Abstract
Benzalacetone synthase (BAS), a plant-specific type III polyketide synthase (PKS), catalyzes a one-step decarboxylative condensation of malonyl-CoA and 4-coumaroyl-CoA to produce the diketide benzalacetone. We solved the crystal structures of both the wild-type and chalcone-producing I207L/L208F mutant of Rheum palmatum BAS at 1.8 Å resolution. In addition, we solved the crystal structure of the wild-type enzyme, in which a monoketide coumarate intermediate is covalently bound to the catalytic cysteine residue, at 1.6 Å resolution. This is the first direct evidence that type III PKS utilizes the cysteine as the nucleophile and as the attachment site for the polyketide intermediate. The crystal structures revealed that BAS utilizes an alternative, novel active-site pocket for locking the aromatic moiety of the coumarate, instead of the chalcone synthase’s coumaroyl-binding pocket, which is lost in the active-site of the wild-type enzyme and restored in the I207L/L208F mutant. Furthermore, the crystal structures indicated the presence of a putative nucleophilic water molecule which forms hydrogen bond networks with the Cys-His-Asn catalytic triad. This suggested that BAS employs novel catalytic machinery for the thioester bond cleavage of the enzyme-bound diketide intermediate and the final decarboxylation reaction to produce benzalacetone. These findings provided a structural basis for the functional diversity of the type III PKS enzymes.
Keywords: biosynthesis, enzyme, polyketide
Benzalacetone synthase (BAS), a member of the plant-specific chalcone synthase (CHS) superfamily of type III polyketide synthases (PKSs) (1–3), catalyzes the one-step decarboxylative condensation of 4-coumaroyl-CoA with malonyl-CoA to produce a diketide benzalacetone, 4-(4-hydroxyphenyl)but-3-en-2-one (Fig. 1A and Fig. S1) (4). BAS is a crucial enzyme in the biosynthesis of the C6-C4 moiety of biologically active phenylbutanoids such as the antiinflammatory glucoside lindleyin in rhubarb, and raspberry ketone, the characteristic aroma of raspberry fruit. In contrast, the typical type III PKSs catalyze iterative condensations of malonyl-CoA with a CoA-linked starter molecule (Fig. S1) (1–3). For example, CHS and stilbene synthase (STS), sharing ≈70% amino acid sequence identity with BAS, catalyze sequential condensations of 4-coumaroyl-CoA and three molecules of malonyl-CoA to produce the tetraketides naringenin chalcone and resveratrol, respectively. Recent crystallographic analyses of the type III PKSs have revealed that the functional diversity of the CHS-superfamily enzymes is principally derived from the small modifications of the active-site architecture (5–12).
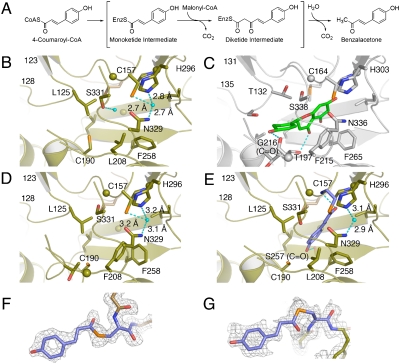
Fig. 1.
Enzyme reaction and active-site structure of R. palmatum BAS. (A) Proposed mechanism for the formation of benzalacetone by BAS. The active-site structures of wild-type BAS apo (B), M. sativa CHS (C), BAS I207L/L208F mutant (D), and wild-type BAS with the covalently bound coumarate (E). The coumarate and naringenin molecules are shown as blue and green stick models, respectively. The water molecule and the hydrogen bonds are indicated with light-blue spheres and dotted lines, respectively. The Fo-Fc density map of the monoketide intermediate covalently bound to the catalytic Cys157 (F) in monomer A, and in monomer B of wild-type BAS, countered at 2.0 s (G).
