Abstract
Crossing over establishes connections between homologous chromosomes that promote their proper segregation at the first meiotic division. However, there exists a backup system to ensure the correct segregation of those chromosome pairs that fail to cross over. We have found that, in budding yeast, a mutation eliminating the synaptonemal complex protein, Zip1, increases the meiosis I nondisjunction rate of nonexchange chromosomes (NECs). The centromeres of NECs become tethered during meiotic prophase, and this tethering is disrupted by the zip1 mutation. Furthermore, the Zip1 protein often colocalizes to the centromeres of the tethered chromosomes, suggesting that Zip1 plays a direct role in holding NECs together. Zip3, a protein involved in the initiation of synaptonemal complex formation, is also important for NEC segregation. In the absence of Zip3, both the tethering of NECs and the localization of Zip1 to centromeres are impaired. A mutation in the MAD3 gene, which encodes a component of the spindle checkpoint, also increases the nondisjunction of NECs. Together, the zip1 and mad3 mutations have an additive effect, suggesting that these proteins act in parallel pathways to promote NEC segregation. We propose that Mad3 promotes the segregation of NECs that are not tethered by Zip1 at their centromeres.
Keywords: spindle checkpoint, Mad3, nondisjunction, Zip3, centromere
Meiotic crossing over leads to the formation of chromatin bridges between homologous chromosomes that persist until metaphase and ensure the proper alignment of chromosome pairs on the meiosis I spindle. Physical associations may also be important for the segregation of nonexchange chromosomes (NECs). NECs are associated with each other in the majority of pachytene nuclei (1, 2), when homologous chromosomes are held together along their lengths by the synaptonemal complex (SC). However, at least in budding yeast, NEC associations do not involve extensive SC formation (1), and they occur specifically at or near centromeres (2).
The Zip1 protein, a major SC building block in yeast, localizes specifically to centromeres early in meiotic prophase (3). Unlike Zip1 polymerization along the arms of chromosomes, centromeric localization of Zip1 is independent of recombination initiation. Furthermore, this early centromeric Zip1 holds chromosomes together in groups of two (3). In wild type, most centromere couples initially involve nonhomologous chromosomes, but eventually all centromeres become homologously coupled (3). Given its ability to couple the centromeres of nonhomologous chromosomes, it was postulated that Zip1 might play a role in the segregation of NECs (3). Recently, we found that the Zip1 protein improves chromosome segregation in msh4 and msh5 mutants, in which homologous chromosomes frequently fail to cross over (4).
Here, we describe a previously unreported role for Zip1 in the segregation of NECs at meiosis I. Zip1 promotes tethering of NECs at their centromeres throughout meiotic prophase, and this tethering correlates with improved segregation at the first meiotic division. We have also uncovered a role for the synapsis initiation protein, Zip3, in this process. In the absence of Zip3, Zip1 localization to the centromeres of NECs, and therefore tethering, are reduced. Finally, we have uncovered a parallel pathway involving the spindle checkpoint protein, Mad3, in the segregation of NECs. Our data suggest that this second pathway aids the segregation of NECs not tethered by Zip1.
Results and Discussion
Zip1 Mutant Displays Increased Nondisjunction of NECs.
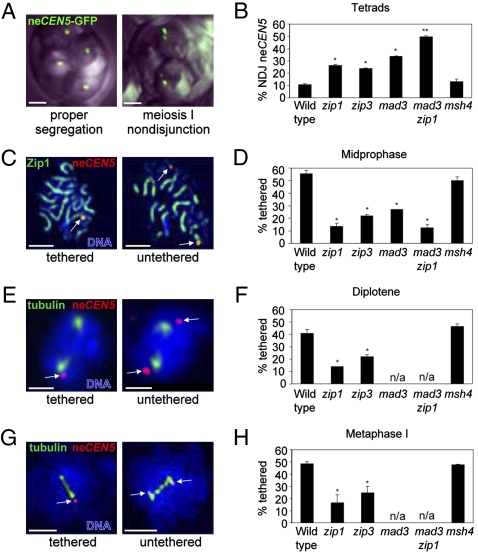
To determine whether Zip1 is involved in NEC segregation, we examined the segregation of a number of different achiasmate chromosome pairs. The first assay used a Saccharomyces cerevisiae diploid in which one copy of chromosome V is derived from Saccharomyces carlsbergensis. The homeologous chromosomes V are 20 to 30% divergent in sequence and fail to recombine in 99% of meioses (5–7). Both homeologs carry LacO operator sequences near their centromeres; these are recognized by the LacI protein fused to GFP (2). Introduction of the LacO repeats does not influence the segregation of these homeologous chromosomes (2, 7). Normal segregation yields a single GFP focus in each of the four haploid spores in a tetrad (Fig. 1A, Left). Meiosis I nondisjunction results in a tetrad in which only two spores contain GFP, and each of these contains two foci (Fig. 1A, Right). In wild type, the NEC pair shows 11% nondisjunction; this frequency is elevated to 27% in the zip1 mutant (Fig. 1B).
