Abstract
Stem cells that adopt distinct lineages cannot be distinguished based on traditional cell shape. This study reports that higher-order variations in cell shape and cytoskeletal organization that occur within hours of stimulation forecast the lineage commitment fates of human mesenchymal stem cells (hMSCs). The unique approach captures numerous early (24 h), quantitative features of actin fluororeporter shapes, intensities, textures, and spatial distributions (collectively termed morphometric descriptors). The large number of descriptors are reduced into “combinations” through which distinct subpopulations of cells featuring unique combinations are identified. We demonstrate that hMSCs cultured on fibronectin-treated glass substrates under environments permissive to bone lineage induction could be readily discerned within the first 24 h from those cultured in basal- or fat-inductive conditions by such cytoskeletal feature groupings. We extend the utility of this approach to forecast osteogenic stem cell lineage fates across a series of synthetic polymeric materials of diverse physicochemical properties. Within the first 24 h following stem cell seeding, we could successfully “profile” the substrate responsiveness prospectively in terms of the degree of bone versus nonbone predisposition. The morphometric methodology also provided insights into how substrates may modulate the pace of osteogenic lineage specification. Cells on glass substrates deficient in fibronectin showed a similar divergence of lineage fates, but delayed beyond 48 h. In summary, this high-content imaging and single cell modeling approach offers a framework to elucidate and manipulate determinants of stem cell behaviors, as well as to screen stem cell lineage modulating materials and environments.
Keywords: biomaterials, differentiation, imaging and modeling, stem cells, actin organization
Stem cells exhibiting pluripotency may be extracted from human embryonic tissues (1), obtained from reprogrammed somatic cells (2) or isolated from niches within adult tissues including the bone marrow. Human mesenchymal stem cells (hMSCs) isolated from adult bone marrow can be steered to differentiate into a multitude of terminal cell types relevant to tissue engineering (3) by manipulating surface chemistry (4), material mechanics (5), and/or applied growth factor mixtures (3).
At present, the guided differentiation of stem cells remains difficult because the mechanisms that govern the passage from uncommitted stem cells toward differentiated cells have yet to be completely identified. Hindering this identification further is the reliance on traditional endpoint assays to assess lineage, which often require weeks to months of culture and provide limited early assessment of cell behavior. Methods such as gene microarrays allow for the classification of stem cell maturation into distinct stages (6), however such methods treat heterogeneous groupings as single populations and are unable to follow the kinetics of mechanisms underlying lineage commitment. These issues slow the pace of screening and discovery of promising material and soluble factor treatments for stem cell tissue regeneration.
During the process of lineage commitment, cells undergo a number of morphological changes relating to differences in the spatial distribution and expression of key cytoskeletal proteins (7, 8). While differentiation is often accompanied by changes in cell shape, studies have demonstrated that cell shape in turn regulates cellular differentiation (9–12). Cytoskeletal components, particularly actin and its downstream effectors, are strong mediators of hMSC differentiation toward the osteoblastic lineage, with early cytoskeletal organization having a large effect on longer-term functions (13, 14). This study presents a “high-content” imaging methodology where quantitative features of actin fluororeporter shapes, intensities, textures, and spatial distributions (morphometric descriptors) are defined and utilized to assess cell behavior. Of central interest to the present study is whether the spatial and structural organization of the cytoskeleton in general, and actin in particular, encode the predilection of cultured stem cells to differentiate toward particular cell fates. If so, high-content analyses of stem cell lineage commitment could provide predictive measures of long-term cell behaviors at early time points for the effective screening of biomaterials; real time measures of stem cell fates and identification of when lineage commitment is determined; cell-by-cell based analysis that is able to capture the heterogeneity of sample populations; and the ability to parse out lineage commitment in stem cells influenced by multiple stimuli.
Results
hMSC Adopt Distinct Differentiation Outcomes That Are Not Apparent from Early Cell Shape/Morphology.