We previously reported that the diketide-forming activity of Rheum palmatum BAS is attributed to the characteristic substitution of the active-site Phe215; the conserved CHS residues 214LF are uniquely replaced by IL in R. palmatum BAS (numbering in Medicago sativa CHS, Fig. S2) (13). The conformationally flexible Phe215, located at the junction between the active-site cavity and the CoA-binding tunnel, is absolutely conserved in all of the known type III PKSs, and is considered to facilitate the decarboxylation of malonyl-CoA by maintaining the orientations of substrates and intermediates during the sequential condensation reactions (5). Indeed, the BAS I207L/L208F mutant (numbering in BAS) restored the chalcone-forming activity, supporting the hypothesis that the replacement of Phe208 in BAS (corresponding to Phe215 in M. sativa CHS) accounts for the interruption of the polyketide chain elongation at the diketide stage (13, 14).
We now present the crystal structures of both the wild-type and chalcone-producing I207L/L208F mutant of R. palmatum BAS at 1.8 Å resolution. In addition, we solved the crystal structure of the wild-type enzyme, in which a monoketide coumarate intermediate is covalently bound to the catalytic cysteine residue, at 1.6 Å resolution. This is direct evidence that a type III PKS utilizes the cysteine as the nucleophile and as the attachment site for the polyketide intermediate. The crystal structures revealed that BAS utilizes an alternative active-site pocket for locking the aromatic moiety of the coumarate, instead of the CHS’s “coumaroyl-binding pocket”, and indicated the presence of a putative nucleophilic water molecule that forms hydrogen bond networks with the Cys-His-Asn catalytic triad. These findings have led to a proposal for a unique mechanism of the enzyme reaction and provided a structural basis for the functional diversity of the type III PKS enzymes.
Results and Discussion
Overall Structure of BAS.
The homodimeric apo-structure of BAS consists of residues 8–383 of monomer A, and residues 8–382 of monomer B (Fig. S3). Both monomers are nearly identical to each other, with root-mean-square deviations (RMSDs) of 0.44 Å. Each monomer binds the other monomer with a twofold axis, thereby forming a biologically active, symmetric dimer. Upon dimerization, each monomer buries 2200 Å2 of its surface, and Met130 protrudes into the other monomer by the formation of a cis-peptide bond between Met130 and Pro131 on a loop, to complete the wall of an active-site cavity in each monomer. The catalytic triad of Cys157, His296 and Asn329 is buried deep within each monomer, and sits at the intersection of a traditional 16 Å-long CoA-binding tunnel and a large internal cavity, in a location and orientation very similar to those of the other plant type III PKSs (5–7, 10). The CoA-binding tunnel is connected to the protein surface, thus facilitating the entrance of the substrate into the catalytic center. The overall structures of BAS are highly homologous to those of the previously reported plant type III PKSs, including M. sativa CHS (5) and Pinus sylvestris STS (7), with RMSDs of 0.66 Å and 0.73 Å, respectively, for the Cα-atoms (Fig. S3).
Active-Site Architecture of BAS.