Fig. 1.
Centromere tethering and NEC segregation are perturbed in zip1, zip3, and mad3 mutants. (A) Nondisjunction (NDJ) of chromosome V at meiosis I was analyzed using the LacO/LacI-GFP system in tetrads containing four GFP foci. (B) The frequencies of NEC nondisjunction are shown. In both wild type and zip1, <5% of tetrads display precocious separation of sister chromatids or meiosis II nondisjunction. (C–H) Centromere tethering was assessed for different stages of meiosis I by staining surface-spread meiotic nuclei with antibodies against tubulin, GFP (neCEN5), and Zip1. (C) Midprophase spreads from wild type and mutants were identified by screening DAPI-stained chromatin for condensed, worm-like chromosomes. [In wild type, 96% of such nuclei exhibit linear Zip1 staining, indicative of the pachytene stage (n = 101).] (E) Diplotene spreads have a single nucleus with two separate tubulin foci. (G) Metaphase I spreads have short spindles (∼2 μm). Arrows indicate the positions of the neCEN5 centromeres. (Scale bars, 2 μm.) (D, F, and H) Shown are the frequencies of tethered neCEN5s for the different stages of meiosis. The asterisks denote P-values < 0.017 compared to wild type. Strains: Y712, Y787, Y790, Y1010, Y784, Y1155 (see Table S1).
We also monitored the segregation of two unrelated chromosomes, each lacking their homolog. In a diploid strain carrying only one copy of chromosome I and one copy of chromosome III, these chromosomes behave as a pair, usually segregating away from each other at meiosis I (8). The nondisjunction frequency is elevated from 13% in wild type to 23% in the zip1 mutant (Fig. S1 and Table S1). Thus, the Zip1 protein improves NEC segregation of both homeologous and heterologous NEC pairs.
The zip1 mutant shows a reduced level of crossing over (9, 10), which leads to an increased number of chromosomes that fail to cross over (11). These homologous NECs could interfere with the segregation of the obligate NECs whose disjunction is being measured (2). To address this possibility, we assessed NEC segregation in the msh4 mutant, which displays a decrease in crossing over of similar magnitude to zip1 (10, 12, 13). The nondisjunction frequency of the homeologous chromosome V pair in the msh4 strain (11%) (see Fig. 1B) is similar to that observed in wild type, arguing that the effect of zip1 on NEC segregation cannot be attributed to its defect in crossing over.
Our results may seem at odds with previous findings concluding that NEC segregation is not affected by the zip1 mutation (2). These differences are likely because of the nature of the assessment; we measured NEC segregation cytologically, whereas Dawson and colleagues (2) monitored NEC segregation genetically by tetrad analysis. Whereas cytological observations allow direct estimates of missegregation to be obtained, genetic estimates rely upon overall spore viability, which is greatly reduced in the zip1 mutant. Indeed, Dawson and colleagues (14) now also report that Zip1 does play a role in NEC segregation, based on the results of a cytological assay.
Zip1 Promotes Centromeric Associations of NECs.
