Abstract
The potential for endogenous remyelination and axonal protection can be an important factor in determining disease outcome in demyelinating diseases like multiple sclerosis. In many multiple sclerosis (MS) patients CNS repair fails or is incomplete whereas in others the disease is accompanied by extensive repair of demyelinated lesions. We have described significant differences in the ability of two strains of mice to repair CNS damage following Theiler’s virus-induced demyelination: FVB/NJ (FVB) mice repair damaged myelin spontaneously and completely, whereas B10.D1-H2q/SgJ (B10.Q) mice are deficient in the repair process. A QTL analysis was performed to identify genetic loci that differentially regulate CNS repair following chronic demyelination in these strains and two QTL were detected: one on chromosome 3 with a LOD score of 9.3 and a second on chromosome 9 with a LOD score of 14.0. The mouse genes for epidermal growth factor (EGF) and Tyk2 are encoded within the QTL on chromosomes 3 and 9, respectively. Sequence polymorphisms between the FVB and B10.Q strains at both the EGF and Tyk2 loci define functional variations consistent with roles for these genes in regulating myelin repair. EGF is a key regulator of cell growth and development and we show a sevenfold increase in EGF expression in FVB compared to B10.Q mice. Tyk2 is a Janus kinase that plays a central role in controlling the TH1 immune response and we show that attenuation of Tyk2 function correlates with enhanced CNS repair.
Keywords: quantitative trait locus, remyelination, Theiler’s murine encephalomyelitis virus, multiple sclerosis
Multiple sclerosis (MS) is a complex disease characterized by CNS inflammation with the subsequent development of focal demyelinated lesions in the white matter of the brain and spinal cord. The disease is heterogeneous in both its pathology and its clinical course (1–4). Remyelination and repair of demyelinated lesions is a well-established phenomenon but the extent and timing of this repair is quite variable among patients (5).
The repair of demyelinated CNS lesions is a complex process that likely requires the proliferation, migration, and differentiation of oligodendrocyte progenitor cells, the maintenance and preservation of axons, axonal recognition by differentiating oligodendrocytes and the formation of new myelin, and the proper regulation of the CNS infiltrating inflammatory response (6, 7). Failure of any one of these processes might result in the general failure of CNS repair but currently little is known about the factors that are important in determining the extent of repair following demyelination.
We have described strain-specific differences in the ability of mice to repair CNS damage following virus-induced demyelination (8). In susceptible strains of mice, CNS infection with Theiler’s murine encephalomyelitis virus (TMEV) induces an inflammatory demyelinating disease of the spinal cord that is similar in clinical and pathological presentation to the spinal form of chronic progressive multiple sclerosis. Demyelination is extensive in all susceptible strains by 90–100 days postinfection. In B10.D1-H2q/SgJ (B10.Q) mice, at 300 days postinfection there is minimal CNS repair with a progressive accumulation of neurologic deficits leading to eventual paralysis and death. In contrast, FVB/NJ (FVB) mice show extensive spontaneous myelin repair with axonal and functional preservation (8). When FVB and B10.Q mice are mated, the (FVB/B10.Q)F1 hybrid progeny show strong repair; the “reparative phenotype” of the FVB strain is therefore inherited as a dominant trait. The large difference in the reparative phenotype between these strains (effect size) and the high penetrance of the phenotype suggested that the mapping of genetic loci that are important for CNS repair might be possible. In this report we characterize the patterns of inheritance of the reparative phenotype between the FVB and B10.Q strains and identify two strong QTL for CNS repair. Furthermore, genetic polymorphisms in the epidermal growth factor (EGF) and Tyk2 genes, which map within these QTL, may define a functional basis for understanding differences in the ability of these animals to repair demyelinated lesions.
Results
CNS Repair After TMEV-Induced Demyelination Is a Quantitative Trait.
In a previous report we characterized the significant differences in spontaneous CNS repair and remyelination that are observed in different strains of mice following TMEV-induced CNS demyelination (8). Infection of B10.Q mice with Theiler’s virus results in chronic demyelination with minimal repair and the progressive accumulation of neurologic deficits. In contrast, a second strain, FVB, shows extensive spontaneous repair with axonal and functional preservation.
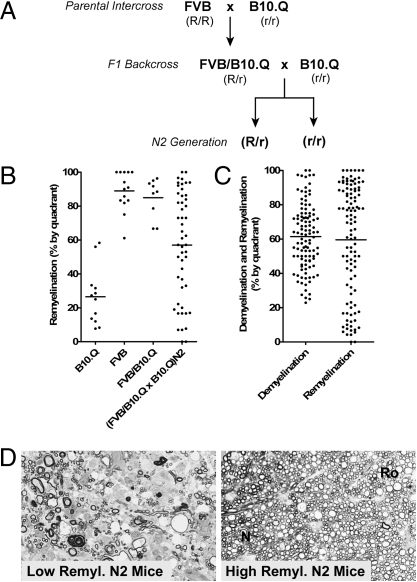
We examined the inheritance of myelin repair by crossing FVB with B10.Q mice to generate (FVB/B10.Q)F1 hybrid mice (Fig. 1A). Infected mice from this cross developed demyelinated lesions followed by extensive remyelination similar to that observed in FVB mice (Fig. 1B, and Fig. S1). Most remyelination was oligodendrocyte mediated, although Schwann cell-mediated remyelination was also present. At 300 days postinfection, remyelination in (FVB/B10.Q)F1 mice averaged 85% of total lesion area (Fig. 1B). F1 hybrids showed few neurologic symptoms of demyelinating disease up to 1 year postinfection. Thus, lesion repair is inherited as a dominant trait in the F1 generation.
Fig. 1.