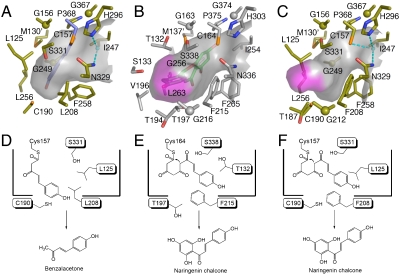
As previously reported (13), the benzalacetone-forming activity of R. palmatum BAS is attributed to the characteristic substitution of the conserved active-site Phe215 (numbering in M. sativa CHS) with leucine. Indeed, a comparison of the active-site structure of R. palmatum BAS with that of M. sativa CHS revealed that both the location and angle of the planar residue Phe215 of M. sativa CHS are well conserved in Leu208 of BAS, but the Cδ carbon of the Leu208 side-chain protrudes into the CHS’s active-site cavity, thereby causing a substantial contraction of the active-site cavity of BAS (Fig. 1B and C). In addition, in R. palmatum BAS, the terminal hydroxyl group of Ser331 is isometric with that of the corresponding Ser338 in M. sativa CHS. The serine residue neighboring the catalytic cysteine is considered to be crucial for modulation of the catalytic activity (7). In fact, we previously reported that the BAS S331 V mutant exhibited a twofold increase in the kcat/KM value for the benzalacetone-producing activity, which suggested that the residue is important for providing steric guidance for the diketide formation reaction (14). In the BAS crystal structure, the hydroxyl group of Ser331 rotates by nearly 120°, thereby blocking the entrance of the CHS’s coumaroyl-binding pocket (5). Moreover, a structural comparison with M. sativa CHS revealed significant backbone changes in the loops corresponding to residues 123–128 in R. palmatum BAS and the residues 131–135 in M. sativa CHS, respectively (RMSDs of 1.00 Å for the Cα-atoms, Fig. 1B and C). This movement is caused by various neighboring amino acid substitutions at the loop, most notably the replacement of Val98 in M. sativa CHS with the bulkier side-chain Gln in BAS, with the result that the backbone torsion angle of Leu125 (-51, -42), as compared with that of Thr132 (-112, 6), is shifted by a ϕ angle of -61° and a ψ angle of +48° toward Leu208 in BAS. A similar situation has also been found in the crystal structure of resveratrol-forming P. sylvestris STS (7), which will be discussed later. The conformational differences of Leu208 and Ser331 cause the loss of the CHS’s coumaroyl-binding pocket (5) from the BAS active-site cavity. As a result, the total cavity volume (350 Å3) of the active site of BAS is much smaller than that ofM. sativa CHS (750 Å3) (Fig. 2A and B), suggesting that the steric contraction leads to the shorter diketide-forming activity of BAS (Fig. 2D and E).
Fig. 2.
Surface and schematic representations of the active-site cavities of wild-type BAS (A and D), M. sativa CHS (B and E) , and BAS I207L/L208F mutant (C and F). The bottoms of the “coumaroyl-binding pocket” are highlighted as purple surfaces. The covalently bound coumarate, the water molecules and the hydrogen bonds are colored as in Fig. 1.
Phe258 in R. palmatum BAS and the corresponding Phe265 in M. sativa CHS are located at the entrance of the active-site, as in the case of the above mentioned Leu208 in BAS and Phe215 in CHS. The backbone torsion angle of Phe258 (-131, 118), in comparison with that of Phe265 (-123, 131), is slightly shifted by a ϕ angle of +8° and a ψ angle of +13°, and the aromatic moiety of Phe258 protrudes more toward Leu208, as compared to that of Phe265 of M. sativa CHS, and forms a hydrophobic interaction with the side-chain of Leu208 (Fig. 1B and C, and Fig. S4). Although positional differences of this gatekeeper phenylalanine are often observed in the type III PKSs (7), this conformational change appears to be caused by the F208L substitution in BAS. The total area of the active-site entrance of BAS is thus ≈34 Å2, which is twice as large as that of M. sativa CHS (17 Å2) (Fig. S4). A similar widening of the active-site entrance has also been reported for the structure of the M. sativa F215S CHS mutant, which accepts the bulky N-methylanthraniloyl-CoA as a starter substrate (15). Noel and coworkers (15) have proposed that the F215S substitution opens the space at the cavity entrance to accommodate the methylamine moiety of N-methylanthraniloyl-CoA and facilitates the positioning of the thioester-carbonyl moiety next to the Cys-His-Asn catalytic triad. These observations may also account for the previous report that R. palmatum BAS readily accepts N-methylanthraniloyl-CoA as a starter substrate to efficiently produce N-methyl-4-hydroxy-2(1H)-quinolone after condensation with one molecule of malonyl-CoA (16).
Restoration of Chalcone-Forming Activity in the BAS I207L/L208F Mutant.