If Zip1 facilitates segregation by holding NECs together at their centromeres (thus defining them as a “pair”), then the association of NEC centromeres observed at pachytene (2) should be dependent on the Zip1 protein. We examined spread meiotic nuclei to determine whether the centromeres of the homeologous chromosomes V are associated at pachytene (Fig. 1C). We found a single focus of GFP staining in 56% of the nuclei examined, indicating that the tagged centromeres are often associated. This frequency of association is reduced approximately fivefold by the zip1 mutation, but not significantly affected by the msh4 mutation (Fig. 1D and Fig. S2). These results indicate that Zip1 plays a role in holding together the centromeres of NECs during pachytene, when all other chromosomes are homologously paired and fully synapsed. Note, however, that Zip1 is not absolutely required for tethering. A small fraction of NECs (14%) are tethered even in the absence of Zip1 (see Fig. 1D).
Zip1 Localizes to the Centromeres of NECs.
Does Zip1 play a direct role in centromere tethering? If so, then Zip1 should localize to the centromeres of the NECs. Indeed, when Zip1 and the GFP-tagged centromeres of the homeologous chromosomes V were visualized in wild-type cells at midprophase, 90% of tethered centromeres had Zip1 associated (Fig. 2 A and B). When the two centromere signals were untethered, Zip1 colocalized with both GFP signals in only half of the cells (Fig. 2E). In addition, both the intensity of Zip1 staining at centromeres and the degree of overlap between the Zip1 signal and the GFP signal were reduced in cells in which centromeres were untethered (Figs. S3–S5). Thus, Zip1 localization to the centromeres of the nonexchange homeologous chromosomes V (neCEN5s) correlates with centromere tethering.
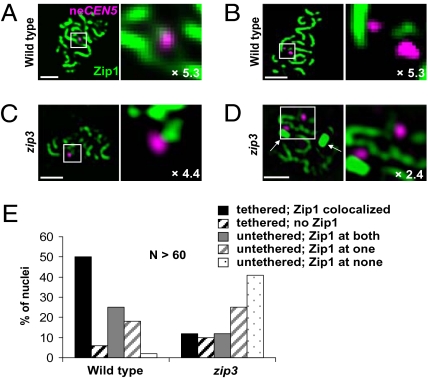
Fig. 2.
Localization of Zip1 to neCEN5s at midprophase. (A–D) Meiotic nuclear spreads were stained for neCEN5 (magenta) and Zip1 (green) at midprophase stages. The boxed areas shown in the left panel are magnified in the right panel; the extent of magnification is indicated. Arrows indicate polycomplexes, which are aggregates of SC proteins unassociated with chromatin. (A and B) Examples of wild-type (strain Y712) (see Table S1) nuclei with tethered neCEN5s and Zip1 colocalized (A) or untethered neCEN5s with no associated Zip1 (B). (C and D) Examples of zip3 (strain Y1010) (see Table S1) nuclei with tethered (C) or untethered (D) neCEN5s and no associated Zip1. (E) Percent of nuclei with Zip1 colocalized, according to whether neCEN5s are tethered or untethered.
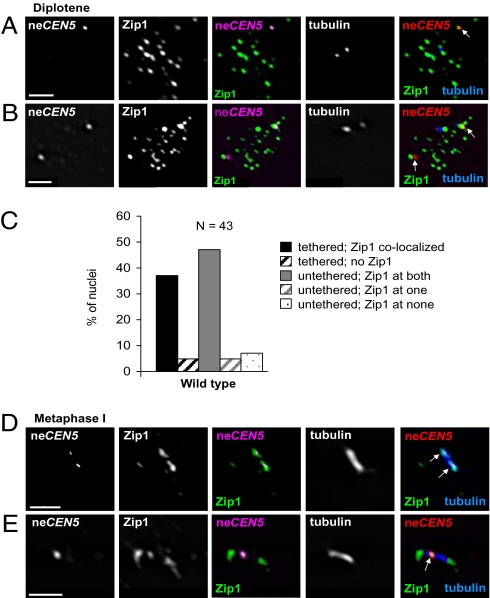
If centromere tethering plays a role in NEC segregation, then the tether should persist until chromosomes are aligned on the metaphase I spindle. To test this possibility, we examined spread nuclei at diplotene and metaphase (Fig. 1 E–H). Similar to pachytene cells, Zip1 localizes to the centromeres of NECs during diplotene (Fig. 3 A–C), and the association of the neCEN5s is strongly dependent on Zip1 (Fig. 1F). At metaphase I, Zip1 is often found distributed along the spindle (27 out of 34 spindles examined), making it impossible to determine if Zip1 colocalizes specifically with the neCEN5s (Fig. 3 D and E). Consistent with the centromeres remaining tethered from pachytene until the metaphase-anaphase I transition, the proportion of live cells with a single GFP focus remained steady throughout a meiotic time course until the onset of the meiotic nuclear divisions (Fig. S6). The persistence of a physical interaction between NECs is analogous to NEC segregation in other organisms where such pairs remain associated until the metaphase I to anaphase I transition (15–17).
