -0.5 | 0 | -1 | -0.5 | -1 |
| Refinement statistics | |||||
| Resolution limits (Å) | 50–2.5 (2.59–2.5) | 50–2.05 (2.12–2.05) | 50–2.4 (2.49–2.4) | 50–2.4 (2.49–2.4) | 50–2.8 (2.9–2.8) |
| Number of reflections | 30311 | 33064 | 110458 | 38741 | 24872 |
| Completeness (%) | 93.2 (74.0) | 95.1 (83.4) | 87.2 (59.8) | 93.5 (70.9) | 93.7 (69.2) |
| Cutoff criteria l/σl | 0 | 0 | 0 | 0 | 0 |
| Atoms: Protein | 6560 | 3280 | 19680 | 6550 | 6550 |
| Nucleic Acid | 234 | 117 | 2856 | 246 | 234 |
| Ligand | 66 | 32 | 192 | 0 | 0 |
| Solvent | 211 | 223 | 424 | 138 | 138 |
| Rcryst† | 0.184 (0.240) | 0.190 (0.254) | 0.197 (0.323) | 0.225 (0.340) | 0.210 (0.295) |
| Rfree (5% of data) | 0.210 (0.254) | 0.223 (0.299) | 0.215 (0.335) | 0.256 (0.348) | 0.265 (0.322) |
| Bonds (Å)/Angles (°)‡ | 0.007/1.2 | 0.006/1.2 | 0.008/1.4 | 0.007/1.1 | 0.008/1.1 |
| Mean B value (Å2) | 30 | 22.3 | 39.8 | 47.1 | 41.4 |
| Coordinate errors (Å)§ | 0.31 | 0.27 | 0.33 | 0.38 | 0.46 |
| Ramachandran plot | |||||
| Most favored (%) | 87.2 | 89.6 | 87.8 | 85.1 | 82.9 |
| Allowed (%) | 12.7 | 10.4 | 12.2 | 15.0 | 16.8 |
| Disallowed (%) | 0 | 0 | 0 | 0 | 0 |
Numbers in parentheses indicate the range of the highest resolution bin and statistics for data in this resolution bin.
*Rmerge = ΣhklΣi|l(hkl)i - 〈l(hkl)〉|lΣhklΣi〈l(hkl)i〉
†Rcryst = Σhkl|Fo(hkl)-Fc(hkl)|Fo(hkl)|, where Fo and Fc are observed and calculated structure factors, respectively.
‡Values indicate rms deviations in bond lengths and bond angles.
§Estimated coordinate errors from cross-validated Luzzati Plot.
ATP Coordination and Active-Site Rearrangement for Catalysis.
Similar to other ATPases composed of RecA-like domains (21), NS3h closes these two domains in the presence of ATP mimics (ADP·BeF3 and 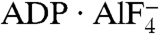 ), which bind in a pocket formed by motifs I, II, III, V, along with a newly defined motif Y (Fig. 1C and Fig. S1). Interestingly, in the absence of structural information on nucleotide binding, no previous sequence or structural analysis had identified motif Y. The aromatic side chain (Y241) and the location of this motif between motif Ia and Ib allowed us to define several related helicases (Fig. S1). Surprisingly, no motif Y exists in the flavivirus helicases, which are thought to be highly similar to HCV NS3h.
), which bind in a pocket formed by motifs I, II, III, V, along with a newly defined motif Y (Fig. 1C and Fig. S1). Interestingly, in the absence of structural information on nucleotide binding, no previous sequence or structural analysis had identified motif Y. The aromatic side chain (Y241) and the location of this motif between motif Ia and Ib allowed us to define several related helicases (Fig. S1). Surprisingly, no motif Y exists in the flavivirus helicases, which are thought to be highly similar to HCV NS3h.
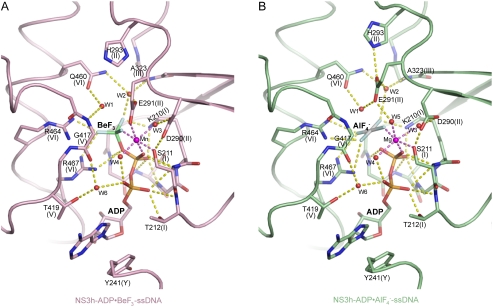
The BeF3 group is tetrahedral, with the beryllium atom approximately 1.7 Å from the bridging oxygen of the ADP β-phosphate and approximately 3.5 Å from the nucleophilic water (W1; Fig. 2A and Fig. S2A and B), indicating that ADP·BeF3 mimics a bound ATP in the ground state. In contrast, the 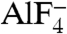 group is planar, mimicking the trigonal planar geometry of a γ phosphate in the transition state, with the aluminum center about equidistant (approximately 2.0 Å) from the two apical oxygens (W1 and ADP β-phosphate; Fig. 2B and Fig. S2C and D). This indicates that
group is planar, mimicking the trigonal planar geometry of a γ phosphate in the transition state, with the aluminum center about equidistant (approximately 2.0 Å) from the two apical oxygens (W1 and ADP β-phosphate; Fig. 2B and Fig. S2C and D). This indicates that 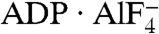 approximates the transition state of ATP hydrolysis.
approximates the transition state of ATP hydrolysis.
Fig. 2.
