Abstract
EBV, a member of the herpes virus family, is a paradigm for human tumor viruses and a model of viral latency amenable for study in vitro. It induces resting human B lymphocytes to proliferate indefinitely in vitro and initially establishes a strictly latent infection in these cells. BZLF1, related to the cellular activating protein 1 (AP-1) family of transcription factors, is the viral master gene essential and sufficient to mediate the switch to induce the EBV lytic phase in latently infected B cells. Enigmatically, after infection BZLF1 is expressed very early in the majority of primary B cells, but its early expression fails to induce the EBV lytic phase. We show that the early expression of BZLF1 has a critical role in driving the proliferation of quiescent naïve and memory B cells but not of activated germinal center B cells. BZLF1’s initial failure to induce the EBV lytic phase relies on the viral DNA at first being unmethylated. We have found that the eventual and inevitable methylation of viral DNA is a prerequisite for productive infection in stably, latently infected B cells which then yield progeny virus lacking cytosine-phosphatidyl-guanosine (CpG) methylation. This progeny virus then can repeat EBV’s epigenetically regulated, biphasic life cycle. Our data indicate that the viral BZLF1 protein is crucial both to establish latency and to escape from it. Our data also indicate that EBV has evolved to appropriate its host’s mode of methylating DNA for its own epigenetic regulation.
Keywords: cytosine-phosphatidyl-guanosine methylation, latency, transcription, herpesvirus, reactivation
EBV infects resting primary human B cells and induces their indefinite proliferation in vitro. Expansion of these B cells gives rise to stable lymphoblastoid cell lines in which the virus resides latently and its genome is maintained extrachromosomally. The infected cells can express two sets of viral genes that relate either to the latent or lytic phases of the EBV life cycle (1). In latently infected B cells, a few viral genes, termed “latent genes,” are expressed that are instrumental for the induction and maintenance of cellular proliferation and viral latency; some of these latent genes also are associated causally with EBV’s being a human tumor virus (2).
Our findings indicate that only latently infected B cells can give rise to progeny virus, a process that requires the induction of a different set of viral genes. During de novo virus synthesis, about 70 different lytic EBV genes are expressed that asynchronously support viral DNA amplification and encode viral structural components to allow maturation of virus and release of progeny virus. The transition from viral latency to productive lytic infection is orchestrated by two ‘immediate-early’ genes (3), BZLF1 and BRLF1, which encode the transcription factors Zta (also called “Z,” “ZEBRA,” or “EB1”) and Rta (also called “R”), respectively. The former is a homolog of the activating protein 1 (AP-1) transcription factor family (4) and is a master regulator of the switch needed to induce the lytic phase of the EBV life cycle in latently infected B cells (5–7).
During latency, the viral lytic genes presumably are repressed by host-driven methylation of viral DNA, heterochromatin formation, and/or cellular transcriptional repressors (8). In the switch from the latent to the lytic phase of the EBV life cycle, Zta overturns this epigenetic silencing of the latent EBV genome. The signals that activate the expression of BZLF1 in latently infected B cells are assumed to be linked to antigen-mediated stimulation of the B-cell receptor signaling pathway (1, 9).
We have examined the fundamental events in the EBV life cycle and find that EBV first establishes a nonproductive, latent infection in B lymphocytes. About 2 weeks postinfection (p.i.) EBV first evolves to support its own virion synthesis in these cells. Central to this finding is the viral gene BZLF1, which can transactivate viral promoters depending on their status of cytosine-phosphatidyl-guanosine (CpG) methylation. Previous work indicated that the BZLF1 protein can bind in a sequence-specific manner to certain DNA motifs with methylated CpG dinucleotides (10). These earlier findings are consistent with Zta’s overcoming a repressed state of latent viral genomes by virtue of their being highly CpG-methylated and thereby inducing the EBV lytic phase in latently infected cells (11, 12). Here we show that Zta performs a second, unexpected, but critical role during the initiation of viral latency in the absence of CpG methylation.
Results
The Majority of Infected B Cells Initially Express EBV Immediate-Early Genes.