Fig. 3.
Zip1 persists on neCEN5s after SC disassembly in wild type. (A and B) Spread nuclei from wild type (strain Y712) (see Table S1) stained for Zip1, tubulin, and neCEN5 at diplotene. (C) Percent of wild-type nuclei with Zip1 colocalized at diplotene, according to whether neCEN5s are tethered or untethered. (D and E) Colocalization of neCEN5 and Zip1 at metaphase I. Zip1 is often found along the entire spindle (27/34 spindles examined). (A, B, D, and E) Arrows indicate neCEN5. (Scale bars, 2 μm.)
Zip3, a Component of the Synapsis Initiation Complex, Regulates Zip1 Function at Centromeres.
How is Zip1 function regulated at the centromeres of NECs? The synapsis initiation complex, which includes the Zip3 protein, promotes SC assembly by triggering the polymerization of Zip1 along chromosomes (18). Zip3 has SUMO E3 ligase activity and may sumoylate substrates along the chromosome cores to which Zip1 binds (19, 20). In zip3 mutants, Zip1 shows severely delayed and incomplete association with meiotic chromosomes. To determine whether Zip3 is important for NEC segregation, we analyzed nondisjunction and neCEN5 tethering in a zip3 mutant. The nondisjunction frequency was increased to 24% in zip3, compared to 11% in wild type (see Fig. 1B). Furthermore, both the level of tethering and the fraction of untethered centromeres associated with Zip1 were reduced more than twofold in zip3 (see Figs. 1 and 2). These results suggest that Zip3’s role in NEC segregation is to facilitate the association (or maintenance) of Zip1 with the centromeres of NECs. This observation was unexpected, because “centromere coupling” (which is also mediated by Zip1) is independent of Zip3 (3). Thus, centromere coupling (before or in the absence of recombination) and NEC tethering (after recombination is initiated) have distinct genetic requirements. Although the Zip3 protein localizes to centromeres, it is dispensable for synapsis initiation at centromeres (21). Our studies suggest a role for centromere-localized Zip3 protein.
Zip2, Zip4, and Spo16, but Not Msh4 or Mer3, Are also Required for Centromere Tethering of NECs.
Extension of Zip1 polymers along homologous chromosome pairs depends upon a complex of proteins that includes Zip2, Zip4, and Spo16 (22–24). In mutants lacking any one of these proteins, Zip1 localizes to foci on chromosomes, but fails to polymerize along the lengths of chromosomes. Although Zip1 associates with the neCEN5s in zip2 and zip4 mutants (Fig. S7), centromere tethering by pachytene is abrogated to a similar extent as in the zip1 mutant (Table S2). This is associated with a concomitant increase in the nondisjunction frequency of the NECs (see Table S2).
Synapsis is also affected in mutants that lack DNA recombination and repair proteins, including the Mer3 helicase and the mismatch repair paralogue, Msh4. During meiosis, Msh4 forms a heterodimer with Msh5 (25), which recognizes double Holliday junctions in vitro (26). Together with Zip3, Zip1, Zip2, Zip4, and Spo16, these proteins promote crossover recombination between homologous chromosomes and are known as the “ZMM” ensemble (27). Both the msh4 and mer3 mutants display wild-type levels of centromere tethering and NEC nondisjunction (see Table S2), suggesting differential requirements for the ZMM proteins in the segregation of NECs.
Zip1 Promotes Segregation of Homologous Chromosome Pairs.