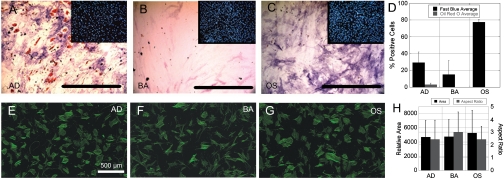
Human bone-marrow-derived MSCs demonstrate differentiation behaviors that are dependent upon both cell seeding density and growth factors. Traditionally, adipogenic differentiation of hMSCs is observed at high density (25,000 cells per cm2) in media supplemented with dexamethasone, indomethacin, insulin and isobutylmethylxanthine; while osteogenic differentiation of hMSC is induced at low density (1,000–3,000 cells per cm2) with supplementation of dexamethasone, ascorbate and β-glyerophosphate (3). By culturing cells at 3,000 cells per cm2 on fibronectin-treated glass, we observed distinct differentiation fates after two weeks in adipogenic (AD) (Fig. 1A), basal (Fig. 1B), and osteogenic (OS) induction media (Fig. 1C). At 3,000 cells per cm2 on fibronectin-coated glass in AD media (Fig. 1A), which is well below the optimal seeding density for adipogenic differentiation, hMSCs displayed evidence of both osteoblastic and adipocytic differentiation at two weeks as assessed by Fast Blue and Oil Red O staining (FBOR) respectively (Fig. 1D). Under the same seeding density with OS media (Fig. 1D), hMSCs exhibited osteoblastic differentiation with greater than 75% of the cells displaying alkaline phosphatase activity and no detectable evidence of lipid accumulation. Finally, under basal control media (Fig. 1B), the vast majority of cells were identifiable neither as osteoblastic nor adipocytic cells. Thus, under these conditions, hMSCs demonstrate robust differentiation behaviors in response to altered soluble cues. Screening for early changes in the “overall” cell morphology (24 h) fails to capture these differences between conditions (Fig. 1E–H). The identification of which cells are committed toward the osteoblastic versus adipocytic lineages is impossible between conditions. In fact, comparison of traditional shape parameters such as total cell area and cell aspect ratio (Fig. 1H) reveals no apparent difference between AD- and OS-treated samples.
Fig. 1.
hMSCs display growth factor dependent differentiation behaviors at 2 weeks that are not apparent at 24 h. hMSCs were cultured on fibronectin-coated glass substrates at 3,000 cells per cm2 for 2 weeks in either (A) AD media, (B) BA or, (C) OS induction media. Differentiation behaviors for OS and AD phenotype were assessed by Fast Blue Staining (Blue stain) for positive alkaline phosphatase activity or Oil Red O staining (Red stain) for intracellular lipid accumulation respectively (FBOR). Inlays represent corresponding images of fluorescently labeled nuclei. (D) Quantitative analysis of FBOR staining demonstrates 75% fast blue positive cells in OS media and significantly (p < 0.05) less fast blue positive cells and almost no oil red O positive cells in AD and BA media. Error bars represent the standard deviation of N = 2 independent experiments. hMSCs were cultured for 18 h in (E) AD media (F) BA media, and (G) OS media. Cells were fixed and then stained for actin at 24 h of total culture time. (H) Traditional cell shape parameters (cell area and aspect ratio) demonstrated no significant differences between AD, BA, and OS-treated hMSCs at 24 h. Error bars represent the standard deviation for N = 350–437 cells.
High-Content Cytoskeletal Imaging and Computational Modeling Yield Morphologically Distinguishable Cell Features.
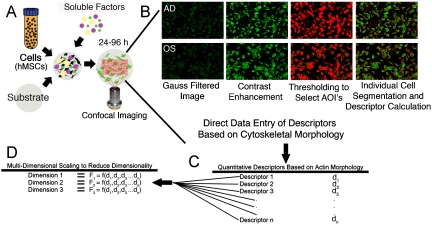
Our premise is that early cytoskeletal morphology of single cells enabled through high-content imaging may hold clues to longer-term hMSC behavior. Tile scanned confocal images of hMSCs cultured under adipogenic, osteogenic, and basal media conditions on various substrates were obtained (Fig. 2A). Images were subjected to a series of image processing steps including Gauss filtering, contrast enhancement, and fluorescence-based thresholding to yield individual cellular objects that were further isolated through segmentation (Fig. 2B). Analysis of each object yielded 43 cytoskeletal shape-, intensity-, and texture-based descriptors of whole cell and individual filament morphology for each cell (Fig. 2C; SI Text, Fig. S1). Using multidimensional scaling (MDS), key combinations of these 43 descriptors were reduced into 3 dimensions (Fig. 2D; SI Text, Fig. S2).
Fig. 2.
The development of descriptor-based computational modeling approaches to parse hMSCs at 24 h in culture: (A) A quantitative approach was undertaken to parse cell fates. Tile scan images for single experiments were obtained and underwent image processing (B) to yield a list of 43 morphometric descriptors that described whole cell shape and cytoskeletal distribution. (B) Individual cell images were processed utilizing a ten pass Gaussian filter and then image enhanced (image contrast, brightness and gamma value adjustment), and a thresholding process employed to segment individual whole cells based on the image intensity histogram. Shape and higher-order moment descriptors for segmented cell elements were then calculated as shown in (C). (D) Utilizing MDS, the 43 dimensional descriptor space (denoted by d1,d2,…dn) was combined in a nonlinear fashion to create a new 3-dimensional space to visualize the data. The new dimensions (referred in Fig. 3 as D1–D3) represent components of all of the original 43 descriptors combined into 3 different nonlinear combinations. In the MDS, each point represents the three-dimensional descriptor coordinates for a single cell cultured under the defined media condition.