Genetics of CNS repair. (A) Crosses used to generate F1 and N2 mice for the analysis of the genetics of CNS repair. FVB/NJ and B10.D1-H2q/SgJ (B10.Q) were the parental strains. The intercross/backcross strategy was designed to identify dominant traits from the FVB strain and the genotype beneath each strain designation indicates the potential dominant (R) or recessive (r) nature of the genotype at any genomic locus with regard to remyelination. In the N2 generation there are only two possible genotypes: FVB/B10.Q (R/r) or B10.Q/B10.Q (r/r). (B) Extent of remyelination in the FVB and B10.Q parental strains, and in F1 and N2 hybrid mice, as measured by quadrant analysis at 300 days postinfection. Mean repair values are indicated by the horizontal lines. (C) Extent of demyelination and remyelination in the 109 (FVB/B10.Q × B10.Q)N2 mice used for the QTL analysis. Mean repair values are indicated by the horizontal lines. (D) Examples of the high and low remyelination phenotypes in (FVB/B10.Q × B10.Q)N2 mice at 300 days postinfection. N, normal myelin; Ro, remyelination.
When (FVB/B10.Q)F1 mice were backcrossed to the B10.Q parent strain to produce (FVB/B10.Q × B10.Q)N2 progeny, we saw that the extent of remyelination in the N2 generation was distributed continuously between mice with very little remyelination at 300 days postinfection, to mice that showed nearly complete remyelination (Fig. 1B). Histologic examples of the high and low remyelination phenotypes are shown in Fig. 1D. Demyelination was characterized by the absence of normal myelin sheaths and by the presence of inflammatory cellular infiltrates and myelin debris. Remyelination (Ro) was easily distinguished from normal myelination (N); the myelin was thinner, with fewer wraps around the axon and consequently stained less intensely with myelin stains. The broad phenotypic distribution in the N2 generation indicates that the reparative phenotype is inherited as a quantitative trait and is determined by the inheritance of allelic variants from multiple genetic loci. We therefore selected an intercross/backcross strategy, using the (FVB/B10.Q)F1 mice and the B10.Q parental strain, for QTL analysis to detect loci that were involved in CNS repair (Fig. 1A).
We mapped genetic determinants of CNS repair in a cohort of 109 (FVB/B10.Q × B10.Q)N2 mice. We examined 10 spinal cord sections from each animal and scored demyelination and remyelination using the quadrant method (SI Methods). The quadrant demyelination and remyelination scores for this cohort are shown in Fig. 1C. Animals with demyelination in <20% of the quadrants on 10 spinal cord cross sections were excluded from the remyelination analysis; if there is no demyelination, CNS repair cannot be scored.
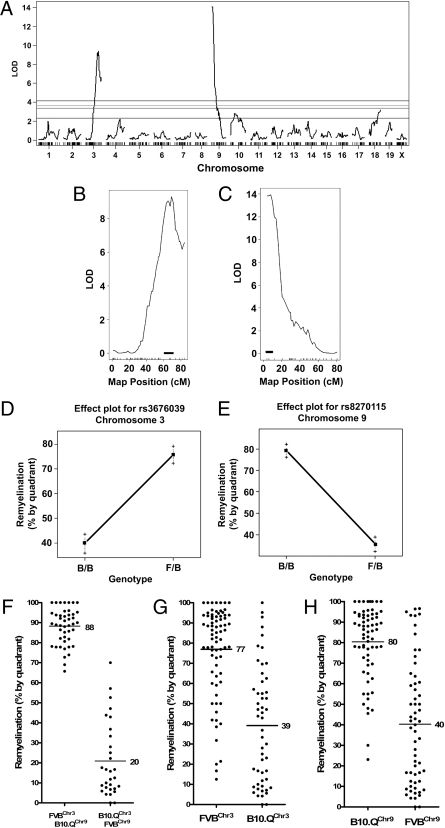
Genomic DNA was isolated from all animals and linkage analysis was preformed using R/qtl software (9). QTL were initially identified from a one-dimensional genomewide scan using a model in which gender, QTL, and gender interaction with QTL were taken into consideration. We detected two highly significant QTL on chromosomes 3 and 9, with LOD scores of 9.3 and 14.0, respectively (Fig. 2 A–C). The LOD scores for significance were determined using 1,000 permutations of the data (10) and significance thresholds of 1% (LOD = 4.15), 5% (LOD = 3.65), 10% (LOD = 3.36), and 63% (LOD = 2.30) were calculated (Fig. 2A). QTL with LOD scores >1% are considered highly significant, those >5% are considered significant, and those >63% are considered suggestive. The QTL on chromosomes 3 and 9 both exceeded the 1% threshold for high significance with genomewide P values of 1.42 × 10−10 and 4.11 × 10−15, respectively. QTL on chromosomes 10 and 18 exceeded the 63% threshold for suggestive significance (LOD = 2.80 and 3.18, respectively). We do not consider these further except to note that the chromosome 10 QTL may show preferential effects in male animals (Fig. S2).
Fig. 2.
QTL for CNS repair. (A) Genomewide one-dimensional QTL scan. The horizontal lines across the plot indicate four confidence thresholds calculated at 1% (top, highly significant), 5%, 10%, and 63% (bottom, suggestive). Highly significant QTL were identified on chromosomes 3 and 9. (B and C) Confidence interval plots for chromosomes 3 and 9. The small horizontal bars at the base of the plots indicate the genomic segment that corresponds to the 95% confidence interval for each QTL. (D and E) Effect plots for QTL on chromosomes 3 and 9. Plots are based on the genotypes detected at a representative SNP located at the peak of LOD score (SNP rs3676039 for chromosome 3 and rs8270115 for chromosome 9). Effect plot for chromosome 3 shows that the FVB allele increases remyelination whereas on chromosome 9 it decreases remyelination. Genotypes: BB, B10.Q/B10.Q; FB, FVB/B10.Q. (F–H) Phenotypic values for remyelination were plotted following stratification of the animals based on their genotypes over the 95% confidence intervals for each QTL. F plots animals with the genotypes that should give the best or the poorest repair. G and H plot animals on the basis of their chromosome 3 or 9 genotypes.