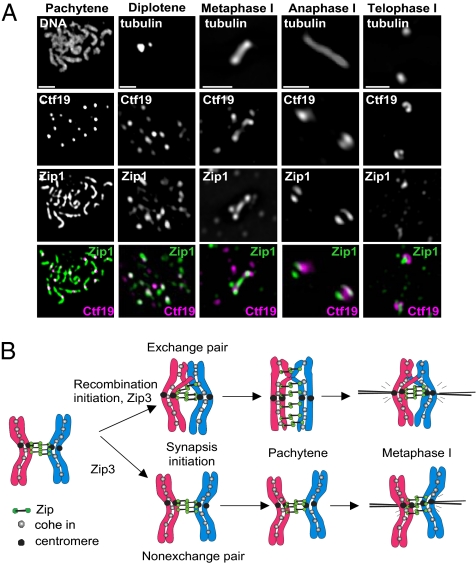
Our data demonstrate that Zip1 promotes the disjunction of both nonexchange homeologous and heterologous chromosome pairs. If Zip1 also plays a role in the segregation of homologous NECs, then the zip1 mutant should show increased nondisjunction of homologous chromosomes compared to an msh4 control strain. To address this possibility, we followed the segregation of chromosome III tagged with LacO repeats (4). Chromosome III is one of the smallest chromosomes and therefore frequently fails to cross over in zip1 and msh4 strains (11). Importantly, the zip1 mutant has similar or slightly increased crossover frequencies compared to msh4 (10, 12). Chromosome III missegregates in 15% of meioses in a zip1 strain, compared to 7% in msh4 (n = 100, P < 0.05, t-test), suggesting that Zip1 does indeed play a role in the segregation of homologous NECs. Consistent with this notion, Zip1 colocalizes with centromeres throughout meiosis I in wild-type nuclei containing only homologous chromosome pairs (Fig. 4A). We propose that Zip1 promotes proper chromosome segregation by directly mediating centromere associations throughout meiosis I of both chiasmate and achiasmate chromosome pairs. Although this function is most apparent for NECs, centromere tethering may lead to improved chromosome segregation in general (Fig. 4B).
Fig. 4.
Zip1 localizes to homologous centromeres throughout meiosis I. (A) Representative images of pachytene, diplotene, metaphase, anaphase, and telophase nuclei from wild type (strain Y636) (see Table S1) stained for DNA, tubulin, Zip1, and centromeres (detected with antibodies to the Myc-tagged kinetochore component, Ctf19). At diplotene, 75% of centromere foci have Zip1 colocalized, 11% have Zip1 juxtaposed (i.e., touching, but not overlapping), and 14% of centromeres are not associated with Zip1 (n = 543 Ctf19 foci, 31 nuclei inspected). Diplotene nuclei displayed 23 ± 5 (SD) Zip1 foci per nucleus, and 59% of these colocalized with centromeres. (Scale bars: 2 μm.) (B) Model of Zip1 function at centromeres. Early in prophase, Zip1 localizes to the centromeres of both chiasmate and achiasmate chromosome pairs, thereby producing a functional tether. After SC disassembly, Zip1 remains localized to the centromeres of both nonexchange and chiasmate chromosomes, thus aiding their correct bipolar attachment to the meiotic spindle.
Mad3, a Component of the Spindle Checkpoint, Acts in Parallel with Zip1 to Promote NEC Segregation.
Comparison of the frequency of tethering and the efficiency of meiosis I disjunction suggests that centromeric tethering by Zip1 is not the only mechanism that promotes proper segregation of NECs. In wild type, ∼55% of NECs are tethered. If the remaining 45% of NEC pairs segregate randomly (half the time going to the same pole, and half the time to opposite poles), then the frequency of meiosis I nondisjunction should be 22.5%. However, only 11% of NECs nondisjoin (see Fig. 1B). Similarly, in zip1, 14% of NECs are tethered, predicting a nondisjunction frequency of 43%; yet only 27% of NECs missegregate at meiosis I.
A recent report suggested that the spindle checkpoint component, Mad3, specifically improves the segregation of NEC pairs, without affecting the disjunction of crossover-proficient homologs (28). In a mad3 mutant, the neCEN5s are associated in 27% of pachytene nuclei (see Fig. 1D). This frequency is lower than in wild type, probably because of the shortened prophase period in mad3 (28). Of the 73% of nuclei in which the neCEN5s are unassociated in the mad3 mutant, 36.5% are expected to missegregate. The observed rate is 34% (see Fig. 1B). Thus, in mad3, there is an excellent correspondence between the frequency of centromere tethering and the frequency of correct disjunction, suggesting that only tethered chromosomes segregate correctly in the absence of Mad3. The nondisjunction frequency we observed is different from another study, where the authors reported ∼50% nondisjunction of the same neCEN5 pair (28). The differences are likely to be attributable to the different assessment methods. Whereas we used the LacO/LacI-GFP system, Cheslock et al. (28) used tetrad dissection, which relies upon the recovery of viable spores. Discrepancies in nondisjunction frequencies using the same homeolog pair, but different assessment methods, have been reported previously for the mad2 mutant (28, 29).