MDS Analysis of Cytoskeletal Descriptors Discerns Bone Precursors Within 24 Hours.
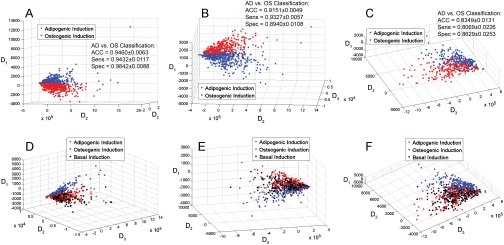
The high-content imaging followed by MDS was used to classify cell subpopulations grown in osteogenic (blue) versus adipogenic (red) conditions (Fig. 3A–C) or versus adipogenic (red) and basal (black) conditions (Fig. 3D–F). Cells grown in OS media locate primarily within a single cluster (OS blue cluster) (Fig. 3A–C). Predicting stem cell lineage outcomes is challenged by inherent variability in the stem cell populations, for example, due to differences in the donor source of cells (15), age of the donor (16), and passage number within a given donor source (17). To examine whether the high-content descriptor-based segmentation of cell populations visualized were donor, or passage dependent, experiments were performed with 2 different donors (age 22 and 36) and with two different passage numbers (Fig. 3D–F). Cells cultured in adipogenic (AD) and basal media (BA) segmented almost entirely into overlapping clusters (red AD/black BA cluster) (Fig. 3D–F), with limited overlapping with the OS cluster. When comparing across different cell sources (compare Fig. 3A–C), clear segmentation of osteogenic treated cell populations from nonosteogenic populations is observed in all cases. Additionally, in all instances, the OS-treated cells segmented from the AD/BA treated cells (Fig. 3E–F). Validation of the OS versus AD segmentation produced by the MDS for the different conditions was performed utilizing a support vector machine (SVM) to classify the different groupings. Statistics from the SVM indicate that the MDS identifies and distinguishes OS-treated cells with high accuracy, specificity, and sensitivity from AD treated cells.
Fig. 3.
Cytoskeleton-based descriptors segment and identify individual osteoblastic cells within hMSCs cultures exposed to soluble factors. MDS, utilizing the morphometric descriptors to create three new dimensions (D1–D3) that each represent nonlinear combinations of the 43 descriptors, demonstrates that individual cells exposed to OS media (Blue) cluster separately from cells in either AD (Red) (A–C) or BA (Black) (D–F) media. All cells were cultured on fibronectin/glass at 3,000 cells per cm2. This segmentation was observed in at least three different cultures consisting of two donors (Donor 1 was a 36-year-old-old male (A, D, B, E); Donor 2 was a 22-year-old female (C, F) and two passages (2 (A, C, D, F) and 4 (B, E)). No segmentation was observed between the AD and BA media conditions (D–F). For (A–C) data is pooled from four different wells for each condition, in (D–F) data are pooled from two wells for each condition. (A–C) Support vector machine (SVM) classification of clustering shows that OS vs. AD classification can be achieved with high accuracy (ACC), sensitivity (Sens), and specificity (Spec). Error reported on SVM classification represents the standard deviation for ACC, Sens, and Spec for N = 50 twofold cross validation pseudoexperiments of MDS data plotted in (A–C).
Forecasting hMSC Osteoblastic Lineage Outcomes via 24 Hour Morphometric Descriptors.