The X-ray crystal structure of the BAS I207L/L208F mutant clearly demonstrated that both the location and orientation of the residues lining the active-site cavity in the mutant are almost perfectly conserved with those in the wild-type enzyme (Fig. 1B and D), whereas those of Ile207, Leu208, Phe258 and Ser331 in the mutant were sterically altered to those of CHS (Fig. 1C and D). Notably, the conformational changes at Phe208 and Ser331 in the mutant are caused by the replacement of Leu208 with the bulkier and planar Phe, which restored the coumaroyl-binding pocket within the active-site of the mutant (Fig. 2B and C). Thus, the Ile207L/Leu208F substitutions open a gate to the buried coumaroyl-binding pocket, thereby increasing the polyketide chain elongation up to three condensations with malonyl-CoA, and leading to the recovery of the chalcone-forming activity in the mutant (Fig. 2E and F). It is remarkable that the functional diversity of the type III PKS evolved from the simple steric modulation of a single, chemically inert, residue lining the active-site cavity.
Crystal Structure of Wild-Type BAS with Covalently Bound Coumarate.
To gain further insights into the polyketide formation machinery, we soaked the apo crystals of BAS with the sole initial substrate, 4-coumaroyl-CoA (see Methods for details). The crystal structure unambiguously revealed that a monoketide intermediate (i.e., 4-coumaroyl thioester) is covalently bound to the SH group of the catalytic Cys157 in each monomer, which had never been observed in any type III PKS structures (Fig. 1F and G, and Fig. S3). This is direct evidence that the type III PKS enzyme utilizes the cysteine residue as the nucleophile and as the attachment site for the polyketide intermediate. The coumarate occupies the CoA-binding tunnel in monomer A, while it lies in the active-site cavity in monomer B (Fig. S3). To accommodate the coumarate in the active-site, the SH group of Cys190 must rotate by about 93°, as compared with that of the BAS apo structure (Fig. 1B and E).
Amazingly, the aromatic moiety of the coumarate in monomer B is locked into an alternative, unique active-site pocket, which protrudes into the “floor” of the conventional CHS active site. The phenolic OH of the coumarate forms a hydrogen bond with the backbone carbonyl oxygen of Ser257 (Fig. 1E). This is in sharp contrast with the active-site structure ofM. sativa CHS in which the coumaroyl moiety of naringenin is locked into the coumaroyl-binding pocket, with its phenolic OH forming a hydrogen bond with the backbone carbonyl oxygen of Gly216, and with the carbonyl oxygen of naringenin interacting with the hydroxyl of Thr197 (Fig. 1C) (5). These observations suggest that BAS utilizes the unique active-site pocket to lock the coumaroyl moiety for the production of benzalacetone, and the chain elongation is interrupted at the diketide stage because of the steric contraction of the active-site cavity (Fig. 2D).
Mechanism of Thioester Bond Cleavage and Decarboxylation.
Considering the conservation of the overall folding and the catalytic triad, it is conceivable that BAS utilizes similar catalytic machinery as that used by CHS for the initiation of the enzyme reaction and the decarboxylative Claisen-type condensation of malonyl-CoA (2, 3). Here, the most interesting points are the catalytic processes of the thioester bond cleavage of the enzyme-bound diketide intermediate and the final decarboxylation reaction that produces the C6-C4 benzalacetone.
It should be noted that the crystal structure of BAS revealed slight conformational changes in the loop between residues 123–128, as compared to those in CHS and STS (Fig. S5). Recent structural analyses ofP. sylvestris STS demonstrated that the backbone change of the loop leads to the subtle displacement of Thr132, resulting in the rearrangement of the so-called “aldol-switch” hydrogen bond network, including Thr132 and a nucleophilic water molecule neighboring the catalytic Cys164 at the active-site center (numbering in M. sativa CHS, Fig. S5) (7). Noel and Schröder (7) thus proposed that the internal aldol cyclization and sequential decarboxylation reactions in STS are principally triggered by the cleavage of the thioester bond of the enzyme-bound intermediate, by the nucleophilic attack of the activated water molecule. In contrast, careful examination of the BAS crystal structure precluded the presence of such hydrogen bond networks, since Thr132 in STS is replaced with the hydrophobic Leu in BAS (Fig. 1B and E, and Fig. S5). Instead, the structure clearly demonstrated the presence of a distinct, activated water molecule which forms hydrogen bond networks with the Cys-His-Asn catalytic triad (Fig. 1B and E, and Fig. S5). Interestingly, the water molecule is also sterically conserved in the structure of the BAS I207L/L208F mutant (Fig. 1E and Fig. S4).