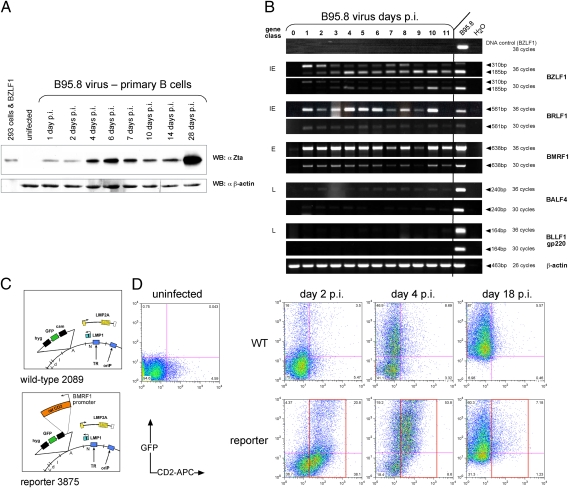
We asked if the expression of lytic viral genes occurred early after infection of primary human B cells with the B95.8 strain of EBV. Immunoblotting indicated that Zta was detectably expressed as early as 24 h p.i. (Fig. 1A), but immunoblotting and the RT-PCR data (Fig. 1B) could not distinguish between all infected B cells and the fraction of them supporting the expression of EBV immediate-early genes. We addressed this uncertainty with an engineered reporter strain of EBV. It monitors the expression of Zta in single infected cells by its expression of the rat CD2 surface receptor gene from the viral BMRF1 promoter (Fig. 1C). BMRF1 is an early viral gene, encodes the EA-D protein, and is directly transactivated by Zta and Rta (13). CD2-positive cells reached a maximum on day 4 p.i. (Fig. 1D). On day 18 p.i. CD2 surface expression was reduced, but all cells continued to express the GFP gene also encoded by the virus (Fig. 1D). Primary B cells infected with the B95.8 strain of EBV support these findings (Fig. S1).
Fig. 1.
The majority of infected B cells express EBV immediate-early genes. (A) Western blot immunodetection of Zta in primary B cells infected with the B95.8 strain of EBV at different time points after infection. HEK293 cells transiently transfected with a BZLF1 expression plasmid serve as positive control. β-actin signals served as loading control. (B) By semiquantitative RT-PCR analysis, B95.8 EBV-infected B cells detectably express the immediate-early genes BZLF1 (a fully spliced and an unspliced transcript) and BRLF1 and the early gene BMRF1 but do not express the late genes BALF4 and BLLF1 essential for virus synthesis during the first 11 days of infection. (C) Construction of the reporter EBV with the rat CD2 gene expressed from the viral BMRF1 promoter. Shown are parts of the wild-type (2089) and reporter (3875) EBV genomes and their functionally relevant genes (26). (D) The majority of GFP-positive B cells infected with the BMRF1 reporter EBV strain express rat CD2 very early after infection.
We assayed representative members of all three classes of lytic viral genes by semiquantitative RT-PCR analysis (Fig. 1B and Table S1). Two members of the class of late genes, BALF4 and BLLF1, encoding the viral glycoproteins gp110 (also called “gp125/VCA”) and gp350/220, respectively, were not expressed detectably. The failure to express both glycoproteins would block the synthesis of infectious virus (14–16).
EBV-Infected Primary B Cells Do Not Release Virus Progeny Early After Infection.
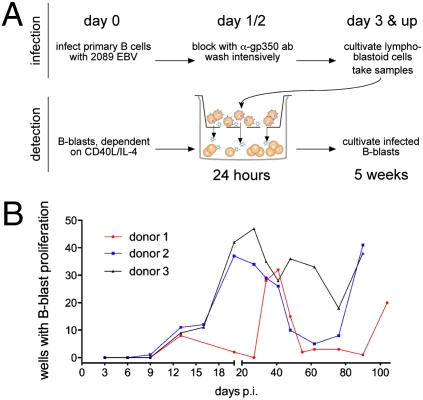
EBV differs from other herpes viruses in its efficient establishment of latent infections in B cells infected in vitro. To understand the events during this establishment, we asked whether in vitro infection of primary human B cells with B95.8 EBV and the early expression of BZLF1 might result in EBV-infected B cells undergoing productive infection despite the failure to detect expression of essential late genes (Fig. 1B). We used an assay to detect progeny virus based on a newly developed cell line (17) that is a sensitive indicator for infectious EBV (Fig. 2A and Fig. S2). Experiments with primary B cells from three donors clearly indicated that no virus is synthesized for up to 9 days p.i., but infectious EBV becomes detectable starting at day 13 and continues for at least 3 months (Fig. 2B).
Fig. 2.