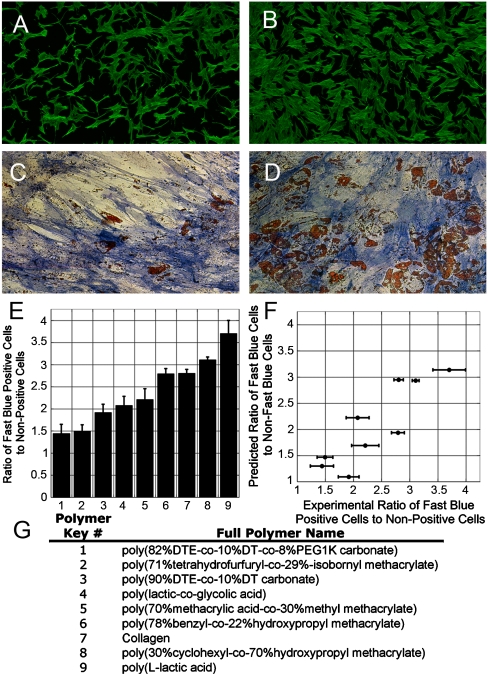
We next examined whether the morphometric approach could be applied to predict osteoblastic differentiation of hMSCs on complex engineered synthetic substrates that promote variable degrees of osteoblastic (∼55–75%) and adipogenic (∼15–47%) differentiation (Fig. 4A–B). Nine polymers with diverse physicochemical properties (Fig. S4) were used in the test case based on their ability to support in excess of 50% of the cells toward the osteoblastic lineage at 2 weeks. High-seeding density (21,000 cells/cm2) and mixed induction media were employed to rank-order and parse out materials that potentiate osteogenic differentiation. Examination of the 24 h high-content images of hMSCs demonstrated no apparent qualitative features (e.g., cell area, aspect ratio) that could differentiate cell response elicited by the materials (Fig. 4C–D). Utilizing the previous outlined MDS approach (Fig. 2) the MDS results for individual polymer samples showed two distinct cell fates within a given sample (Fig. S5). Calculating the ratio of cells that cluster in the osteoblastic regime (identified by the larger of the two clusters) allowed for the prediction of the ratio of cells that will become osteoblasts versus those that will not (Fig. 4F). This prediction was compared with the experimentally observed FBOR results obtained at 2 weeks for the same polymers (Fig. 4E). Polymer chemistry appeared to have a large effect on hMSC differentiation with the ratio of cells that are fast blue positive to those that are not ranging from approximately 1.50 to almost 3.75 for the tested materials (Fig. 4E). The 24 h morphometric-based prediction was compared with the experimentally captured FBOR staining. A high correlation (Pearson correlation coefficient = 0.87) results between the MDS-based 24 h predictions and the 2 week experimental values (Fig. 4F).
Fig. 4.
Altered hMSC osteoblastic commitment resulting from substrate may be predicted based on MDS at 24 h. (A–B) At constant seeding density (21,000 cells per cm2) and applied soluble cues (MX media) synthetic substrates alter the percentage of cells that differentiate toward the osteoblastic lineage at 2 weeks as assessed by FBOR staining. FBOR images are shown for polymer 8 (A) and 1 (B). (C–D) While the FBOR staining in (A–B) shows differences in the ratio of osteoblastic versus nonosteoblastic cells on different substrates, no obvious qualitative morphologic parameters of cytoskeletal morphology are seen that can distinguish these outcomes in the 24 h tile scan actin-stained images shown for the same polymers (example shown for polymers 8 (C) and 1 (D)). (E) Quantification of the ratio of fast blue positive cells to nonfast blue positive cells for each substrate shows ratios ranging from approximately 1.5 to 3.75. Error bars represent the standard deviation of N = 2–4 experiments per substrate. (F) Scatter plot with the X-axis representing the ratio of 2 week osteoblastic cells to nonosteoblastic cells, versus the 24 h morphometric/MDS prediction for the same ratio (Y-axis). A high degree of correlation between the prediction and observed percent of osteoblasts was seen (Pearson correlation coefficient = 0.87). The X error bars represent the standard deviation for N = 2–4 independent experiments. (G) Table listing full polymer names with corresponding keyed system shown in (F).
Cytoskeletal Descriptors Embody Substrate-Governed Time Evolution of Lineage Commitment.
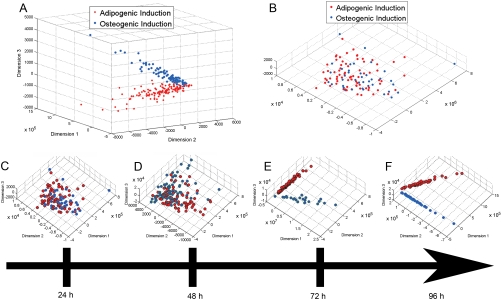
Unlike their fibronectin-coated counterparts, hMSCs cultured on glass substrates without fibronectin pretreatment showed no definable clusters for either the OS or AD treated cells at 24 h (Fig. 5A–B). Time sequence analysis (Fig. 5C–F) of the MDS for cells cultured in OS or AD conditions indicate that on untreated glass, these characteristic morphometric features developed between 48 and 72 h. With increasing time (96 h; Fig. 5F), the segmentation was more pronounced, indicating that within the time frame examined, the cytoskeletal based morphometrics of these two cell populations become progressively divergent.
Fig. 5.