A genomewide two-dimensional scan found no evidence of epistatic interactions between the identified QTL. The QTL identified from the one-dimensional scan using the 63% significance threshold were fit into multiple regression analysis models. These models indicate that the QTL on chromosome 3 results in ∼18.0% of the observed variability in the remyelination data whereas the QTL on chromosome 9 represents ∼30.0%. Therefore, whereas the two identified QTL explain a significant fraction of the genetic effect on CNS repair, there are clearly other loci that also affect this phenotype.
An effect plot for the QTL on chromosome 3 shows that the FVB allele increases repair whereas on chromosome 9 it decreases repair (Fig. 2 D and E). The QTL on chromosome 3 is therefore inherited as a dominant allele from the FVB strain whereas the QTL on chromosome 9 is inherited as a recessive allele from B10.Q.
Confidence interval plots indicate that the 95% confidence interval for the chromosome 3 QTL covers a 10-cM interval from 61.09 to 71.09 cM (Fig. 2B, horizontal bar) or ≈126.4–139.7 Mbp. The confidence interval for the chromosome 9 QTL covers 6 cM from 3.99 to 9.99 cM (Fig. 2C, horizontal bar) or 17.8–27.0 Mbp.
Having identified strong QTL on chromosomes 3 and 9, we stratified the animals on the basis of their genotypes over the 95% confident intervals for these loci and to ask how such stratification is reflected in the repair phenotype. The animals that repair the best should carry the FVB genotype at the chromosome 3 QTL and the B10.Q genotype on chromosome 9 (FVBChr3/B10.QChr9). Those that repair the worst should be B10.QChr3/FVBChr9. These two classes are shown in Fig. 2F, with average repair in FVBChr3/B10.QChr9 at 88% and in B10.QChr3/FVBChr9 at 20%. The overall effect size is therefore ≈68%.
When we stratified the animals only on the basis of the genotype at the chromosome 3 locus, disregarding the chromosome 9 genotype, animals with FVBChr3 show an average repair value of 77% whereas those with B10.QChr3 are at 39%, a difference of 38% (Fig. 2G). Stratification based on chromosome 9 alone gave Fig. 2H. B10.QChr9 animals show 80% repair whereas FVBChr9 animals show 40%, a difference of 40%. These results support a model in which both the chromosome 3 and the chromosome 9 QTL exert major effects on the remyelination phenotype and in which these effects are additive but with only minimal genetic interaction between the loci. The large standard deviations in these data sets are also consistent with the involvement of genetic loci other than the two identified QTL on the remyelination phenotype.
Identification of Candidate Genes.
In the chromosomal regions that lie within the 95% confidence intervals on chromosomes 3 and 9, there are 112 and 171 identified genes, respectively (Table S1). An Ingenuity Pathways Analysis of genes from the chromosome 3 and 9 confidence intervals (Table S2) reveals a number of reasonable candidate genes from within these regions. Caspase 6 lies in the chromosome 3 confidence interval and has recently been shown to play a role in axonal degeneration (11). The erythropoietin receptor lies in the chromosome 9 confidence interval and its ligand, erythropoietin, has neuroprotective effects in the rat experimental autoimmune encephalitis model of MS (12) and has been tested as a potential MS therapeutic (13). We were particularly interested in the Ugt8a gene on chromosome 3. The Ugt8a enzyme, also known as UDP-galactose:ceramide galactosyltransferase (CGT), is responsible for the synthesis of galactocerebroside (GalC), a major lipid component of myelin. CGT mutant mice have relatively normal appearing myelin but develop a generalized tremor, mild ataxia, and progressive hindlimb paralysis (14). We cloned and sequenced the cDNA for the Ugt8a protein and examined the expression of Ugt8a transcripts in FVB and B10.Q spinal cord but found no differences in the coding sequence or expression of the gene.
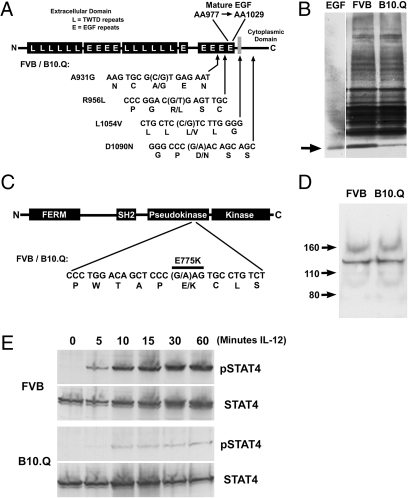
EGF is one of the most broadly active genes at the QTL on chromosome 3. We sequenced the cDNA for the EGF precursor protein from both FVB and B10.Q. There are four nonsynonymous single-nucleotide polymorphisms between the precursor proteins in the two strains (Fig. 3A). Although none of these differences affects the sequence of the mature EGF growth factor peptide, they all lie near the sites of cleavage that produce the peptide. Western blots for EGF from salivary glands of FVB and B10.Q mice demonstrate a 7.4-fold reduction in the amount of mature EGF peptide in B10.Q mice compared to FVB (Fig. 3B, arrow). The higher molecular weight bands on this blot are also cleavage products from the EGF precursor protein; in EGF knockout mice all of these bands disappear (15). A densitometric scan of the blot in Fig. 3B shows a significant difference in the band intensity for the mature EGF peptide between the strains but relatively little difference in the higher molecular weight bands (Fig. S3A). If amino acid polymorphisms in the EGF precursor protein result in variant protein processing for the mature EGF peptide, a change in the intensity of one of the high molecular weight bands might also have been observed but the small intensity differences and variability in these bands make this difficult to detect. Different rates of EGF peptide turnover between the strains might also explain the differences in steady-state EGF level but if this is the case, the differing rates are not due to differences in the EGF peptide itself because the mature peptide sequence is identical in both strains.