In the mad3 zip1 double mutant, NEC centromeres are tethered in 13% of cells, similar to the number observed in the zip1 single mutant. If the tethered chromosomes segregate correctly and the remaining 87% segregate randomly, then a nondisjunction rate of 43.5% is expected. In fact, however, 50% of NEC pairs missegregate, which is the frequency expected for completely random segregation. The observed 50% nondisjunction rate (169 nondisjunction events out of a total of 339) is significantly different from the 43.5% expected (147 nondisjunction events out of 339), as determined using a χ2 goodness-of-fit test (P < 0.017). Thus, the centromeric associations observed in the mad3 zip1 double mutant appear to be ineffective in ensuring disjunction, suggesting that Zip1 activity is required for tethers to be functional in segregation.
Based on these observations, we propose that two distinct mechanisms ensure the segregation of NECs: one requiring Zip1-mediated tethering, the other requiring Mad3. We suggest that Mad3 facilitates proper disjunction of those chromosome pairs that are not tethered by Zip1.
Why Multiple Mechanisms?
Why are multiple mechanisms necessary to ensure accurate segregation in an organism where crossovers are plentiful (∼90 crossovers for 16 chromosome pairs)? A number of observations suggest that not all crossovers ensure proper disjunction; the location of a crossover relative to the centromere is also important. Mad2, another spindle checkpoint component, improves the reorientation of kinetochores when chromosome pairs fail to have a chiasma within ∼180 kb of the centromere (estimated at 32% of cells for the largest chromosome) (29, 30). In humans, trisomy 21 and other aneuploidies increase with maternal age. However, often the maternal chromosomes, from which the majority of aneuploidies are derived, either lack a crossover or display a crossover near chromosome ends, far from the centromere (31, 32). In theory, placing a crossover near all centromeres would be a solution. However, crossovers in very close proximity to centromeres are correlated with increased frequencies of precocious sister-chromatid separation in yeast (33) and meiosis II nondisjunction in humans (34) and flies (35). Indeed, a Zip1-dependent mechanism operates to limit crossing over specifically near centromeres in budding yeast (11). This dichotomy–limiting crossovers at the centromere but requiring crossovers within a certain distance of the centromere– may be resolved by employing multiple mechanisms to ensure proper segregation. Centromere tethering (mediated by Zip1) together with kinetochore reorientation (mediated by Mad2, and perhaps Mad3) could compensate for the absence of a crossover or for inappropriate chiasma position.
Materials and Methods
Strain Construction and Sporulation Conditions.
Strains were constructed using standard molecular procedures, standard yeast media, and lithium acetate transformation. All transformants were verified by PCR or Southern blotting. A list of strains is given in Table S1. Details of the sporulation conditions for the BR, S228C, and SK1, strains are given in SI Materials and Methods.
Cytology.
Meiotic nuclear spreads, indirect immunofluorescence, and fluorescent in situ hybridization were all carried out as described previously (22). Details are given in SI Materials and Methods., which also contains information of all antibodies and concentrations used.
Statistics.
All statistical comparisons were carried out using R (www.r-project.org). Tests used were the Fisher exact test, two-sample t test for proportions, χ2 goodness-of-fit test, and for distribution-free analysis, two-sample Kolmogorov-Smirnov test. The Shapiro-Wilk test was used to test for normality.