Cytoskeletal based morphometric segmentation kinetics occur in a ligand-dependent manner. hMSCs were cultured in either AD or OS media on glass substrates with (A) and without (B) fibronectin pretreatment for 24 h and the time course of descriptors was obtained. At 24 h the fibronectin-pretreated substrates demonstrate a clear segmentation of the hMSCs treated with AD or OS induction whereas glass alone conditions do not. On nontreated glass, the cytoskeletal based segmentation process becomes progressively more pronounced with culture time (C–F). The MDS of cells taken at 24 h (C), 48 h (D), 72 h (E), and 96 h (F) are shown. Cells on untreated glass showed no segmentation until 72 h of culture.
Discussion
High-content cell morphology-based screening approaches have been developed for the screening of potential pharmaceuticals and biologically active small molecules, but these strategies have yet to become widespread for stem cell and tissue engineering applications. We demonstrate here that classifications of individual cell fates accurately predict long-term cellular behaviors days to weeks before they are experimentally detected. Previous work links global cell shape parameters and MSC differentiation behavior (13). However, this study finds that first moment shape descriptors alone and gene transcription events (Fig. S6) fail to capture the early heterogeneity and segregation of hMSCs. To capture these hallmark cytoskeletal “fingerprints” of osteoblastic committed precursors, we propose the use of a high-content methodology.
The cytoskeletal descriptors, which underlie the classification proposed in our study, reinforce the notion that the actin cytoskeleton plays a dominant role in regulating hMSC differentiation via cell shape, Rho, Rho-associated, coiled-coil containing protein kinase (ROCK), and cytoskeletal tension mechanisms, specifically promoting osteoblastic induction (13, 14, 18). The total activity of ROCK may be correlated with the actin fiber thickness and density (19). Additionally, transient expression of a glycerophosphodiester phosphodiesterase localized to actin triggers morphologic changes and increases in alkaline phosphatase activity (20). Thus, it follows that the cell osteogenic lineage decision-making process may be encoded within the cytoskeleton itself.
The methodology of morphometric profiling of cell behaviors could be refined in the future to address discrimination of different lineage types (beyond osteogenic vs. nonosteogenic, for example), and also temporal changes in differentiation within dynamic conditions. Currently, our predictions appear valid when the induction stimuli (i.e., substrata or growth factors) remain unchanged for the entire period of culture, but could be examined when hMSCs transdifferentiate into other lineages (21). The questions to address in the future are which cytoskeletal descriptor combinations, if any, are unique to specific lineages, and whether such descriptors can help parse multiple lineages. Our MDS-based segmentation correctly parses cells cultured under osteogenic conditions, but given its current one-reporter (actin)-based parsing process, it is unable to parse adipogenic cells from those under basal conditions (Fig. 3D–F). This is possibly due to lack of robust adipogenic differentiation under the sparse seeding conditions we employed or because our imaging did not extend to the formative time course of cytoskeletal rearrangement indicative of adipocytic lineage commitment. Thus, the actin-based profiling could not be extended to parse cells near confluence in adipogenic versus basal media, but newer promising methods could be developed for near confluent conditions based on high-content imaging of the nuclear morphology (22).
Another limitation of the current MDS approach is the inability to identify individual descriptors or groups of descriptors that represent lineage commitment. While the MDS approach alone does not allow such identification, complementary computational modeling methods may be used to accomplish this task. For example, decision tree analysis (23) can be used in conjunction with high-content imaging to highlight the most meaningful descriptors (Fig. S7). Utilizing this multimodal computational approach we could not only identify lineage-committed cells at early time points, but also extract features of the cytoskeleton that may be correlated to specific cellular processes.
We report that extracellular matrix preconditioning by fibronectin had a marked effect on accelerating the kinetics of hMSC lineage commitment. In addition, as small changes in substrate chemistry can significantly affect matrix ligand conditioning (24, 25), we examined whether our high-content stem cell profiling approach could help discriminate hMSC commitment to distinct lineages on nine different polymeric substrates. The physicochemical properties of such substrates themselves play a critical role in steering the percentage of cells that commit to specific fates (Fig. 4E). For the nine polymers assessed in this study, a high degree of correlation between the 24 h predictions and the experimentally observed 2 week FBOR was observed (0.87). Despite the seeming complexity of the large number of varied polymer surface properties that could affect stem cell differentiation, we believe cells integrate such signals into early intracellular morphological cues within 24 h which predict the behavior out at two weeks. Thus lineage fates may be driven by rapid deterministic differentiation events early on and not necessarily by different growth kinetic behaviors (13).