Fig. 3.
Identification of candidate genes. (A) Structure of the EGF precursor protein showing the relative sizes of the extracellular, transmembrane, and cytoplasmic domains and the positions of the polymorphisms that differentiate the FVB and B10.Q alleles. The sequence below the structure shows the four nonsynonymous single-nucleotide polymorphisms and the resulting amino acid substitutions. (B) Western blot for EGF on protein from salivary gland from FVB and B10.Q mice. The arrow indicates the position of mature EGF growth factor that was run as a standard. (C) Structure of the Tyk2 protein. The sequence below the structure indicates the single nonsynonymous single-nucleotide polymorphism that differentiates the FVB and B10.Q alleles and the resulting amino acid substitution. (D) Western blot for Tyk2 on protein from the CNS. The positions of three molecular weight standards are indicated. The predominant Tyk2 band runs at ∼130 kDa. (E) Western blots for STAT4 and pSTAT4 from ConA-activated splenocytes from FVB and B10.Q animals after stimulation for varying times with IL-12.
Salivary gland is frequently used as a source of EGF because of the high level of EGF expression in this tissue. EGF is not highly expressed in the spinal cord and we were unable to detect it by Western blot. However, the difference in expression is not specific to salivary gland as we were able to detect significant differences in urinary EGF content between FVB and B10.Q mice (P = 0.003, Fig. S3B). EGF transcript levels were similar in spinal cord tissue from FVB and B10.Q mice (P = 0.297, Fig. S3C), consistent with the notion that the differences in EGF level might occur at the level of protein processing.
The Tyk2 Janus kinase lies in the genomic segment at the site of the chromosome 9 QTL. A naturally occurring mutation in Tyk2 has previously been described in the B10.Q strain (16) and when we sequenced the cDNA for the Tyk2 gene from both FVB and B10.Q, we identified the same difference that was previously reported (Fig. 3C). Initially it was reported that this mutation resulted in reduced Tyk2 protein levels in B10.Q animals but Western blots for Tyk2 from FVB and B10.Q spinal cord protein showed identical amounts of Tyk2 (Fig. 3D).
Because the polymorphism in the Tyk2 coding sequence did not affect the amount of Tyk2 protein, we examined Tyk2 function in FVB and B10.Q mice. The immediate downstream effect of activated Tyk2 is the phosphorylation of STAT4. IL-12 stimulation of activated splenocytes from FVB mice resulted in a rapid and robust phosphorylation of STAT4 whereas stimulation of activated splenocytes from B10.Q mice showed almost no STAT4 phosphorylation (Fig. 3E). This result is consistent with the production of a minimally active Tyk2 gene product in B10.Q mice.
Discussion
Genetic analysis of MS has taken two distinctly different approaches. One approach has focused on disease resistance and susceptibility, asking whether linkage exists between specific genetic loci and the presence or absence of disease (17, 18). A second approach has looked within the population of MS patients to ask whether linkage exists between specific loci and differential measures of disease outcome such as disease severity, time of disease onset, or rate of disease progression (19, 20). Although both approaches have shown increasing promise, except for the very strong linkage of the major histocompatibility haplotypes to disease susceptibility there have been few strong genetic correlations relevant to either disease outcome or susceptibility.
Histologic studies describing remyelination in MS lesions were first reported >40 years ago (21, 22). These initial reports were confirmed when Prineas and coworkers described remyelination in patients with either chronic (23) or early stage MS lesions (24). More recently, a systematic analysis of the frequency and distribution of remyelinated lesions in autopsies from 51 patients found extensive remyelination in ∼20% of patients whereas in others repair was almost nonexistent (5). Although remyelination tended to be more common in patients after a longer disease duration, little correlation was found between the extent of remyelination and patient gender, the clinical subtype of the disease (i.e., relapsing vs. progressive forms), or the pathologic subtype (i.e., subtypes I–IV). This study demonstrates that extensive repair can and often does occur but it also demonstrates the variability in remyelination between patients and underscores our lack of understanding with regard to the factors that determine the extent of CNS repair that can occur in MS.
Investigators have recognized that there exists a subgroup of MS patients that show little or no disease progression over time. This subgroup is often referred to as having “benign MS” (25–27). In general, the benign course of MS is defined by having a low disability score over long disease duration but within this definition there is considerable latitude for interpretation. The estimated frequency of the benign course of MS therefore varies significantly depending on the MS population studied and the particular defining criteria used by different investigators. Despite the uncertainty, the frequency is probably quite high; an estimated average over 17 studies was 25.6% (27). For reasons unknown, patients with benign MS do not experience the full range of pathology or are able to repair CNS damage before axonal loss and permanent disability occurs. In this regard these patients display a phenotype that is similar to mice such as the FVB strain that repairs CNS damage and maintains neurologic ability. Understanding the genetics, cell biology, and physiology of repair in these mice might therefore have relevance to our understanding of the variability of disease course in MS and might direct us toward the most important pathways to target for therapeutic intervention and for the development of diagnostics for the prediction of disease course.