(d)NTPase Active-Site Views. (A) Coordination of ADP·BeF3. (B) Coordination of 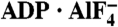 . NS3h-ADP·BeF3-ssDNA(dT6) is pink and
. NS3h-ADP·BeF3-ssDNA(dT6) is pink and 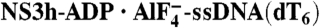 is green. The non-carbon atoms are color coded according to elements. Protein main chains are simplified as tubes. The main-chain atoms and side chains involved in the coordination of nucleotides are represented by sticks. These protein residues are labeled with single-letter codes, residue numbers and motif codes. The metal ions (magenta) and water molecules (red) are spheres. The interactions are represented by dashed lines.
is green. The non-carbon atoms are color coded according to elements. Protein main chains are simplified as tubes. The main-chain atoms and side chains involved in the coordination of nucleotides are represented by sticks. These protein residues are labeled with single-letter codes, residue numbers and motif codes. The metal ions (magenta) and water molecules (red) are spheres. The interactions are represented by dashed lines.
Y241, in the newly defined motif Y, and T419, in motif V, sandwich the ADP adenine base, but the helicase recognizes neither the identity of the base nor the 2′ ribose oxygen, explaining how NS3h can utilize all types of nucleotides (NTP and dNTP) (4). The α- and β-phosphate groups are coordinated by two water molecules (W4 and W6), as well as by motif I main-chain nitrogens (from G207 to T212) and side-chain atoms (K210 [Nζ] and T212 [Oγ]; Fig. 2). These interactions likely stabilize the ADP moiety for catalysis. The γ-phosphate groups, mimicked by BeF3 and 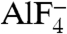 , are coordinated by three positively charged side chains (K210, R464, and R467) and by water (W2)-mediated interactions (E291 [Oϵ2], A323 [main-chain N], and Q460 [Nϵ]) (Fig. 2). The importance of the positively charged residues is supported by previous mutagenesis and arginine methylation studies (22–25). The arginine residues may play a catalytic role analogous to that of the “arginine finger” in the GTPase-activating proteins (26). The metal ion (Mn/Mg), which is essential for catalysis (5, 6), sits at the center of an octahedral coordination complex, making contacts with S211 (Oγ), E291 (Oϵ1) (or W5), two water molecules (W3 and W4), a fluorine atom in
, are coordinated by three positively charged side chains (K210, R464, and R467) and by water (W2)-mediated interactions (E291 [Oϵ2], A323 [main-chain N], and Q460 [Nϵ]) (Fig. 2). The importance of the positively charged residues is supported by previous mutagenesis and arginine methylation studies (22–25). The arginine residues may play a catalytic role analogous to that of the “arginine finger” in the GTPase-activating proteins (26). The metal ion (Mn/Mg), which is essential for catalysis (5, 6), sits at the center of an octahedral coordination complex, making contacts with S211 (Oγ), E291 (Oϵ1) (or W5), two water molecules (W3 and W4), a fluorine atom in 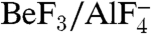 , and a nonbridging oxygen atom in the β-phosphate group (Fig. 2 and Fig. S2).
, and a nonbridging oxygen atom in the β-phosphate group (Fig. 2 and Fig. S2).
Compared with previously characterized SF1 and SF2 helicases (15), NS3h shows similar active-site architecture, but clear distinctions are seen (see Discussion). Indeed, a major feature that distinguishes NS3h from SF1 helicases, such as UvrD (17), is the rearrangement of the motif II residues and the active-site interactions as ATP hydrolysis proceeds from the ground state to the transition state (Fig. 2 and Fig. S3). D290 reorients while maintaining interactions with two metal ligands (S211 [Oγ] and W3). E291 coordinates the metal ion and W2 in the ground state, but makes different interactions in the transition state, as its Oϵ1 atom establishes a bifurcated hydrogen bond with W1 and W5, and Oϵ2 forms an additional interaction with H293 (Nϵ). The E291 (Oϵ2)-H293 (Nϵ) interaction is probably important for the substrate-stimulated (d)NTPase activity, as a H293A mutation dramatically reduces the catalytic activity associated with substrate binding (25) without impacting the basal ATPase activity (23). The nucleophilic water (W1) loses its interaction with G417 (main-chain N) and is contacted by E291 (Oϵ1), as well as Q460 (Oϵ) in the transition state, mimicking an in-line attack to the γ-phosphorus. The importance of these motif II residues and motif VI Q460 is supported by previous mutagenesis studies (24, 25, 27). Our structures suggest that E291 and Q460 stabilize the positive charge developing on the attacking water, whereas the positively charged residues (K210, R464, and R467) and the metal ion likely neutralize the negative charge emerging on the γ-phosphate (Fig 2B).
Three Distinct Conformational States.
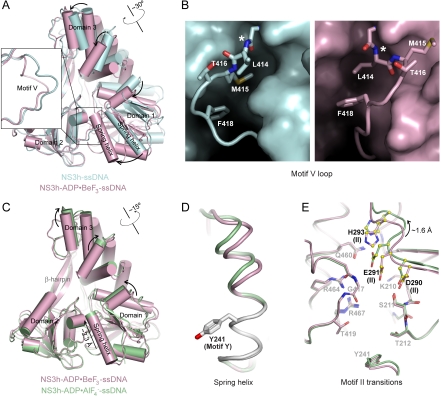
Structural alignment of domain 2 in NS3h-ssDNA and NS3h-ADP·BeF3-ssDNA reveals a structural transition in motif V along with a rotation of domains 1 and 3 (Fig. 3A). In this process, M415 gives up its binding pocket and moves into an adjacent groove, as L414 rotates to the pocket initially occupied by M415 (Fig. 3B). This transition positions the main-chain atoms of G417 and side chain of T419 for nucleotide binding (Fig. 2), and moves the main-chain nitrogen of L414 away from ssDNA (Figs. 3B and 5).
Fig. 3.