In summary, we have shown that through high-content imaging, it is possible to identify the differentiation behavior of stem cells early within culture periods. This methodology applies to the differential behavior of these cells resulting from altered soluble factors, ligand conditioning, or synthetic/biomimetic substrates. These findings are significant because they first show that surface chemistry plays a critical role in the kinetics of cellular differentiation. Additionally, the ability to identify cellular fates early on may allow us to establish a time line for cellular differentiation and identify critical points in cellular evolution for further assaying. The ability to obtain reasonably accurate predictions of cellular functions early opens up unique approaches for the screening of different polymers for directed stem cell differentiation; separating heterogeneous populations of cells; and increasing the fidelity of cellular differentiation assays. Finally, the MDS-based stem cell identification could be utilized as an approach to rapidly screen and identify lead biomaterials for use in regenerative medicine.
Materials and Methods
Cell Culture.
hMSCs were obtained from commercial sources (Lonza). Cells were cultured in a humidity-controlled environment under 5% CO2 and 37 °C and fed every 3–4 d with growth media (basal media) supplemented with commercial SingleQuot’s© (Catalog number PT-3001, Lonza). Cells were received at passage 1 and used for up to 4 passages. Cells were subcultured upon reaching 90% confluence. OS, AD induction media were reconstituted as per manufacturer (Lonza). Adipogenic media were cycled with 3 d induction followed by 1 d maintenance. Mixed media (MX) conditions consisted of 50∶50 osteogenic:adipogenic induction/maintenance media.
hMSC Differentiation on Fibronectin-Coated Glass Substrates.
Fibronectin (BD Biosciences) coated glass substrates were created via the application of 400 μL of 25 μg/mL fibronectin/PBS solution to each well of a glass bottom 24-well plate for 1 h at room temperature. hMSCs (Lot number 7F3458 (36 yr-old male) and Lot number 7F3675 (22 yr-old female) were applied at 3,000 cells per cm2 and allowed to attach in basal media for 6 h. After the 6 h, the basal media was replaced with appropriate induction media. For differentiation assays, cells were cultured for 2 weeks and then stained for alkaline phosphatase activity and lipid production with Fast Blue RR/napthol (Kit number 85, Sigma-Aldrich) and 30 mg/mL Oil Red O (Sigma), respectively, as described previously (13, 17). For total cell counts, cell nuclei were stained with 1 μg/mL DAPI (Sigma).
High-Content Imaging of hMSC on Glass and Fibronectin-Coated Glass.
hMSCs (Lot number: 7F3458 (Donor 1) and 7F3675 (Donor 2), (Lonza) were applied at 3,000 cells per cm2 for 24 h morphology and 1,000 cells per cm2 for time course studies and allowed to adhere to treated and untreated glass surfaces for 6 h in basal media. After a 6-h attachment period, basal medium was replaced with appropriate induction media. After 18–90 (as indicated) additional hours cells were fixed and stained with AlexaFluor 488 Phalloidin (Invitrogen). Cells were imaged under a 63X objective (NA = 1.3) with a Leica TCS SP2 system (Leica Microsystems Inc.) and excited at 488 nm with a 500–700 nm detection band-pass filter. All images represent a mosaic of 60 individual images (10 × 6 grid). Raw images of subconfluent cells were exported to Image Pro Plus Version 5.1 (Media Cybernetics) and equalized for image intensity histograms, segmented and processed to yield shape, intensity and higher-order moment parameters as output to a text file prior to modeling, a process referred to as high-content imaging (Fig. 2).
Computational Methods to Parse Cytoskeletal Descriptors.
Forty-three associated features (called descriptors) were archived for each cell. These features represent quantifiable measures of the whole cell and actin filament morphology and organization throughout a single cell. Features that were defined include; shape features, intensity-based features that relate to the degree of actin filament formation, and textural and cytoskeleton organizational features that describe the spatial distribution of actin filaments and their location within a given cell. A complete list of the 43 descriptors calculated and how they describe the actin cytoskeleton is shown in Fig. S1. MDS was then implemented via Matlab’s (Mathworks) statistical toolbox to regraph cellular descriptor coordinates based on a reduced three-dimensional representation where each point represents the combined features of individual cells (Fig. 2). MDS simply represents a dimensionality reduction technique that allows the 43 descriptors described in Fig. S1 for each cell to be plotted in 3D. The MDS algorithm aides in identifying similarities as well as dissimilarities in groupings of cells based on the 43 individual cell cytoskeletal descriptors (Fig. S2). These groupings are defined via a nonlinear combination of the descriptors, which effectively reduce the 43-dimensional complex representation down to three dimensions, such that the observed similarities and dissimilarities between cells are maximized. Ultimately, the individual cells are graphed in three-dimensions, with the proximity of points indicating their similarity. To ensure that reduced datasets were valid, a synthetic dataset was generated based on the variance of each of the individual classifiers (Fig. S3) and compared to MDS results for sample conditions. Classification of MDS-based cell coordinates was performed utilizing a support vector machine (26). Classification accuracy, sensitivity and specificity were performed with a randomized twofold cross validation run 50 times on individual datasets utilizing a radial distribution function.