In this report we have identified linkage groups on chromosomes 3 and 9 that determine differences in the capacity to spontaneously repair damaged myelin and have presented both physical and functional evidence for specific QTL candidate genes within these linkage groups. We have identified polymorphisms between the FVB and B10.Q strains in the protein-coding sequences of the genes encoding EGF and Tyk2. The polymorphisms in the EGF protein appear to affect the steady-state level of accumulation of the mature growth factor whereas the polymorphisms in the Tyk2 protein greatly reduce its function as a kinase.
Both EGF and EGF receptor (EGFR; ErbB-1) are widely expressed in the rodent CNS both during development and in the adult (28). EGF plays a central role in controlling the proliferation, survival, and migration of neural stem cells both in culture and in vivo (29–32) and likely plays a similar role in the development of glial stem cells (33). In general, EGF appears to maintain these stem cells in a proliferative state and may therefore play a role in determining the number of stem cells that form during development or during disease.
Much of the work on EGF has focused on its effects on mitotic cells but it also has diverse effects on postmitotic cells. Treatment of differentiated oligodendrocytes with EGF in culture promotes process formation and regrowth following injury (34). EGF treatment of oligodendrocytes down-regulates differentiation-specific antigens, allowing them to reenter a proliferative state (35). In aggregated rat brain cultures, the addition of EGF enhances remyelination following antibody- and complement-induced demyelination (36). These observations indicate that EGF has direct effects on oligodendrocytes that might affect how these cells respond following disease and during CNS repair.
Whereas EGF knockout mice are healthy and fertile with no obvious behavioral or neurologic abnormalities (15), inactivation of EGFR is lethal (37, 38). This dramatic difference in phenotype is undoubtedly because the EGF receptor is activated by a small family of ligands and its removal therefore affects a significantly larger number of pathways than the removal of EGF alone. Several recent studies have demonstrated a direct role for signaling through EGFR during oligodendrocyte development and during remyelination (33, 39). The examples cited above demonstrate that EGF signaling has pleiotropic effects on several CNS cell types including neurons, oligodendrocytes, and oligodendrocyte precursors, and it is easy to imagine a variety of potential roles for EGF and EGFR in CNS repair.
Many of the cytokine receptors have no intrinsic kinase activity and they activate their downstream pathways by first activating members of the Janus kinase family (40). Tyk2 interacts with the IL-12 receptor and plays a central role in regulating the TH1 immune response that is essential for an organism’s ability to fight intracellular pathogens but is also associated with autoimmune disease and tissue damage. A natural mutation in Tyk2 is known to exist in the B10.Q strain (16), which diminishes Tyk2 function and would certainly be inherited as a recessive trait similar to the chromosome 9 QTL. The presence of this mutation in B10.Q attenuates the TH1 immune response (16, 41) and limits susceptibility to autoimmune responses (42). One might imagine that this mutation results in an immune response that is strong enough for the organism to survive the initial viral infection but that once the demyelinating phase of the disease is complete the mutation limits the extent of tissue damage caused by CNS-infiltrating immune cells allowing the later repair phase to proceed.
Although the role that IL-12 and Tyk2 play in the development of the TH1 response has been well studied, Tyk2 also interacts with a number of other cytokine receptors including the receptors for IL-23, IL-10, IL-6, and interferons α and β; the effects that we see on CNS repair might alternatively occur through these pathways (43, 44). Whichever pathways are involved, it is worth noting that a recent genomewide association study identified 17 potential candidate genes for increased MS susceptibility in humans. On extended analysis of these genes, evidence for association increased only for Tyk2 (45). These results suggest that our animal model is relevant to the study of human MS genetics and that the continued genetic analysis of the QTL from our study will be relevant to human disease.
Our QTL screen for CNS repair has identified two loci with strong effects on CNS repair and remyelination and we have identified candidate genes within both of these QTL. A systematic genetic dissection of the chromosome 3 and chromosome 9 intervals, such as through the assessment of CNS repair in interval-specific congenic mouse strains (46) or in EGF and Tyk2 knockout mice (15, 47), will be necessary to more firmly establish these genes as responsible for the reparative phenotypes that we observe. It is the nature of QTL screens that only genetic loci that have allelic variation between the two parental strains can be detected and there are undoubtedly many other genes that are important for CNS repair that were not detected in this screen because the genes are identical in FVB and B10.Q mice. However, the strength of the QTL and the unbiased nature of the screen, together with the large effect size of the reparative phenotype, should provide a unique opportunity for the study of these QTL and perhaps for the identification of additional QTL in future screens using other models of demyelinating disease and CNS repair.
Methods
Mice and Viral Infection.
FVB/NJ (FVB) and B10.D1-H2q/SgJ (B10.Q) mice were obtained from The Jackson Laboratories. F1 hybrid mice and N2 backcross mice were bred at the Mayo Clinic. To induce demyelinating disease, 6- to 8-week-old mice were injected intracerebrally with 2.0 × 106 pfu of Daniel’s strain of Theiler’s murine encephalomyelitis virus. All animal protocols were approved by the Mayo Institutional Animal Care and Use Committee and are consistent with National Institutes of Health guidelines for the care and use of animals.
Tissue Preparation and Histopathology Analyses.
Quantitation of demyelination and remyelination was conducted on cross sections of spinal cord. Spinal cord tissue was fixed by transcardiac perfusion with Trump’s fixative followed by postfixation with osmium tetroxide and embedding in araldite plastic. One-micrometer-thick cross sections were cut from each block and stained with 4% p-phenylenediamine (PPD) to visualize the myelin sheaths.