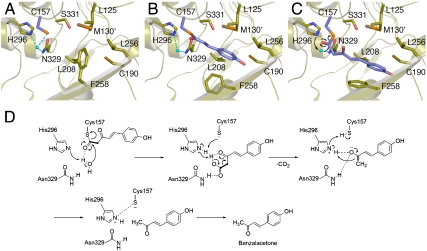
Since the chalcone-forming I207L/L208F mutant still produces benzalacetone (13), both the wild-type and mutant enzymes could possibly maintain the same catalytic machinery for the formation of benzalacetone. Furthermore, as described above, the BAS S331V mutant, in which the corresponding Ser338 in the aldol-switch hydrogen bond network is substituted with the hydrophobic Val, exhibited a twofold increase in the kcat/KM value for the benzalacetone-producing activity (14). In addition, we have recently successfully detected the 4-coumaroyl β-keto acid, 5-(4-hydroxyphenyl)-3-oxopent-4-enoic acid, in the reaction products of BAS (Fig. S6). All of these observations strongly suggest that BAS utilizes alternative and unique catalytic machinery, which is distinct from that of STS, for the thioester bond cleavage of the enzyme-bound intermediate and the final decarboxylation reaction, and that the β-keto acid produced by the nucleophilic attack of the water molecule, presumably activated by His296, subsequently undergoes decarboxylation to yield the C6-C4 benzalacetone (Fig. 3D). Thus, we propose that the decarboxylation of the β-keto acid proceeds via proton abstraction by His296, reactivated by the Cys157 thiolate, and formation of an enolate anion presumably stabilized by the His296-Asn329 oxyanion hole, just as in the case of the decarboxylation of malonyl-CoA (2). Finally, tautomerization to the keto form produces benzalacetone and restores the Cys157-His296 thiolate-imidazolium ion pair (Fig. 3D).
Fig. 3.
Proposed mechanism for the BAS enzyme reaction. The active-site structures of the apo (A) and the coumarate-bound BAS (B), respectively. Three-dimensional model for the diketide intermediate covalently bound to the catalytic cysteine. The putative nucleophilic water molecule is shown as a light-blue sphere. The dotted lines indicate the hydrogen bond (C). Proposed mechanism for the enzyme catalyzed hydrolysis and decarboxylation reaction (D).
Recently, 2′-oxoalkylresorcylic acid synthase (ORAS), which catalyzes the condensation of four molecules of malonyl-CoA with a long-chain acyl-CoA thioester (C16, C18 and C20) as a starter substrate to produce the pentaketide alkylresorcylic acid and resorcinol, has been characterized from Neurospora crassa (Fig. S1) (12, 17). The stable resorcinol acid is considered to be an intermediate which is further enzymatically converted into the final alkylresorcinol product by a decarboxylation reaction. In spite of the similar catalytic process to that of STS, the crystallographic analysis of ORAS revealed that, unlike STS, the enzyme structure does not share the aldol switch in the active-site cavity (12). In contrast, our structural analyses revealed that the location of the putative nucleophilic water molecule in BAS is nearly identical to that of the sulfinic oxygen of the oxidized catalytic cysteine in ORAS (Fig. S7). Therefore, it is quite probable that the sulfinic oxygen in ORAS can be regarded as an analog of the nucleophilic water molecule in BAS, and that the resorcinol acid is released from ORAS, as in the case of the β-keto acid in BAS.