Preparation of Spin-Coated Polymeric Substrates.
Tyrosine-derived polycarbonates (27), polymethacrylates (28, 29), poly(D,L-lactide-co-glycolide) (Resomer 506) and poly(L-lactic acid) (Resomer L-206) (Boehringer Ingelheim) were dissolved in a 1.5% (v/v) methanol in methylene chloride solution yielding a 1% (w/v) polymer solution. Polymer solutions were then spin-coated onto a 15 mm glass coverslip. Spin coating was conducted at 4,000 RPM for 30 s. Collagen coated coverslips were made by incubating a glass coverslip in a 7 μg/mL collagen solution at 37 °C overnight.
hMSC Morphometrics and Differentiation on Polymeric Substrates.
Polymer-coated glass coverslips were placed in the bottom of a 24-well glass bottom tissue culture plate and secured with a silicon O-ring (Catolog number 111, Molding Solution). The setup was sterilized with a UV light applied at 5,500 to 6,500μW per cm2 for 900 s. hMSCs (Lot number’s 6F4392 (18 yr-old male) and 7F3458) were applied at 21,000 cells per cm2 and allowed to attach in basal media for 6 h,, switched to induction media, processed and imaged as described above. 21,000 cells per cm2 was chosen so as to provide significant amounts of both osteogenic and adipogenic (maximum 47%) differentiation while keeping the cell cultures subconfluent to allow individual cell segmentation and descriptor analysis. For differentiation assays, cells were stained for alkaline phosphatase activity and lipid production as described previously. Only substrates that elicited greater than 50% alkaline phosphatase activity at 2 weeks were examined within this study.
Supplementary Material
Acknowledgments.
The efforts of Drs. Abraham Joy and Das Bolikal with synthesis of polymers, and of Shannon Agner and Dr. Anant Madabhushi with data processing, are gratefully acknowledged. This study was partially supported by National Institute of Health EB001046 (RESBIO, Integrated Resources for Polymeric Biomaterials), Rutgers University Academic Excellence Fund, National Science Foundation DGE IGERT 0333196 and DGE IGERT 0801620.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909597107/DCSupplemental.
References
- 1.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451(7175):141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 3.Pittenger MF, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284(5411):143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 4.Curran JM, Chen R, Hunt JA. The guidance of human mesenchymal stem cell differentiation in vitro by controlled modifications to the cell substrate. Biomaterials. 2006;27(27):4783–4793. doi: 10.1016/j.biomaterials.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 6.Kulterer B, et al. Gene expression profiling of human mesenchymal stem cells derived from bone marrow during expansion and osteoblast differentiation. BMC Genomics. 2007;8:70. doi: 10.1186/1471-2164-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pockwinse SM, Stein JL, Lian JB, Stein GS. Developmental stage-specific cellular responses to vitamin D and glucocorticoids during differentiation of the osteoblast phenotype: Interrelationship of morphology and gene expression by in situ hybridization. Exp Cell Res. 1995;216(1):244–260. doi: 10.1006/excr.1995.1031. [DOI] [PubMed] [Google Scholar]
- 8.Sikavitsas VI, Temenoff JS, Mikos AG. Biomaterials and bone mechanotransduction. Biomaterials. 2001;22(19):2581–2593. doi: 10.1016/s0142-9612(01)00002-3. [DOI] [PubMed] [Google Scholar]
- 9.Carvalho RS, Schaffer JL, Gerstenfeld LC. Osteoblasts induce osteopontin expression in response to attachment on fibronectin: Demonstration of a common role for integrin receptors in the signal transduction processes of cell attachment and mechanical stimulation. J Cell Biochem. 1998;70(3):376–390. doi: 10.1002/(sici)1097-4644(19980901)70:3<376::aid-jcb11>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez Fernandez JL, Ben-Ze’ev A. Regulation of fibronectin, integrin and cytoskeleton expression in differentiating adipocytes: Inhibition by extracellular matrix and polylysine. Differentiation. 1989;42(2):65–74. doi: 10.1111/j.1432-0436.1989.tb00608.x. [DOI] [PubMed] [Google Scholar]
- 11.Spiegelman BM, Ginty CA. Fibronectin modulation of cell shape and lipogenic gene expression in 3T3-adipocytes. Cell. 1983;35(3 Pt 2):657–666. doi: 10.1016/0092-8674(83)90098-3. [DOI] [PubMed] [Google Scholar]