Stained plastic sections were used to quantify demyelination and remyelination using a quadrant technique (SI Methods). Demyelination was characterized by the absence of normal myelin sheaths, denuded axons, inflammatory cellular infiltrates, and glial hypertrophy. Demyelinated areas with remyelination were characterized by myelin sheaths that are thinner and stain more lightly than normal myelin. Any quadrant that showed significant demyelination or remyelination was scored positive for that histopathology. Completely remyelinated quadrants were also scored positive for demyelination to reflect the previous pathologic state of the tissue. Percentage of demyelination was calculated as (total quadrants with demyelination)/(total quadrants examined) × 100. Percentage of remyelination was calculated as (total quadrants with remyelination)/(total quadrants with demyelination) × 100 (48).
Refer to SI Methods for a more detailed description of tissue preparation and histopathology analyses.
QTL Linkage Analysis.
FVB mice and B10.Q mice were mated to generate F1 progeny. We backcrossed (FVB/B10.Q)F1 progeny to the B10.Q parental strain to obtain N2 animals for QTL linkage analysis.
Immediately before transcardiac perfusion for the fixation of CNS tissue, liver tissue was removed and frozen for the isolation of genomic DNA. Genomic DNA for SNP typing was isolated using a phenol-chloroform extraction procedure. DNA concentration was assessed by reading the absorbance at 260 nm and adjusted to 70 ng/μL for SNP analysis.
The Illumina Mouse Medium-Density Linkage Array was used for SNP analysis. This array types each experimental animal for 1,449 SNPs; of these 1,449, a set of 588 SNPs was identified that distinguishes between the FVB and the B10.Q genomes and for which both physical and genetic positions are known on the basis of the mapping studies of Shifman et al. (49). Details regarding these 588 SNPs are presented in Table S3. Fig. S4 graphically displays their relative chromosomal positions.
Linkage analysis tests were performed using R/qtl software (version 1.08–56) (50) with linkage analysis results expressed as LOD scores. Thresholds for significant detection were determined using 1,000 permutations of the dataset (10). Four confidence thresholds were calculated at 1, 5, 10, and 63%. QTL with LOD scores >1% were considered highly significant, those >5% were considered significant, and those >63% were considered suggestive (51). For initial QTL identification a genomewide one-dimensional scan was performed using a full model that considered gender, QTL, and gender interaction with QTL. A genomewide two-dimensional scan was also performed to detect epistatic effects. Multiple regression analysis was performed from the one-dimensional model using the 63% confidence threshold to fit the model. The 95% confidence intervals for the chromosomal positions of putative QTL were approximated by the Bayesian credible intervals (9).
Refer to SI Methods for a more detailed description of procedures used for DNA isolation, SNP detection, and QTL analysis.
Splenocyte Activation Assays.
Spleen cells were isolated from FVB and B10.Q mice, plated at 1 × 106 cells/mL in RPMI culture medium (RPMI 1640, 10% FCS, 10 mM Hepes, 100 units/mL Pen/Strep), and activated by the addition of 2.5 μg/mL Con A for 48 h. After Con A treatment, the cells were washed twice with acidified RPMI 1640 (pH 6.4) and then replated in starvation medium (RPMI 1640, 1% FCS, 1% BSA, 10 mM Hepes, 100 units/mL Pen/Strep) at 1 × 105 cells/mL and cultured overnight. The cells were stimulated by the addition of IL-12 at 20 ng/mL for treatment periods of 0, 5, 10, 30, and 60 min. Cells were harvested by centrifugation at the appropriate times after IL-12 addition and frozen on dry ice. Cells were then prepared for SDS/PAGE and analyzed by Western blotting. Refer to SI Methods for a more detailed description of the splenocyte activation assay and analysis.
Western Blotting.
Proteins from mouse spinal cord, salivary gland, and cultured splenocytes were analyzed by SDS/PAGE and Western blotting according to standard procedures. The primary antibodies used for Western blotting were rabbit anti-mouse Tyk2 EP1127Y (Abcam), rabbit anti-mouse pSTAT4 Y693 (Zymed), rabbit anti-mouse STAT4 H-119 (Santa Cruz Biotechnology), and goat anti-mouse EGF (R&D Systems). Following incubation with primary antibody the blots were incubated with an appropriate peroxidase-conjugated secondary antibody and signal was visualized using the Pierce SuperSignal West Pico Chemiluminescent System. Refer to SI Methods for a detailed description of procedures used for sample preparation, SDS/PAGE, and Western blotting.
Statistics.
All statistical comparisons (other than those directly involved in the QTL analysis) were made by nonparametric Mann–Whitney rank sum tests. P values <0.05 were considered indicative of statistical significance. Variances are reported as standard errors of the mean.
Supplementary Material
Acknowledgments
We thank Eugene and Marcia Applebaum and the families of Merwyn and Dolly Dan and Ron and Susan Bernstein for their generous support. We acknowledge the help of the Mayo Biospecimens Accessioning Processing Core for the preparation of genomic DNA for genotyping and the Mayo Genotyping Shared Resource for performing the SNP genotyping. This work was supported by Grants NS048357 (to A.B.) and NS24180 (to M.R.) from the National Institutes of Health and Grants RG 3481A and CA1011A8 (to A.B.) and RG 3172A and CA1011A8 (to M.R.) from the National Multiple Sclerosis Society. K.S. was supported as a McDonald Fellow of the Multiple Sclerosis International Federation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906589107/DCSupplemental.