In conclusion, the crystal structures revealed that BAS utilizes an alternative, unique active-site pocket for locking the aromatic moiety of the coumarate, instead of the coumaroyl-binding pocket, which is lost in the active site of the wild-type enzyme and restored in the I207L/L208F mutant. Furthermore, the crystal structures indicated the presence of a putative nucleophilic water molecule which forms hydrogen bond networks with the Cys-His-Asn catalytic triad. This suggested that BAS employs unique catalytic machinery for the thioester bond cleavage of the enzyme-bound diketide intermediate and the final decarboxylation reaction to produce benzalacetone. These findings provide a structural basis for the functional diversity of the type III PKS enzymes.
Methods
Chemicals.
4-coumaroyl-CoA was synthesized as described previously. Oligonucleotides were obtained from Hokkaido System Science Co., Ltd. Standard chemicals were obtained from Sigma-Aldrich and Hampton Research.
Expression and Purification.
Wild-type BAS was expressed in E. coli M15 and purified as described previously (18). The L207L/L208F mutant expression plasmid was constructed with a QuikChange Site-Directed Mutagenesis Kit (Stratagene), according to the manufacturer’s protocol, by using 5′-CCATGATAGGCCAA GCATTATTCGGCGATGGGGCTGC-3′ as the sense primer, 5′-GCAGCCCCATCGCC GAATAATGCTTGGCCTATCATGG-3′ as the antisense primer, and the wild-type BAS expression plasmid as the template. The mutant protein was expressed and purified by the same procedure used for the wild-type BAS, and was concentrated to 20 mg/mL in 20 mM HEPES-NaOH (pH7.5) buffer, containing 100 mM NaCl and 2 mM DTT.
Crystallization.
Crystallization of the wild-type BAS was performed as previously reported (18). The I207L/L208F mutant crystals were also obtained by the same crystallization methods as those used for the wild-type BAS. Both crystals were independently transferred to a reservoir solution with 20% (v/v) glycerol as a cryoprotectant, and were then flash-cooled at 100 K in a nitrogen-gas stream. For the monoketide intermediate-complexed crystal of the wild type, a single crystal, as described above, was captured in a nylon loop and transferred to the reservoir solution containing 2 mM 4-couamroyl-CoA. After an incubation at 20° for 2 d, the crystal was transferred to the same cryoprotectant as that used for the wild-type and mutant apo crystals, except that it also contained 4-coumaroyl-CoA.
Structure Determination.
Both X-ray diffraction datasets from the wild-type BAS crystals were collected at 100 K at BL24XU of SPring-8 (wavelength, 0.82656 Å), by using a Rigaku R-AXIS V imaging plate. The X-ray diffraction dataset from the mutant crystal was collected at 100 K at BL41XU of SPring-8 (wavelength, 1.00000 Å), by using an ADSC Quantum 210 CCD detector. All data were indexed, integrated, and scaled with the HKL2000 program (19).
The initial phases of the wild-type apo structure were determined by molecular replacement by using the BAS structure model generated by the SWISS-MODEL package (http://expasy.ch/spdpv/) based on the crystal structure of M. sativa CHS [Protein Data Bank (PDB) entry 1BQ6], as a search model. The molecular replacement was performed with the program Crystallography and NMR System (CNS) (20). Crystallographic refinement and model building were performed with CNS and XTALVIEW (21), respectively. Each refinement cycle was followed by model building using the σA-weighted 2Fo-Fc and Fo-Fc electron density maps. The water molecules were automatically placed into the difference electron density maps with XTALVIEW, and were retained or rejected on the basis of geometric criteria as well as their refined B-factors. After several rounds of model building and refinement, the final model was obtained. The other structures were solved by the same procedure as used for the model refinement of the BAS wild-type apo structure, except for the use of either the entire or I204L/L208F-substituted final models of the wild-type apo structure as the search model in the molecular replacement methods. Both the 2Fo-Fc and Fo-Fc maps indicated the presence of a portion of the monoketide intermediate covalently bound to the catalytic cysteine of each monomer in the intermediate-complexed structure, and the intermediate manually fits into the visible electron density. Each model consists of resides 8–383 of monomer A, and residues 8–382 of monomer B. The qualities of the final models were assessed with PROCHECK (22). Details of the data collection, processing, and structure refinement are summarized in Table S1. The cavity volume and the active-site entrance area were calculated by the program CASTP (http://cast.engr.uic.edu/cast/). All crystallographic figures were prepared with PyMOL (DeLano Scientific, http://www.pymol.org).