Nucleotide-Dependent Conformational Changes of NS3h. (A) Structural comparison of NS3h in the NS3h-ssDNA(dA6) (cyan) and NS3h-ADP·BeF3-ssDNA(dT6) (pink) complexes, aligned through domain 2. (B) Motif V in the nucleotide-free state (cyan) and the ground state (pink). The complexes are aligned through domain 2, and presented in the same direction. The asterisks mark the L414 main-chain nitrogen atom, which moves away from the substrate-binding groove upon nucleotide binding. (C) Structural comparison of NS3h in the NS3h-ADP·BeF3-ssDNA(dT6) and 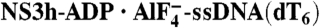 (green) complexes, aligned through their ADP moieties. (D) The spring helices from the ground-state and transition-state complexes are aligned through motif Y and the adjacent residues colored in white. Y241 is shown in sticks. (E) Structural remodeling of the (d)NTPase active site from the ground state to the transition state. The complexes are aligned as those in (B). Motif II is highlighted with yellow spheres (carbon atoms).
(green) complexes, aligned through their ADP moieties. (D) The spring helices from the ground-state and transition-state complexes are aligned through motif Y and the adjacent residues colored in white. Y241 is shown in sticks. (E) Structural remodeling of the (d)NTPase active site from the ground state to the transition state. The complexes are aligned as those in (B). Motif II is highlighted with yellow spheres (carbon atoms).
Fig. 5.
Structural Features Recapitulating the Ratchet Mechanism. (A) ssDNA binding in the NS3h-ssDNA(dA6) complex. (B) ssDNA binding in the NS3h-ADP·BeF3-ssDNA(dT12) complex. (C) ssDNA binding in the 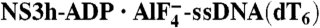 complex. The carbon elements of the complexes are color coded as in Fig 3. Protein main chains are simplified as tubes and ribbons. The residues involved in ssDNA binding are shown with sticks and color coded according to elements. Side chains are omitted for those residues that interact with ssDNA only through their main-chain atoms. The DNA strands are modeled with sticks and color coded according to elements. The orientations of the DNA bases are noted as either anti or syn. The deoxyribose groups labeled B are in a C2′-endo, whereas the ones labeled A are in C3′-endo. The yellow dashed lines represent hydrogen bonds. The location of the spring helix is noted.
complex. The carbon elements of the complexes are color coded as in Fig 3. Protein main chains are simplified as tubes and ribbons. The residues involved in ssDNA binding are shown with sticks and color coded according to elements. Side chains are omitted for those residues that interact with ssDNA only through their main-chain atoms. The DNA strands are modeled with sticks and color coded according to elements. The orientations of the DNA bases are noted as either anti or syn. The deoxyribose groups labeled B are in a C2′-endo, whereas the ones labeled A are in C3′-endo. The yellow dashed lines represent hydrogen bonds. The location of the spring helix is noted.
We aligned the two ternary complexes (NS3h-ADP·BeF3-ssDNA and 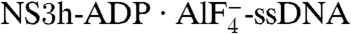 ) through their ADP moieties and observed a further conformational change as ATP hydrolysis proceeds. This change occurs mainly in domain 3 and the top of domain 1, which rotate about 15° along the axis noted in Fig. 3C. In the transition state, the lower (C-terminal) part of the α-helix containing motif Y is locked by the stacking interaction between Y241 and the ADP adenine base (Fig. 2), whereas the main-chain hydrogen bonds in the upper (N-terminal) region are extended (Fig. 3D). As a result, this α-helix is tilted and stretched like a spring, positioning the N-terminal region approximately 2.3 Å (main-chain nitrogen atom of V232) closer to the ssDNA-binding groove in the transition state versus the ground state. We call this α-helix the spring helix for convenience. The conformational change toward the transition state is coupled to the structural alterations in the (d)NTPase active site. The most obvious rearrangement occurs in motif II, which moves approximately 1.6 Å (between the Cα atoms of C292) along the nucleotide-binding site as the side chains are reoriented as described above (Fig. 3E). This probably optimizes the active-site interaction network to stabilize the transition state of nucleotide hydrolysis (Fig. 2B). The conformational difference between the ground state and transition state is supported by our ssDNA-binding assay, in which these two types of complexes, reconstituted with the same metal ion (Mn), show different substrate binding affinities. No analogous feature is observed for the UvrD helicase (17), suggesting that the coupled motion of NS3h is distinct from that of the SF1 enzymes (see Discussion).
) through their ADP moieties and observed a further conformational change as ATP hydrolysis proceeds. This change occurs mainly in domain 3 and the top of domain 1, which rotate about 15° along the axis noted in Fig. 3C. In the transition state, the lower (C-terminal) part of the α-helix containing motif Y is locked by the stacking interaction between Y241 and the ADP adenine base (Fig. 2), whereas the main-chain hydrogen bonds in the upper (N-terminal) region are extended (Fig. 3D). As a result, this α-helix is tilted and stretched like a spring, positioning the N-terminal region approximately 2.3 Å (main-chain nitrogen atom of V232) closer to the ssDNA-binding groove in the transition state versus the ground state. We call this α-helix the spring helix for convenience. The conformational change toward the transition state is coupled to the structural alterations in the (d)NTPase active site. The most obvious rearrangement occurs in motif II, which moves approximately 1.6 Å (between the Cα atoms of C292) along the nucleotide-binding site as the side chains are reoriented as described above (Fig. 3E). This probably optimizes the active-site interaction network to stabilize the transition state of nucleotide hydrolysis (Fig. 2B). The conformational difference between the ground state and transition state is supported by our ssDNA-binding assay, in which these two types of complexes, reconstituted with the same metal ion (Mn), show different substrate binding affinities. No analogous feature is observed for the UvrD helicase (17), suggesting that the coupled motion of NS3h is distinct from that of the SF1 enzymes (see Discussion).