Supplementary Material
Acknowledgments
We thank members of the Hoffmann and Roeder laboratories for insightful comments on the manuscript, Dean Dawson for communicating data prior to publication, and Dean Dawson (Oklahoma Medical Research Foundation, Oklahoma City, OK), Amy MacQueen (Wesleyan University, Middletown, CT), Angelika Amon (Massachusetts Institute of Technology, Cambridge, MA), and David Kaback (UMDNJ-New Jersey Medical School, Newark, NJ) for yeast strains. This work was funded by the Howard Hughes Medical Institute (G.S.R. laboratory), the Biotechnology and Biological Sciences Research Council (E.H. laboratory), a Medical Research Council studentship to (to L.N.), and a European Molecular Biology Organization Short-Term Fellowship as well as a Royal Society Dorothy Hodgkin Fellowship (to E.H.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913435107/DCSupplemental.
References
- 1.Loidl J, Scherthan H, Kaback DB. Physical association between nonhomologous chromosomes precedes distributive disjunction in yeast. Proc Natl Acad Sci USA. 1994;91:331–334. doi: 10.1073/pnas.91.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kemp B, Boumil RM, Stewart MN, Dawson DS. A role for centromere pairing in meiotic chromosome segregation. Genes Dev. 2004;18:1946–1951. doi: 10.1101/gad.1227304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsubouchi T, Roeder GS. A synaptonemal complex protein promotes homology-independent centromere coupling. Science. 2005;308:870–873. doi: 10.1126/science.1108283. [DOI] [PubMed] [Google Scholar]
- 4.Chan A, Borts RH, Hoffmann ER. Temperature-dependent modulation of chromosome segregation in msh4 mutants of budding yeast. PLoS One. 2009;4:e7284. doi: 10.1371/journal.pone.0007284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nilsson-Tillgren T, Gjermansen C, Kielland-Brandt MC, Peterson JGL, Holmberg S. Genetic differences between Saccharomyces carlsbergensis and Saccharomyces cerevisiae. Analysis of chromosome III by single chromosome transfer. Carlsberg Res Commun. 1981;46:65–76. [Google Scholar]
- 6.Nilsson-Tillgren T, Gjermansen C, Holmberg S, Petersen JGL, Kielland-Brandt MC. Analysis of chromosome V and the ILV1 gene from Saccharomyces carlsbergensis. Carlsberg Res Commun. 1986;51:309–326. [Google Scholar]
- 7.Maxfield Boumil R, Kemp B, Angelichio M, Nilsson-Tillgren T, Dawson DS. Meiotic segregation of a homeologous chromosome pair. Mol Genet Genomics. 2003;268:750–760. doi: 10.1007/s00438-002-0796-9. [DOI] [PubMed] [Google Scholar]
- 8.Guacci V, Kaback DB. Distributive disjunction of authentic chromosomes in Saccharomyces cerevisiae. Genetics. 1991;127:475–488. doi: 10.1093/genetics/127.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sym M, Roeder GS. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell. 1994;79:283–292. doi: 10.1016/0092-8674(94)90197-x. [DOI] [PubMed] [Google Scholar]
- 10.Börner GV, Kleckner N, Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- 11.Chen SY, et al. Global analysis of the meiotic crossover landscape. Dev Cell. 2008;15:401–415. doi: 10.1016/j.devcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novak JE, Ross-Macdonald PB, Roeder GS. The budding yeast Msh4 protein functions in chromosome synapsis and the regulation of crossover distribution. Genetics. 2001;158:1013–1025. doi: 10.1093/genetics/158.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann ER, Borts RH. Meiotic recombination intermediates and mismatch repair proteins. Cytogenet Genome Res. 2004;107:232–248. doi: 10.1159/000080601. [DOI] [PubMed] [Google Scholar]
- 14.Stewart MN, Obeso D, Chuong H, Dawson DS. The synaptonemal complex protein, Zip1, promoties bi-orientation of centromeres at meiosis I. PLoS Genet. 2009 doi: 10.1371/journal.pgen.1000771. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas SE, et al. Identification of two proteins required for conjunction and regular segregation of achiasmate homologs in Drosophila male meiosis. Cell. 2005;123:555–568. doi: 10.1016/j.cell.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 16.Rasmussen SW. The transformation of the Synaptonemal Complex into the ‘elimination chromatin’ in Bombyx mori oocytes. Chromosoma. 1977;60:205–221. doi: 10.1007/BF00329771. [DOI] [PubMed] [Google Scholar]