References
- 1.Kantarci OH, Weinshenker BG. Natural history of multiple sclerosis. Neurol Clin. 2005;23:17–38, v. doi: 10.1016/j.ncl.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Lucchinetti C, et al. Heterogeneity of multiple sclerosis lesions: Implications for the pathogenesis of demyelination. Ann Neurol. 2000;47:707–717. doi: 10.1002/1531-8249(200006)47:6<707::aid-ana3>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 3.Lucchinetti CF, Parisi J, Bruck W. The pathology of multiple sclerosis. Neurol Clin. 2005;23:77–105, vi. doi: 10.1016/j.ncl.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Pittock SJ, Lucchinetti CF. The pathology of MS: New insights and potential clinical applications. Neurologist. 2007;13:45–56. doi: 10.1097/01.nrl.0000253065.31662.37. [DOI] [PubMed] [Google Scholar]
- 5.Patrikios P, et al. Remyelination is extensive in a subset of multiple sclerosis patients. Brain. 2006;129:3165–3172. doi: 10.1093/brain/awl217. [DOI] [PubMed] [Google Scholar]
- 6.Franklin RJ. Why does remyelination fail in multiple sclerosis? Nat Rev Neurosci. 2002;3:705–714. doi: 10.1038/nrn917. [DOI] [PubMed] [Google Scholar]
- 7.Franklin RJ, Ffrench-Constant C. Remyelination in the CNS: From biology to therapy. Nat Rev Neurosci. 2008;9:839–855. doi: 10.1038/nrn2480. [DOI] [PubMed] [Google Scholar]
- 8.Bieber AJ, Ure DR, Rodriguez M. Genetically dominant spinal cord repair in a murine model of chronic progressive multiple sclerosis. J Neuropathol Exp Neurol. 2005;64:46–57. doi: 10.1093/jnen/64.1.46. [DOI] [PubMed] [Google Scholar]
- 9.Broman KW, Sen S. A Guide to QTL Mapping with R/qtl. Berlin: Springer; 2009. [Google Scholar]
- 10.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nikolaev A, McLaughlin T, O’Leary DD, Tessier-Lavigne M. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature. 2009;457:981–989. doi: 10.1038/nature07767. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Sättler MB, et al. Neuroprotective effects and intracellular signaling pathways of erythropoietin in a rat model of multiple sclerosis. Cell Death Differ. 2004;11(Suppl 2):S181–S192. doi: 10.1038/sj.cdd.4401504. [DOI] [PubMed] [Google Scholar]
- 13.Diem R, et al. Combined therapy with methylprednisolone and erythropoietin in a model of multiple sclerosis. Brain. 2005;128:375–385. doi: 10.1093/brain/awh365. [DOI] [PubMed] [Google Scholar]
- 14.Coetzee T, et al. Myelination in the absence of galactocerebroside and sulfatide: Normal structure with abnormal function and regional instability. Cell. 1996;86:209–219. doi: 10.1016/s0092-8674(00)80093-8. [DOI] [PubMed] [Google Scholar]
- 15.Luetteke NC, et al. Targeted inactivation of the EGF and amphiregulin genes reveals distinct roles for EGF receptor ligands in mouse mammary gland development. Development. 1999;126:2739–2750. doi: 10.1242/dev.126.12.2739. [DOI] [PubMed] [Google Scholar]
- 16.Shaw MH, et al. A natural mutation in the Tyk2 pseudokinase domain underlies altered susceptibility of B10.Q/J mice to infection and autoimmunity. Proc Natl Acad Sci USA. 2003;100:11594–11599. doi: 10.1073/pnas.1930781100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hafler DA, et al. International Multiple Sclerosis Genetics Consortium. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 18.Oksenberg JR, Baranzini SE, Sawcer S, Hauser SL. The genetics of multiple sclerosis: SNPs to pathways to pathogenesis. Nat Rev Genet. 2008;9:516–526. doi: 10.1038/nrg2395. [DOI] [PubMed] [Google Scholar]
- 19.Kantarci OH, de Andrade M, Weinshenker BG. Identifying disease modifying genes in multiple sclerosis. J Neuroimmunol. 2002;123:144–159. doi: 10.1016/s0165-5728(01)00481-7. [DOI] [PubMed] [Google Scholar]
- 20.Ramagopalan SV, Deluca GC, Degenhardt A, Ebers GC. The genetics of clinical outcome in multiple sclerosis. J Neuroimmunol. 2008;201-202:183–199. doi: 10.1016/j.jneuroim.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 21.Feigin I, Popoff N. Regeneration of myelin in multiple sclerosis. The role of mesenchymal cells in such regeneration and in myelin formation in the peripheral nervous system. Neurology. 1966;16:364–372. doi: 10.1212/wnl.16.4.364. [DOI] [PubMed] [Google Scholar]
- 22.Périer O, Grégoire A. Electron microscopic features of multiple sclerosis lesions. Brain. 1965;88:937–952. doi: 10.1093/brain/88.5.937. [DOI] [PubMed] [Google Scholar]
- 23.Prineas JW, Connell F. Remyelination in multiple sclerosis. Ann Neurol. 1979;5:22–31. doi: 10.1002/ana.410050105. [DOI] [PubMed] [Google Scholar]
- 24.Prineas JW, Barnard RO, Kwon EE, Sharer LR, Cho ES. Multiple sclerosis: Remyelination of nascent lesions. Ann Neurol. 1993;33:137–151. doi: 10.1002/ana.410330203. [DOI] [PubMed] [Google Scholar]
- 25.Pittock SJ, et al. Clinical implications of benign multiple sclerosis: A 20-year population-based follow-up study. Ann Neurol. 2004;56:303–306. doi: 10.1002/ana.20197. [DOI] [PubMed] [Google Scholar]
- 26.Pittock SJ, Rodriguez M. Benign multiple sclerosis: A distinct clinical entity with therapeutic implications. Curr Top Microbiol Immunol. 2008;318:1–17. doi: 10.1007/978-3-540-73677-6_1. [DOI] [PubMed] [Google Scholar]