Structural Transitions in Associated ssDNA.
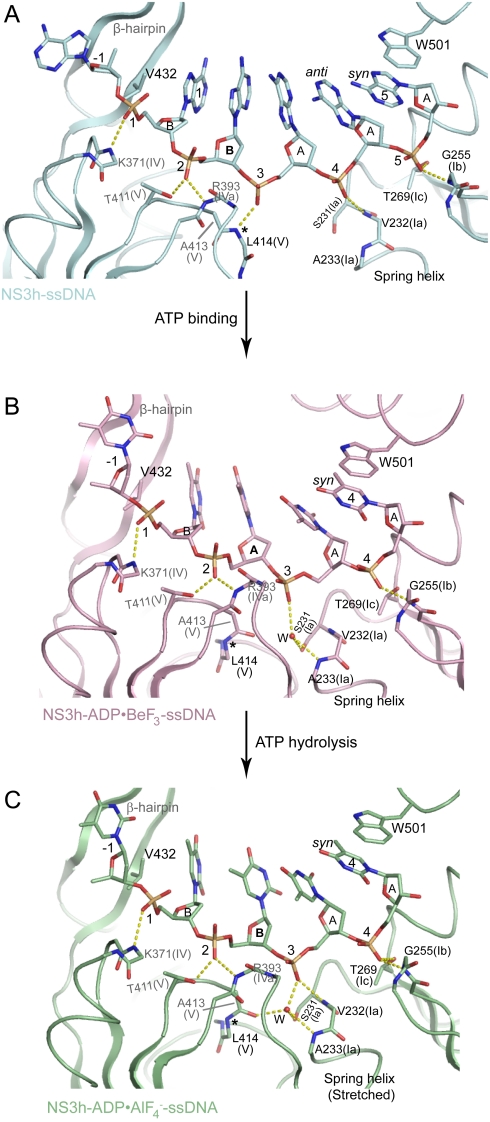
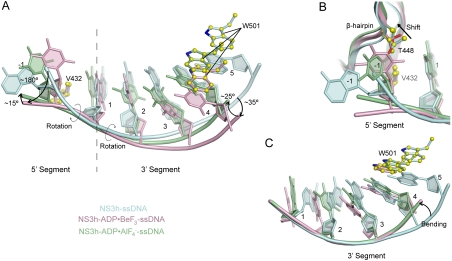
Our structures reveal that nucleotide-dependent conformational changes are accompanied by structural transitions within the bound DNA strand (Fig. 4), indicating that efficient translocation requires coordinated actions of NS3h and its substrate. Upon nucleotide binding, the 5′ and 3′ ssDNA segments flanking deoxynucleoside 1 rotate approximately 180° and 35° (Fig. 4A), through torsions of the ϵ and ζ angles in deoxynucleoside -1 and of the α, β, and γ angles in deoxynucleoside 2 and 3, respectively. As ATP hydrolysis proceeds into the transition state, the 5′ and 3′ fragments further rotate approximately 15° and 25°, respectively (Fig. 4A). Rotation of the 5′ segment moves the -1 base closer to the base-stacking row, whereas T448 in the domain 2 β-hairpin progressively shifts away (Fig. 4B). Alignment of the 3′ ssDNA segments through phosphates 1 to 3 reveals an additional bending in response to closure of the nucleotide-binding site (Fig. 4C); torsion of the α, β, and γ angles in deoxynucleoside 4 places the backbone between motifs Ib and Ic as base 4 makes its stacking interaction with W501 (Figs. 4C and 5B and C).
Fig. 4.
Rotation and Bending of ssDNA. (A) Structural comparison of ssDNA in the NS3h-ssDNA(dA6), NS3h-ADP·BeF3-ssDNA(dT12), and 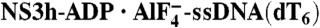 complexes. NS3h-ssDNA(dA6) and NS3h-ADP·BeF3-ssDNA(dT12) are aligned through domain 2. NS3h-ADP·BeF3-ssDNA(dT12) and
complexes. NS3h-ssDNA(dA6) and NS3h-ADP·BeF3-ssDNA(dT12) are aligned through domain 2. NS3h-ADP·BeF3-ssDNA(dT12) and 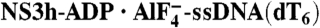 are aligned through the ADP moieties. (B) A closer view of the 5′ segments of ssDNA and the β-hairpin. The structures are aligned through domain 2. (C) Structural comparison of the 3′ segments of ssDNA. The structures are aligned through phosphate 1 to 3 of the DNA strands. The complexes are color coded as in Fig 3. The DNA bases and deoxyribose groups are represented by sticks and numerically labeled in each panel. The phosphodiester backbones are simplified as tubes. Protein side chains are modeled with sticks. The carbon atoms in protein residues are highlighted by yellow spheres.
are aligned through the ADP moieties. (B) A closer view of the 5′ segments of ssDNA and the β-hairpin. The structures are aligned through domain 2. (C) Structural comparison of the 3′ segments of ssDNA. The structures are aligned through phosphate 1 to 3 of the DNA strands. The complexes are color coded as in Fig 3. The DNA bases and deoxyribose groups are represented by sticks and numerically labeled in each panel. The phosphodiester backbones are simplified as tubes. Protein side chains are modeled with sticks. The carbon atoms in protein residues are highlighted by yellow spheres.