- 17.Wolf KW. How meiotic cells deal with non-exchange chromosomes. Bioessays. 1994;16:107–114. doi: 10.1002/bies.950160207. [DOI] [PubMed] [Google Scholar]
- 18.Agarwal S, Roeder GS. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell. 2000;102:245–255. doi: 10.1016/s0092-8674(00)00029-5. [DOI] [PubMed] [Google Scholar]
- 19.Hooker GW, Roeder GS. A Role for SUMO in meiotic chromosome synapsis. Curr Biol. 2006;16:1238–1243. doi: 10.1016/j.cub.2006.04.045. [DOI] [PubMed] [Google Scholar]
- 20.Cheng CH, et al. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 2006;20:2067–2081. doi: 10.1101/gad.1430406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsubouchi T, MacQueen AJ, Roeder GS. Initiation of meiotic chromosome synapsis at centromeres in budding yeast. Genes Dev. 2008;22:3217–3226. doi: 10.1101/gad.1709408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chua PR, Roeder GS. Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell. 1998;93:349–359. doi: 10.1016/s0092-8674(00)81164-2. [DOI] [PubMed] [Google Scholar]
- 23.Tsubouchi T, Zhao H, Roeder GS. The meiosis-specific zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with zip2. Dev Cell. 2006;10:809–819. doi: 10.1016/j.devcel.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 24.Shinohara M, Oh SD, Hunter N, Shinohara A. Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat Genet. 2008;40:299–309. doi: 10.1038/ng.83. [DOI] [PubMed] [Google Scholar]
- 25.Hollingsworth NM, Ponte L, Halsey C. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 1995;9:1728–1739. doi: 10.1101/gad.9.14.1728. [DOI] [PubMed] [Google Scholar]
- 26.Snowden T, Acharya S, Butz C, Berardini M, Fishel R. hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol Cell. 2004;15:437–451. doi: 10.1016/j.molcel.2004.06.040. [DOI] [PubMed] [Google Scholar]
- 27.Lynn A, Soucek R, Börner GV. ZMM proteins during meiosis: crossover artists at work. Chromosome Res. 2007;15:591–605. doi: 10.1007/s10577-007-1150-1. [DOI] [PubMed] [Google Scholar]
- 28.Cheslock PS, Kemp BJ, Boumil RM, Dawson DS. The roles of MAD1, MAD2 and MAD3 in meiotic progression and the segregation of nonexchange chromosomes. Nat Genet. 2005;37:756–760. doi: 10.1038/ng1588. [DOI] [PubMed] [Google Scholar]
- 29.Lacefield S, Murray AW. The spindle checkpoint rescues the meiotic segregation of chromosomes whose crossovers are far from the centromere. Nat Genet. 2007;39:1273–1277. doi: 10.1038/ng2120. [DOI] [PubMed] [Google Scholar]
- 30.Shonn MA, McCarroll R, Murray AW. Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science. 2000;289:300–303. doi: 10.1126/science.289.5477.300. [DOI] [PubMed] [Google Scholar]
- 31.Hassold T, Hall H, Hunt P. The origin of human aneuploidy: where we have been, where we are going. Hum Mol Genet. 2007;16 Spec No. 2(R2):R203–R208. doi: 10.1093/hmg/ddm243. [DOI] [PubMed] [Google Scholar]
- 32.Hassold T, Hunt P. Rescuing distal crossovers. Nat Genet. 2007;39:1187–1188. doi: 10.1038/ng1007-1187. [DOI] [PubMed] [Google Scholar]
- 33.Rockmill B, Voelkel-Meiman K, Roeder GS. Centromere-proximal crossovers are associated with precocious separation of sister chromatids during meiosis in Saccharomyces cerevisiae. Genetics. 2006;174:1745–1754. doi: 10.1534/genetics.106.058933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamb NE, et al. Susceptible chiasmate configurations of chromosome 21 predispose to non-disjunction in both maternal meiosis I and meiosis II. Nat Genet. 1996;14:400–405. doi: 10.1038/ng1296-400. [DOI] [PubMed] [Google Scholar]
- 35.Koehler KE, et al. Spontaneous X chromosome MI and MII nondisjunction events in Drosophila melanogaster oocytes have different recombinational histories. Nat Genet. 1996;14:406–414. doi: 10.1038/ng1296-406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.