Enzyme Reaction.
The reaction mixture contained 54 μM of 4-coumaroyl-CoA, 108 μM of malonyl-CoA, and 20 μg of the purified wild-type enzyme in a final volume of 500 μL of 100 mM Tris-HCl buffer (pH 8.0) and 1 mM EDTA. Incubations were performed at 30 °C for 1 hr, and were stopped by the addition of 50 μL of 20% HCl. The products were then extracted with 3 mL of ethyl acetate. The products were separated by reverse-phase HPLC (JASCO 880) on a TSK-gel ODS-80Ts column (4.6 ÅE 150 mm, TOSOH), at a flow rate of 0.8 mL/ min. Gradient elution was performed with H2O and MeOH, both containing 0.1% TFA: 0–5 min, 30% MeOH; 5–17 min, linear gradient from 30 to 60% MeOH; 17–25 min, 60% MeOH; 25–27 min, linear gradient from 60 to 70% MeOH. Elutions were monitored by a multichannel UV detector (MULTI 340, JASCO) at 280 nm. UV spectra (198–400 nm) were recorded every 0.4 s. Online LC-ESIMS spectra were measured with an Agilent Technologies series 1100 HPLC coupled to a Bruker Daltonics esquire4000 ion-trap mass spectrometer fitted with an ESI source. HPLC separations were performed under the same conditions as described above. The ESI capillary temperature and the capillary voltage were 320 °C and 4.0 V, respectively. The tube lens offset was set at 20.0 V. All spectra were obtained in the positive mode over a mass range of m/z 50–500, and at a range of one scan every 0.2 s. The collision gas was helium, and the relative collision energy scale was set at 30.0% (1.5 eV).
Spectroscopic Data for (E)-5-(4-Hydroxyphenyl)-3-Oxopent-4-Enoic Acid (4-Coumaroyl Diketide β-Keto Acid).
UV λmax 283 nm; LC-ESIMS: MS, m/z 207 [M + H]+, MS/MS (precursor ion at m/z 207), m/z 147 [M + H-CO2-CH2]+; HRMS (FAB) found for [C11H11O4]+ 207.0668, calculated value 207.0657.
Supplementary Material
Acknowledgments.
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (I.A. and H.M.), by grants from The Naito Foundation (I.A.) and Takeda Science Foundation (H.M.), and from the National Project on Protein Structural and Functional Analyses (S.S. and T.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909982107/DCSupplemental.