- 27.Ramsaransing GS, De Keyser J. Benign course in multiple sclerosis: A review. Acta Neurol Scand. 2006;113:359–369. doi: 10.1111/j.1600-0404.2006.00637.x. [DOI] [PubMed] [Google Scholar]
- 28.Wong RW, Guillaud L. The role of epidermal growth factor and its receptors in mammalian CNS. Cytokine Growth Factor Rev. 2004;15:147–156. doi: 10.1016/j.cytogfr.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 29.Benoit BO, et al. Neurotrophin channeling of neural progenitor cell differentiation. J Neurobiol. 2001;46:265–280. [PubMed] [Google Scholar]
- 30.Fricker-Gates RA, et al. EGF infusion stimulates the proliferation and migration of embryonic progenitor cells transplanted in the adult rat striatum. Exp Neurol. 2000;165:237–247. doi: 10.1006/exnr.2000.7482. [DOI] [PubMed] [Google Scholar]
- 31.Mokry J, et al. Differentiation of neural stem cells into cells of oligodendroglial lineage. Acta Medica. 2007;50:35–41. [PubMed] [Google Scholar]
- 32.Ohta Y, et al. Intrathecal injection of epidermal growth factor and fibroblast growth factor 2 promotes proliferation of neural precursor cells in the spinal cords of mice with mutant human SOD1 gene. J Neurosci Res. 2006;84:980–992. doi: 10.1002/jnr.21017. [DOI] [PubMed] [Google Scholar]
- 33.Ivkovic S, Canoll P, Goldman JE. Constitutive EGFR signaling in oligodendrocyte progenitors leads to diffuse hyperplasia in postnatal white matter. J Neurosci. 2008;28:914–922. doi: 10.1523/JNEUROSCI.4327-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knapp PE, Adams MH. Epidermal growth factor promotes oligodendrocyte process formation and regrowth after injury. Exp Cell Res. 2004;296:135–144. doi: 10.1016/j.yexcr.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 35.Crang AJ, Gilson JM, Li WW, Blakemore WF. The remyelinating potential and in vitro differentiation of MOG-expressing oligodendrocyte precursors isolated from the adult rat CNS. Eur J Neurosci. 2004;20:1445–1460. doi: 10.1111/j.1460-9568.2004.03606.x. [DOI] [PubMed] [Google Scholar]
- 36.Matthieu JM, Comte V, Tosic M, Honegger P. Myelin gene expression during demyelination and remyelination in aggregating brain cell cultures. J Neuroimmunol. 1992;40:231–234. doi: 10.1016/0165-5728(92)90138-b. [DOI] [PubMed] [Google Scholar]
- 37.Sibilia M, Steinbach JP, Stingl L, Aguzzi A, Wagner EF. A strain-independent postnatal neurodegeneration in mice lacking the EGF receptor. EMBO J. 1998;17:719–731. doi: 10.1093/emboj/17.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sibilia M, Wagner EF. Strain-dependent epithelial defects in mice lacking the EGF receptor. Science. 1995;269:234–238. doi: 10.1126/science.7618085. [DOI] [PubMed] [Google Scholar]
- 39.Aguirre A, Dupree JL, Mangin JM, Gallo V. A functional role for EGFR signaling in myelination and remyelination. Nat Neurosci. 2007;10:990–1002. doi: 10.1038/nn1938. [DOI] [PubMed] [Google Scholar]
- 40.Yamaoka K, et al. The janus kinases (JAKs) Genome Biol. 2004;5:253. doi: 10.1186/gb-2004-5-12-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tokumasa N, et al. Expression of Tyk2 in dendritic cells is required for IL-12, IL-23, and IFN-gamma production and the induction of Th1 cell differentiation. Blood. 2007;110:553–560. doi: 10.1182/blood-2006-11-059246. [DOI] [PubMed] [Google Scholar]
- 42.Spach KM, et al. A single nucleotide polymorphism in Tyk2 controls susceptibility to experimental allergic encephalomyelitis. J Immunol. 2009;182:7776–7783. doi: 10.4049/jimmunol.0900142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minegishi Y, et al. Human tyrosine kinase 2 deficiency reveals its requisite roles in multiple cytokine signals involved in innate and acquired immunity. Immunity. 2006;25:745–755. doi: 10.1016/j.immuni.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 44.Watford WT, O’Shea JJ. Human tyk2 kinase deficiency: Another primary immunodeficiency syndrome. Immunity. 2006;25:695–697. doi: 10.1016/j.immuni.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 45.Ban M, et al. Wellcome Trust Case-Control Consortium (WTCCC) Replication analysis identifies TYK2 as a multiple sclerosis susceptibility factor. Eur J Hum Genet. 2009;17:1309–1313. doi: 10.1038/ejhg.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Darvasi A. Interval-specific congenic strains (ISCS): An experimental design for mapping a QTL into a 1-centimorgan interval. Mamm Genome. 1997;8:163–167. doi: 10.1007/s003359900382. [DOI] [PubMed] [Google Scholar]
- 47.Shimoda K, et al. Tyk2 plays a restricted role in IFN alpha signaling, although it is required for IL-12-mediated T cell function. Immunity. 2000;13:561–571. doi: 10.1016/s1074-7613(00)00055-8. [DOI] [PubMed] [Google Scholar]
- 48.McGavern DB, Murray PD, Rodriguez M. Quantitation of spinal cord demyelination, remyelination, atrophy, and axonal loss in a model of progressive neurologic injury. J Neurosci Res. 1999;58:492–504. doi: 10.1002/(sici)1097-4547(19991115)58:4<492::aid-jnr3>3.0.co;2-p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shifman S, et al. A high-resolution single nucleotide polymorphism genetic map of the mouse genome. PLoS Biol. 2006;4:e395. doi: 10.1371/journal.pbio.0040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 51.Lander E, Kruglyak L. Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.