Further analysis reveals different sugar puckers and base orientations among the bound DNA residues that had not been observed previously (7, 8) (Fig. 5 and Figs. S4 and S5). In the NS3h-ssDNA complex, the sugar rings of deoxynucleoside 1 and 2 have a C2′-endo pucker, whereas those of 3–5 adopt a C3′-endo pucker. In addition, the fifth base, which stacks with W501, is in a syn orientation, whereas the other bases are in anti orientations (Fig. 5A). In the ATP-bound state, the fifth base is presumably released from the stacking row whereas the fourth base takes its place to stack with W501 in a syn orientation (Fig. 5B). This orientation is likely preferred for binding, and we suggest that the base rotates as it is released from the stacking row and placed next to W501. Similarly, the sugar pucker of deoxynucleoside 2 is changed from C2′-endo to C3′-endo on nucleotide binding and further alternates back to C2′-endo in the transition state (Fig. 5B and C). After the release of ADP and Pi, one DNA base, newly separated from the duplex section, will presumably join the stacking row and the second deoxynucleoside will become the third. As shown in the ATP-free state, the sugar pucker of the third deoxynucleoside is C3′-endo (Fig. 5A). The pucker alteration seems to be coupled to the relocation of the ssDNA phosphate groups, such as phosphate 3, and thus may facilitate movement of nucleic acid substrates (Fig. 5).
Although the structural details of ssRNA binding remain to be described, the bound ssDNA visualized here suggests that the 2′ oxygen atoms in ssRNA would sterically restrict the backbone ribose pucker conformations, and thus limit ssRNA binding to HCV NS3h. The NS3 protease domain, which is positioned to capture the C-terminal tail of the helicase in the structure of an engineered single-chain NS3-4A protein (28), may rearrange its interactions with NS3h to enhance RNA substrate binding (29, 30).
Intradomain Structural Transitions Propel Helicase Motion.
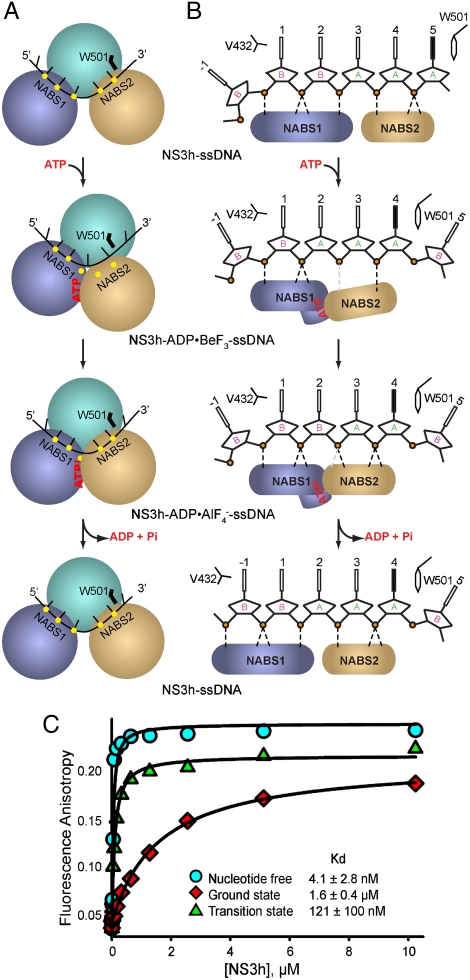
We name two nucleic acid binding surfaces (NABS1 and NABS2) to describe translocation. NABS1 is formed by motifs IV, IVa [or “arginine clamp” (31)] and V in domain 2, and NABS2 is made up of motifs Ia, Ib, and Ic in domain 1 (Figs. 1C and 5 and Fig. S1). Initially, NABS1 coordinates phosphates 1 to 3, while NABS2 captures phosphates 4 and 5. Following ATP binding, NABS1 appears to maintain association with phosphates 1 and 2 but loses contact with phosphate 3, as L414 (main-chain N) is moved away by the transition of motif V (Fig. 3B). As NABS2 moves toward NABS1, phosphate 3 is located near the spring helix through water-mediated interactions, and phosphate 4 interacts with G255 (main-chain N), forming the single direct contact between NABS2 and the phosphodiester backbone (Figs. 5 and 6). In the transition state, distances between NABS2 and DNA are rearranged and new hydrogen bonds appear. Phosphate 3 is directly coordinated by the stretched spring helix (V232 [main-chain N]), and phosphate 4 establishes an additional interaction with T269 (Oγ) as the distance shortens from approximately 3.5 Å to 2.8 Å (a change larger than the estimated coordinate errors; Figs. 5 and 6 and Table 1). The changes in coordination of phosphate 3 suggest that the structural transitions in motif V and the spring helix accompany the overall domain rotations to accomplish a ratchet action, releasing phosphate 3 from NABS1 and recapturing it in NABS2 to secure the unidirectional motion. The loss and recovery of helicase-DNA interactions suggest alternating weak and strong substrate associations along the unwinding cycle. This is supported by the observation that the ssDNA-binding affinity is significantly decreased in the presence of ATP or ground-state mimics (20). We observed an approximately 400-fold increase of the dissociation constant Kd with the addition of ground-state mimics, and an approximately 30-fold increase of Kd with transition-state mimics (Fig. 6C). The actions predicted by these structures are expected to be followed by reset of the spring helix and motif V as NABS1 slides and regrips ssDNA to return to the initial state (Fig. 6 and Figs. S7A and Movie S1).
Fig. 6.