EBV-infected primary B cells do not release virus progeny early after infection. (A) Experimental flowchart. (Upper) Primary B cells were infected with wild-type EBV (2089) on day 0 with an MOI of 0.1 and were cultivated for 24 h. One and 2 days p.i., the infected cells were incubated with a neutralizing α-gp350 antibody (ab) (Fig. S2), washed, and cultivated further. Samples were taken beginning on day 3 p.i. (Lower) The infected B cells were placed on a porous sieve above indicator B blasts for 24 h. B-blast proliferation is dependent on either CD40L/IL-4 stimulation or infection with EBV. The B blasts were cultured in 96-well cluster dishes in the absence of CD40L/IL-4. After 5 weeks the number of wells with proliferating B blasts was counted. (B) Detection of infectious progeny virus indicates a failure of early de novo virus synthesis in EBV-infected primary B cells. Progeny virus first became detectable at day 13 p.i. and were detectable for up to 3 months but varied cyclically in the infected B-cell samples from three different donors.
Analysis of CpG Methylation of Virion and Genomic EBV DNA.
In eukaryotes, transcriptional regulation of genes involves epigenetic changes such as DNA methylation and modifications of histone tails. We explored the possibility that such epigenetic changes underlie the seemingly contradictory activities of BZLF1 early and late in infection. The extensive methylation of viral genomes such as EBV during latent infection has been associated with a repression of their transcription (see refs.18 and 19 for a recent review). However, as with herpes simplex virus (20), the EBV virion DNA likely does not carry methylated CpG sites (21). We therefore assessed the levels of methylation of EBV DNA in B cells infected in vitro with B95.8 EBV via methylation-sensitive restriction enzymes and Southern blot hybridization. These analyses indicated that virion DNA was not detectably methylated but became increasingly methylated in infected B cells over the following 4 weeks (Fig. S3A).
The use of methyl-sensitive endonucleases and Southern blot hybridizations provides very detailed data for single CpGs but cannot practically cover the whole EBV genome, given its size of about 170 kbp. We therefore also used methylated DNA immunoprecipitation (MeDIP) analysis to survey the entire EBV genome (22). One week p.i. with B95.8 virus, little if any enrichment of CpG-methylated EBV DNA was detected by MeDIP, but after 2 weeks, there was a low, general increase in the ratio of methylated DNA. This trend continued and yielded a reproducible pattern with some regions of EBV DNA showing a higher level of CpG methylation than others (Fig. S3B). Interestingly, the degree of preferred CpG methylation does not correlate with the occurrence of CpG dinucleotide motifs in the EBV sequence (Fig. S3C). Only the Cp promoter, the region encoding the noncoding, highly expressed EBER RNA, the plasmid origin of DNA replication, oriP, and a region upstream of nucleotide coordinate 150,000 with no assigned function are spared eventually being methylated (Fig. S3B). The epigenetic patterns seen in lymphoblastoid cells infected in vitro differ from those of B cells infected with EBV in vivo (23), as found by others (24) and in this study.
Virion Synthesis Is a Function of the Epigenetic State of EBV DNA.
These experiments indicated that methylation of genomic EBV DNA is a gradual process that progresses during latency. Confirming an earlier report (25), our initial experiments indicated that only after a lag phase of about 2 weeks did cells early in latent infection support de novo virus synthesis, which subsequently increased markedly (Fig. 2B). We therefore asked whether these two processes might be functionally linked. To assess this possibility, we used HEK293 cells. They can be stably transfected with recombinant EBV DNA, establish a tightly latent infection similar to that in infected B cells, and support productive infection when BZLF1 is expressed in them (26). We therefore asked if transient cotransfection of Escherichia coli-derived, recombinant EBV DNA (p2089) with an ectopically expressed BZLF1 gene yields a productive infection in HEK293 cells. Surprisingly, it did not (see below).