References
- 1.Schröder J. Comprehensive Natural Products Chemistry. Vol 1. Oxford: Elsevier; 1999. pp. 749–771. [Google Scholar]
- 2.Austin MB, Noel JP. The chalcone synthase superfamily of type III polyketide synthases. Nat Prod Rep. 2003;20:79–110. doi: 10.1039/b100917f. [DOI] [PubMed] [Google Scholar]
- 3.Morita H, Abe I, Noguchi H. Comprehensive Natural Products Chemistry. Oxford, in press: Elsevier; 2009. [Google Scholar]
- 4.Abe I, Takahashi Y, Morita H, Noguchi H. Benzalacetone synthase. A novel polyketide synthase that plays a crucial role in the biosynthesis of phenylbutanones in Rheum palmatum. Eur J Biochem. 2001;268:3354–3359. doi: 10.1046/j.1432-1327.2001.02255.x. [DOI] [PubMed] [Google Scholar]
- 5.Ferrer JL, Jez JM, Bowman ME, Dixon RA, Noel JP. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat Struct Biol. 1999;6:775–784. doi: 10.1038/11553. [DOI] [PubMed] [Google Scholar]
- 6.Jez JM, et al. Structural control of polyketide formation in plant-specific polyketide synthases. Chem Biol. 2000;7:919–930. doi: 10.1016/s1074-5521(00)00041-7. [DOI] [PubMed] [Google Scholar]
- 7.Austin MB, Bowman ME, Ferrer JL, Schröder J, Noel JP. An aldol switch discovered in stilbene synthases mediates cyclization specificity of type III polyketide synthases. Chem Biol. 2004;11:1179–1164. doi: 10.1016/j.chembiol.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 8.Austin MB, et al. Crystal structure of a bacterial type III polyketide synthase and enzymatic control of reactive polyketide intermediates. J Biol Chem. 2004;279:45162–45174. doi: 10.1074/jbc.M406567200. [DOI] [PubMed] [Google Scholar]
- 9.Sankaranarayanan R, et al. A novel tunnel in mycobacterial type III polyketide synthase reveals the structural basis for generating diverse metabolites. Nat Struct Mol Biol. 2004;11:894–900. doi: 10.1038/nsmb809. [DOI] [PubMed] [Google Scholar]
- 10.Morita H, et al. Structural insight into chain-length control and product specificity of pentaketide chromone synthase from Aloe arborescens. Chem Biol. 2007;14:359–369. doi: 10.1016/j.chembiol.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Goyal A, et al. Structural insights into biosynthesis of resorcinolic lipids by a type III polyketide synthase in Neurospora crassa. J Struct Biol. 2008;162:411–421. doi: 10.1016/j.jsb.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Rubin-Pitel SB, et al. Distinct structural elements dictate the specificity of the type III pentaketide synthase from Neurospora crassa. Chem Biol. 2008;15:1079–1090. doi: 10.1016/j.chembiol.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abe I, Sano Y, Takahashi Y, Noguchi H. Site-directed mutagenesis of benzalacetone synthase. The role of the Phe215 in plant type III polyketide synthases. J Biol Chem. 2003;278:25218–25226. doi: 10.1074/jbc.M303276200. [DOI] [PubMed] [Google Scholar]
- 14.Abe T, et al. Structure function analysis of benzalacetone synthase from Rheum palmatum. Bioorg Med Chem Lett. 2007;17:3161–3166. doi: 10.1016/j.bmcl.2007.03.029. [DOI] [PubMed] [Google Scholar]
- 15.Jez JM, Bowman ME, Noel JP. Expanding the biosynthetic repertoire of plant type III polyketide synthases by altering starter molecule specificity. Proc Natl Acad Sci USA. 2002;99:5319–5324. doi: 10.1073/pnas.082590499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abe I, Abe T, Wanibuchi K, Noguchi H. Enzymatic formation of quinolone alkaloids by a plant type III polyketide synthase. Org Lett. 2006;8:6063–6065. doi: 10.1021/ol0625233. [DOI] [PubMed] [Google Scholar]
- 17.Funa N, Awakawa T, Horinouchi S. Pentaketide resorcylic acid synthesis by type III polyketide synthase from Neurospora crassa. J Biol Chem. 2007;282:14476–14481. doi: 10.1074/jbc.M701239200. [DOI] [PubMed] [Google Scholar]
- 18.Morita H, et al. Crystallization and preliminary crystallographic analysis of a plant type III polyketide synthase that produces benzalacetone. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;64:304–306. doi: 10.1107/S1744309108006775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 20.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 21.McRee DE. A visual protein crystallographic software system for X11/Xview. J Mol Graphics Modell. 1992;10:44–46. [Google Scholar]
- 22.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.