Schematic Presentation of Helicase Motion. (A) Schematic view of conformational changes between NS3h subdomains. The NS3h complexes are simplified as spherical modules (NS3h) and black lines (ssDNA). The two DNA-binding surfaces (NABS1 and NABS2) are noted. The yellow dots represent the sites involved in the coordination of phosphate groups of ssDNA. The W501 side chain is simplified as a black line. Nucleotides are noted as red letters. (B) Schematic view of ssDNA in the substrate-binding groove. Individual DNA residues are presented. The deoxyribose groups in C2′-endo pucker are labeled B, whereas the others in C3′-endo pucker are labeled A. The solid-black DNA bases are in syn orientation. The two DNA-binding surfaces are simplified as blue and pink modules. The black dashed lines represent hydrogen bonds between NS3h and the phosphodiester backbone of DNA, whereas the gray dashed lines are water-mediated interactions. (C) Fluorescence anisotropy titration in the absence and presence of ATP mimics. Data were fit to a quadratic equation to obtain dissociation constants (Kd).
Previous studies detected various kinetic steps of the HCV helicase along the unwinding process and suggested that they represent periodic release of tension accumulated by several minimum elemental steps (9, 10, 12). Our structural snapshots define the minimum translocation step of one base per ATP hydrolysis (Figs. 5 and 6 and Movie S1), consistent with the previous prediction based on the analysis of 3-bp kinetic unwinding steps (10).
These results provide structural details to demonstrate the translocation mechanism for a SF2 helicase, revealing how coordinated structural transitions asymmetrically alter the substrate contacts of the two DNA-binding sites and propel the helicase in a 3′-5′ direction along the DNA strand (Figs. 5 and 6). Although the NS3 protease domain likely modulates nucleic acid substrate binding to increase duplex unwinding, it is not essential for the helicase activity (29, 32). Thus, the fundamental mode of structural transitions involved in the motion of the HCV helicase is likely independent of the protease.
Discussion
Ratchet-Type Unidirectional Translocation.
Taken together, the three conformational snapshots of the HCV helicase reveal a series of structural transitions within the two motor domains and the DNA substrate in the process of ATP binding and active-site remodeling toward the transition state. Like the actions of a ratchet, the successive transitions of motif V and the spring helix release phosphate 3 from NABS1 and recapture it in NABS2, presumably preventing phosphate 3 from walking in the opposite direction (Figs. 6A and B). The mechanism illustrated here is likely adopted by other SF2 DExH members with similar motifs, especially the newly defined motif Y (Fig. S1), although divergence among different SF2 subgroups cannot be neglected. Whereas we believe the ratchet is one significant element involved in unidirectional motion, multiple factors probably combine to define directionality. For example, the base rotation around W501 (a potential “gatekeeper”) probably allows NABS2 to slide and initiate the translocation, and DNA pucker alternations along with the backbone rotations may facilitate the motion. No analogous intradomain structural transitions have been observed in the SF1 enzymes, although ATP-induced rigid-body rotations of the motor domains are seen (16–18). Indeed, structural comparison of the representative SF1 and SF2 members reveals explicit differences in catalyzing nucleotide hydrolysis and motion (Figs. S6 and S7), reflecting the fact that these helicases have evolved to adopt divergent mechanisms and act in different biological processes.
Comparison of Helicase Mechanisms.
To compare the catalytic features of SF1 and SF2 enzymes, we aligned the nucleotide-binding sites of HCV NS3h and three other helicases as ground-state ternary complexes. DENV (dengue virus) NS3h represents flavivirus-encoded SF2 DExH helicases (33), Vasa represents SF2 DEAD-box helicases (34), and UvrD represents SF1 helicases (17). Although the nucleotide-binding pockets of these helicases have similar architecture, notable differences are seen (Fig. S6). First, the AMP moieties are contacted by differing structural/motif elements. The orientations of the adenosine groups in HCV and DENV NS3h are similar, but almost orthogonal to those in Vasa and UvrD. It is also notable that the ribose group in HCV NS3h adopts a C3′-endo pucker, whereas those in the other three helicases are in C2′-endo. Another major difference involves the coordination of the nucleophilic water. In HCV NS3h, motif II E291 does not coordinate the nucleophilic water in the ground-state structure, whereas the corresponding residues in the other helicases are involved in coordination. HCV and DENV NS3h use a glutamine in motif VI to coordinate the nucleophilic water, whereas Vasa uses a motif VI histidine and UvrD uses a motif III glutamine. In addition, an interaction between the nucleophilic water and the main-chain nitrogen atom of the motif V glycine is seen in all SF2 helicases, whereas this is not observed in SF1 UvrD. It is also noteworthy that the DEAD-box Vasa establishes an interaction network among motif II, III, and VI, whereas no such interactions exist in the viral SF2 or UvrD SF1 helicases.
Our structural snapshots of HCV NS3h show a series of nucleotide-dependent inter- and intradomain structural transitions (Fig. 3). Similar to NS3h, SF1 enzymes show closure of the two RecA-like domains upon ATP-analog binding (16–18). No intradomain 1 and 2 structural transitions analogous to those in HCV NS3h, however, have been observed. Indeed, the ground-state and transition-state complexes of UvrD show superimposable overall conformations and active-site arrangements (17). Although reorganization of a short region (part of motif IV) upon ATP-analog binding has been reported for the RecD2 helicase, the overall structural transitions are similar to those of other SF1 enzymes (16). In addition, UvrD shows a reoriented “gating helix” in domain 2B in some crystal forms (17), yet no intradomain transition in the analogous domain 3 of NS3h was observed. These differences reflect the divergent features of helicase translocation, which allow distinct mechanisms of substrate recognition for different biological processes. Compared with NS3h, the PcrA and UvrD helicases alternate stacking contacts through base flipping actions (18), and the RecD2 helicase shifts contacts along the phosphodiester backbone along with base flipping around motif Ia (16) (Fig. S7).