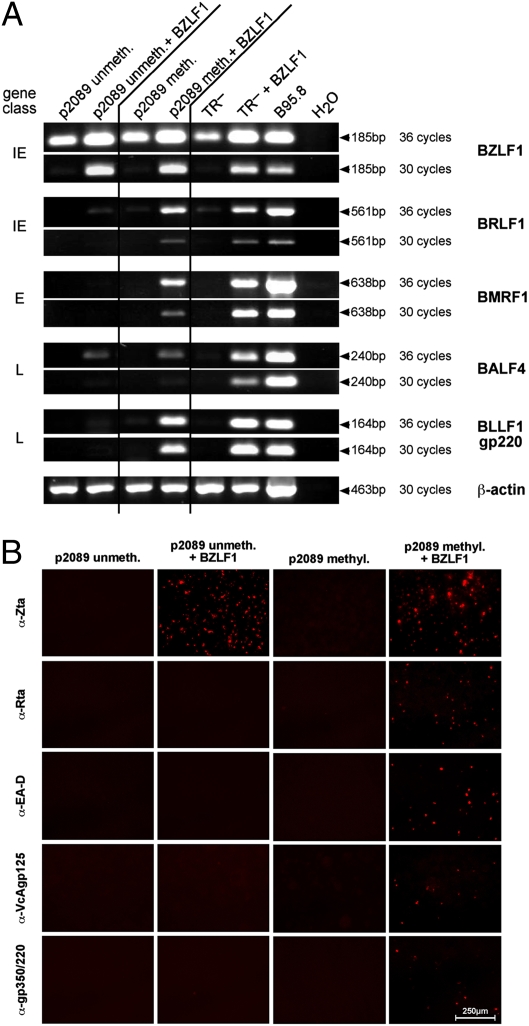
We analyzed HEK293 cells transiently cotransfected with BZLF1 and p2089 EBV DNA by RT-PCR and immunostaining for selected viral transcripts and proteins (Fig. 3; controls are shown in Fig. S4 and Fig. S5). Three days postinfection none of the tested viral lytic genes were detectably expressed, whether or not the BZLF1 expression plasmid was cotransfected (Fig. 3). The supernatants of the transfected HEK293 cells were probed for progeny virus using the B-blast indicator cell line as in Fig. 2. No progeny virus could be detected (Table 1).
Fig. 3.
Virion synthesis is a function of the epigenetic state of EBV DNA. (A) RT-PCR analysis of RNA prepared from HEK293 cells that had been transiently transfected for 3 days with EBV DNA (p2089) prepared from E. coli (unmeth.) or treated in vitro with the methyltransferase M.SssI (meth.). As indicated, p2089 DNA was cotransfected with a BZLF1 expression plasmid. Transcripts of essential viral lytic genes (BRLF1, BMRF1, and BLLF1) are detectable only when p2089 DNA is CpG-methylated before transfection. The genes are indicated on the right; the gene classes are given on the left. E, early; IE, immediate-early L, late; TR–, HEK293 cells stably transfected with EBV, which served as a positive control for the induction of the lytic phase. (B) Immunostaining of transiently transfected HEK293 cells with antibodies detecting Zta, Rta, EA-D, VcAgp125, and gp350/220 encoded by the viral genes BZLF1, BRLF1, BMRF1, BALF4, and BLLF1, respectively. Controls are found in Fig. S5.
Table 1.
Detection of progeny virus upon transient transfection of unmethylated or in vitro CpG-methylated genomic EBV DNA isolated from E. coli
| p2089 maxi-EBV DNA | BZLF1 expression plasmid | Median # wells (range) |
| Unmethylated* | – | 0 (0/0) |
| + | 0 (0/0) | |
| CpG methylated† | – | 0 (0/2) |
| + | 18 (2/48) |
*Data from 12 independent experiments.
†Data from 10 independent experiments.
We therefore determined whether the addition of methylated CpG residues to the E. coli–derived EBV DNA might allow a productive infection, given that the BZLF1 gene product Zta can bind to its cognate sequence motifs when methylated at CpGs (10, 11, 27, 28).
The EBV DNA (p2089) harvested from E. coli was partially CpG-methylated in vitro by the de novo methyltransferase M.SssI and was transfected into HEK293 cells alone or in combination with the BZLF1 expression plasmid. RT-PCR analysis and immunostaining of the transfected cells indicated that they readily supported the expression of key viral lytic genes of all classes (Fig. 3 A and B). Only the supernatants of HEK293 cells cotransfected with BZLF1 and M.SssI-methylated EBV DNA (p2089) contained infectious EBV resulting in the reproducible proliferation of EBV-infected and transformed B blasts that expressed GFP from the recombinant EBV genome (Table 1).
Similarly, an expression M.SssI plasmid (p3663) was cotransfected with unmethylated EBV DNA (p2089) and BZLF1 into HEK293 cells. Infection of B blasts proved de novo synthesis of transformation-competent EBV from transiently transfected HEK293 cells expressing M.SssI (Table S2).