- 12.Thomas CH, Collier JH, Sfeir CS, Healy KE. Engineering gene expression and protein synthesis by modulation of nuclear shape. Proc Natl Acad Sci USA. 2002;99(4):1972–1977. doi: 10.1073/pnas.032668799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6(4):483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez JP, Gonzalez M, Rios S, Cambiazo V. Cytoskeletal organization of human mesenchymal stem cells (MSC) changes during their osteogenic differentiation. J Cell Biochem. 2004;93(4):721–731. doi: 10.1002/jcb.20234. [DOI] [PubMed] [Google Scholar]
- 15.Sekiya I, et al. Expansion of human adult stem cells from bone marrow stroma: Conditions that maximize the yields of early progenitors and evaluate their quality. Stem Cells. 2002;20(6):530–541. doi: 10.1634/stemcells.20-6-530. [DOI] [PubMed] [Google Scholar]
- 16.Moerman EJ, Teng K, Lipschitz DA, Lecka-Czernik B. Aging activates adipogenic and suppresses osteogenic programs in mesenchymal marrow stroma/stem cells: The role of PPAR-gamma2 transcription factor and TGF-beta/BMP signaling pathways. Aging Cell. 2004;3(6):379–389. doi: 10.1111/j.1474-9728.2004.00127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruder SP, Jaiswal N, Haynesworth SE. Growth kinetics, self-renewal, and the osteogenic potential of purified human mesenchymal stem cells during extensive subcultivation and following cryopreservation. J Cell Biochem. 1997;64(2):278–294. doi: 10.1002/(sici)1097-4644(199702)64:2<278::aid-jcb11>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 18.Morrison SJ, White PM, Zock C, Anderson DJ. Prospective identification, isolation by flow cytometry, and in vivo self-renewal of multipotent mammalian neural crest stem cells. Cell. 1999;96(5):737–749. doi: 10.1016/s0092-8674(00)80583-8. [DOI] [PubMed] [Google Scholar]
- 19.Watanabe N, Kato T, Fujita A, Ishizaki T, Narumiya S. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat Cell Biol. 1999;1(3):136–143. doi: 10.1038/11056. [DOI] [PubMed] [Google Scholar]
- 20.Yanaka N, et al. Novel membrane protein containing glycerophosphodiester phosphodiesterase motif is transiently expressed during osteoblast differentiation. J Biol Chem. 2003;278(44):43595–43602. doi: 10.1074/jbc.M302867200. [DOI] [PubMed] [Google Scholar]
- 21.Baksh D, Song L, Tuan RS. Adult mesenchymal stem cells: Characterization, differentiation, and application in cell and gene therapy. J Cell Mol Med. 2004;8(3):301–316. doi: 10.1111/j.1582-4934.2004.tb00320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McBride SH, Knothe Tate ML. Modulation of stem cell shape and fate A: The role of density and seeding protocol on nucleus shape and gene expression. Tissue Eng PT A. 2008;14(9):1561–1572. doi: 10.1089/ten.tea.2008.0112. [DOI] [PubMed] [Google Scholar]
- 23.Murthy SK. Automatic Construction of Decision Trees from Data: A Multi-Disciplinary Survey. Data Min and Knowl Disc. 1998;2(4):345–389. [Google Scholar]
- 24.Bae YH, Johnson PA, Florek CA, Kohn J, Moghe PV. Minute changes in composition of polymer substrates produce amplified differences in cell adhesion and motility via optimal ligand conditioning. Acta Biomater. 2006;2(5):473–482. doi: 10.1016/j.actbio.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Tziampazis E, Kohn J, Moghe PV. PEG-variant biomaterials as selectively adhesive protein templates: Model surfaces for controlled cell adhesion and migration. Biomaterials. 2000;21(5):511–520. doi: 10.1016/s0142-9612(99)00212-4. [DOI] [PubMed] [Google Scholar]
- 26.Cortes C, Vapnik V. Support-vector networks. Mach Learning. 1995;20(3):273–297. [Google Scholar]
- 27.Bourke SL, Kohn J. Polymers derived from the amino acid L-tyrosine: polycarbonates, polyarylates and copolymers with poly(ethylene glycol) Adv Drug Deliver Rev. 2003;55(4):447–466. doi: 10.1016/s0169-409x(03)00038-3. [DOI] [PubMed] [Google Scholar]
- 28.Briggs T, et al. Osteogenic differentiation of human mesenchymal stem cells on poly(ethylene glycol)-variant biomaterials. J Biomed Mater Res A. 2008;91(4):975–984. doi: 10.1002/jbm.a.32310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kholodovych V, et al. Prediction of biological response for large combinatorial libraries of biodegradable polymers: Polymethacrylates as a test case. Polymer. 2008;49(10):2435–2439. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.