Our study is the first to visualize a substrate-bound SF2 helicase in different conformational states. Structures of SF2 DEAD-box helicases are available for a few helicase-AMPPNP-ssRNA complexes and helicases alone (some are in complex with ligands such as sulfate ions, ADP, and AMP) (34–36), but no panel of helicase-substrate binary and transition-state ternary complexes has previously been solved. The recently reported set of DENV NS3h structures shows ssRNA-bound complexes under various nucleotide-binding conditions. The complexes are, however, superimposable, suggesting that one or more altered conformational states could not be trapped (33). Mechanistic comparison of NS3h to other SF2 enzymes therefore awaits the solution of additional structures. It is, however, predictable that the fundamental framework of action illustrated here for NS3h is adopted by other SF2 DExH members (Fig. S1).
NS3 Functions as a Translocase.
Our results suggest that destabilization of the ssDNA-and-dsDNA junction is primarily mediated by the nucleotide-dependent movement of NS3h, indicating that the HCV helicase should be able to act as a ssDNA/ssRNA translocase. The NS3h translocase activity may be responsible for stripping proteins from nucleic acid substrates for protein–nucleic acid complex remodeling, as has been described for various helicases (15). In fact, the NS3 helicase can displace streptavidin from 5′-biotinylated oligonucleotides (37). Translocase activity may allow NS3h to act in various steps of the viral life cycle, indeed the helicase domain has been implicated in virus assembly, although the role of the enzymatic activity in this process is still unclear (38).
Sites for Rational Drug Design.
As an essential enzymatic activity of the replicase, the HCV helicase is an attractive, yet largely unexplored, drug target. Our structures suggest that nucleotide-triphosphate analogs with modified bases and/or ribose groups may inhibit NS3h without affecting cellular ATPases or GTPases, as these groups are solvent exposed and not specifically recognized by the helicase. The structures show that the DNA substrate undergoes a series of transitions within its backbone to accompany the structural transitions occurring in the helicase, suggesting that nucleic acid analogs with adjusted backbone flexibility may efficiently compete for the substrate-binding groove. The advantage of targeting the (d)NTPase active site and/or the substrate-binding groove is that the virus should have difficulty generating resistance through simple point mutations. In addition, small molecules binding to the sites adjacent to motif V or the spring helix can be envisioned to interfere with the transitions of these essential elements and lock the helicase in a certain conformation. Potent inhibitors derived from these strategies could lead to new therapeutic options for chronically infected individuals, and may be utilized to study the functional details of many SF2 helicases.
Experimental Procedures
The HCV NS3 helicase domain (NS3h, Con1 genotype 1b) was expressed in E. coli with N-terminal His6-SUMO fusion. The protein was sequentially purified through Ni-Sepharose, ion exchange, and gel-filtration columns (GE Healthcare). Crystals were obtained with the hanging drop technique. The crystals were screened using Rigaku/MSC MicroMax-007HF in the Rockefeller University Structural Biology Resource Center made possible by Grant 1S10RR022321-01 from the National Center for Research Resources of the National Institutes of Health. Datasets were collected with a synchrotron x-ray source at the National Synchrotron Light Source (Brookhaven National Laboratory). Structures were solved with molecular replacement followed by iterative structural refinement and model building (SI Text).
NS3h in the presence or absence of ATP mimics was added to 20 nM 5′ fluorescein-labeled poly (dT8). Fluorescence anisotropy was measured and plotted as a function of NS3h concentration. The data were fit to a quadratic equation to determine the apparent dissociation constants (SI Text).
Supplementary Material
Acknowledgments.
We thank the staff of the Rockefeller Structural Biology Resource Center and the X29 beamline (National Synchrotron Light Source). We are grateful to Seth Darst, David N. Frick, Agnidipta Ghosh, and Christopher D. Lima for critical input, and to Catherine Murray for editing. M.G. was supported in part by a Marie-Josée and Henry R. Kravis Fellowship at The Rockefeller University. Additional financial support came from NIH Grant CA057973, and generous gifts from the Greenberg Medical Research Institute, the Starr Foundation, the Richard Salomon Family Foundation, the Ronald A. Shellow MD Memorial Fund, Paul Nash and the MGM Mirage Voice Foundation, and Gregory F. Lloyd Memorial contributions.
Footnotes
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected in 2005.
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913380107/DCSupplemental.
References
- 1.De Francesco R, Migliaccio G. Challenges and successes in developing new therapies for hepatitis C. Nature. 2005;436:953–960. doi: 10.1038/nature04080. [DOI] [PubMed] [Google Scholar]
- 2.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 3.Frick DN. The hepatitis C virus NS3 protein: A model RNA helicase and potential drug target. Curr Issues Mol Biol. 2007;9:1–20. [PMC free article] [PubMed] [Google Scholar]
- 4.Suzich JA, et al. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J Virol. 1993;67:6152–6158. doi: 10.1128/jvi.67.10.6152-6158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim DW, Gwack Y, Han JH, Choe J. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem Biophys Res Commun. 1995;215:160–166. doi: 10.1006/bbrc.1995.2447. [DOI] [PubMed] [Google Scholar]
- 6.Tai CL, Chi WK, Chen DS, Hwang LH. The helicase activity associated with hepatitis C virus nonstructural protein 3 (NS3) J Virol. 1996;70:8477–8484. doi: 10.1128/jvi.70.12.8477-8484.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim JL, et al. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: The crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 8.Mackintosh SG, et al. Structural and biological identification of residues on the surface of NS3 helicase required for optimal replication of the hepatitis C virus. J Biol Chem. 2006;281:3528–3535. doi: 10.1074/jbc.M512100200. [DOI] [PubMed] [Google Scholar]
- 9.Dumont S, et al. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porter DJ, et al. Product release is the major contributor to kcat for the hepatitis C virus helicase-catalyzed strand separation of short duplex DNA. J Biol Chem. 1998;273:18906–18914. doi: 10.1074/jbc.273.30.18906. [DOI] [PubMed] [Google Scholar]
- 12.Serebrov V, Pyle AM. Periodic cycles of RNA unwinding and pausing by hepatitis C virus NS3 helicase. Nature. 2004;430:476–480. doi: 10.1038/nature02704. [DOI] [PubMed] [Google Scholar]
- 13.Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. A conserved NTP-motif in putative helicases. Nature. 1988;333:22. doi: 10.1038/333022a0. [DOI] [PubMed] [Google Scholar]
- 14.Hodgman TC. A new superfamily of replicative proteins. Nature. 1988;333:22–23. doi: 10.1038/333022b0. [DOI] [PubMed] [Google Scholar]
- 15.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 16.Saikrishnan K, Powell B, Cook NJ, Webb MR, Wigley DB. Mechanistic basis of 5′-3′ translocation in SF1B helicases. Cell. 2009;137:849–859. doi: 10.1016/j.cell.2009.03.036. [DOI] [PubMed] [Google Scholar]
- 17.Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 19.Kwong AD, Rao BG, Jeang KT. Viral and cellular RNA helicases as antiviral targets. Nat Rev Drug Discovery. 2005;4:845–853. doi: 10.1038/nrd1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levin MK, Gurjar MM, Patel SS. ATP binding modulates the nucleic acid affinity of hepatitis C virus helicase. J Biol Chem. 2003;278:23311–23316. doi: 10.1074/jbc.M301283200. [DOI] [PubMed] [Google Scholar]
- 21.Ye J, Osborne AR, Groll M, Rapoport TA. RecA-like motor ATPases—lessons from structures. Biochim Biophys Acta. 2004;1659:1–18. doi: 10.1016/j.bbabio.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Duong FH, et al. Upregulation of protein phosphatase 2Ac by hepatitis C virus modulates NS3 helicase activity through inhibition of protein arginine methyltransferase 1. J Virol. 2005;79:15342–15350. doi: 10.1128/JVI.79.24.15342-15350.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heilek GM, Peterson MG. A point mutation abolishes the helicase but not the nucleoside triphosphatase activity of hepatitis C virus NS3 protein. J Virol. 1997;71:6264–6266. doi: 10.1128/jvi.71.8.6264-6266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim DW, Kim J, Gwack Y, Han JH, Choe J. Mutational analysis of the hepatitis C virus RNA helicase. J Virol. 1997;71:9400–9409. doi: 10.1128/jvi.71.12.9400-9409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tai CL, et al. Structure-based mutational analysis of the hepatitis C virus NS3 helicase. J Virol. 2001;75:8289–8297. doi: 10.1128/JVI.75.17.8289-8297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheffzek K, et al. The Ras-RasGAP complex: Structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 27.Wardell AD, Errington W, Ciaramella G, Merson J, McGarvey MJ. Characterization and mutational analysis of the helicase and NTPase activities of hepatitis C virus full-length NS3 protein. J Gen Virol. 1999;80:701–709. doi: 10.1099/0022-1317-80-3-701. [DOI] [PubMed] [Google Scholar]
- 28.Yao N, Reichert P, Taremi SS, Prosise WW, Weber PC. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Structure. 1999;7:1353–1363. doi: 10.1016/s0969-2126(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 29.Beran RK, Serebrov V, Pyle AM. The serine protease domain of hepatitis C viral NS3 activates RNA helicase activity by promoting the binding of RNA substrate. J Biol Chem. 2007;282:34913–34920. doi: 10.1074/jbc.M707165200. [DOI] [PubMed] [Google Scholar]
- 30.Brass V, et al. Structural determinants for membrane association and dynamic organization of the hepatitis C virus NS3-4A complex. Proc Natl Acad Sci USA. 2008;105:14545–14550. doi: 10.1073/pnas.0807298105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lam AM, Keeney D, Frick DN. Two novel conserved motifs in the hepatitis C virus NS3 protein critical for helicase action. J Biol Chem. 2003;278:44514–44524. doi: 10.1074/jbc.M306444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frick DN, Rypma RS, Lam AM, Gu B. The nonstructural protein 3 protease/helicase requires an intact protease domain to unwind duplex RNA efficiently. J Biol Chem. 2004;279:1269–1280. doi: 10.1074/jbc.M310630200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo D, et al. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 2008;27:3209–3219. doi: 10.1038/emboj.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 35.Andersen CB, et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 36.Collins R, et al. The DEXD/H-box RNA Helicase DDX19 Is Regulated by an α-Helical Switch. J Biol Chem. 2009;284:10296–10300. doi: 10.1074/jbc.C900018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morris PD. Hepatitis C virus NS3 and simian virus 40 T antigen helicases displace streptavidin from 5′-biotinylated oligonucleotides but not from 3′-biotinylated oligonucleotides: Evidence for directional bias in translocation on single-stranded DNA. Biochemistry. 2002;41:2372–2378. doi: 10.1021/bi012058b. [DOI] [PubMed] [Google Scholar]
- 38.Ma Y, Liang Y, Lemon SM, Yi M. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J Virol. 2008;82:7624–7639. doi: 10.1128/JVI.00724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.