EBV with a Mutated BRLF1 Promoter Reveal a Block of Lytic Gene Expression Downstream of BRLF1 in Primary B Cells.
Zta binds to and activates the CpG-methylated form of the BRLF1 promoter more efficiently (10, 27); an observation interpreted to mean that escape from viral latency by the induction of the EBV lytic phase depends on the methylated status of this promoter. A corollary of this notion is that the unmethylated promoter could limit the expression of BRLF1 and thus prevent the onset of the EBV lytic phase immediately after infection of B cells. To test this possibility, two of three Zta-responsive elements (ZRE) within the BRLF1 promoter were replaced by CpG-free ZREs to which Zta binds strongly. Primary B cells were infected with this EBV mutant (4023) or wild-type EBV (2089) at the same MOI. Neither BRLF1 expression nor the levels of an early (BMRF1) and two late (BALF4, BLLF1) viral genes were affected by the point mutations in this promoter (Fig. S6B). In experiments parallel to those described in Fig. 3, HEK293 cells were cotransfected with BZLF1 and DNAs of the BRLF1 promoter EBV mutant (p4023) or wild-type EBV (p2089). The levels of expression of BRLF1 in these cells did not differ, nor did their supernatants contain detectable progeny virus (SI Results).
These data indicate that the methylation of EBV DNA necessary for induction of the lytic cycle occurs at sites other than those in the ZREs in the BRLF1 promoter. Therefore, viral genes downstream of BRLF1 must be responsible for blocking the switch to lytic gene expression in unmethylated EBV DNA.
A Differential Screen Identifies Putative Methylation-Dependent Binding Sites for Zta.
Thus far, four viral binding sites for Zta with CpGs that might be regulated by their methylation have been identified (10, 27). When we replaced the binding sites in the promoter of the essential BRLF1 gene with binding sites having no CpGs in a recombinant EBV, we found the EBV to be wild-type in its support of the EBV lytic cycle (Fig. S6). We therefore developed a general method to identify binding sites for Zta in the EBV genome potentially regulated by methylation of CpGs.
We wanted to identify the sequence elements that favor the binding of Zta when the EBV genome carries methylated CpGs. To do so, the BZLF1 DNA binding and dimerization domain was fused to GFP, resulting in a chimeric GFP:BZLF1 protein that was synthesized in HEK293 cells, immobilized (29), and used to select randomly fragmented EBV DNA. The initial EBV DNA either was purified directly from E. coli and thus was free of methylated CpGs or was used after full CpG methylation in vitro with the de novo methyltransferase M.SssI. DNA was selectively bound by GFP:BZLF1 in the presence of competitor DNA, washed, eluted, purified, labeled, and hybridized to an EBV tiled microarray. Unselected input DNA served as control. The data of three independent experiments were averaged and normalized as shown in Fig. S7.
Individual ZRE-containing DNA fragments were enriched more than 100-fold above background levels by immunoprecipitation of the BZLF1 DNA-binding moiety with cloned EBV DNA either unmethylated or methylated at CpGs with M.SssI methyltransferase. The methylated viral DNA was the preferred target of BZLF1 binding, with multiple sites being selectively enriched with CpG-methylated DNA (Fig. S7). For example, the fragments that likely contain the promoters of the early genes BMRF1, BSLF2/BMLF1, BSRF1, and BBLF4 were preferentially or exclusively enriched with CpG-methylated DNA. They encode the viral DNA polymerase processivity factor, the RNA export factor SM, a viral tegument protein with unknown function, and the DNA helicase of EBV, respectively. With the exception of BSRF1, these genes are directly or indirectly involved in DNA amplification of virion DNA during productive infection, and BSLF2/BMLF1, BMRF1, and BBLF4 are essential (30, 31). Nothing is known about the Zta-dependent regulation of BSRF1 and BBLF4, but BMRF1 and BSLF2/BMLF1 contain identified but CpG-free ZRE elements in their promoters (4, 32–34). Our data indicate that Zta selectively targets a number of previously unknown methylated ZRE sites in the promoters of viral genes essential for the EBV lytic cycle.
BZLF1 Expression During the Early Phase of Infection Supports the Proliferation of Resting B Cells.
Although BZLF1 was expressed early in EBV-infected primary B cells (Fig. 1) (35), this expression appeared paradoxical because it failed to lead to the de novo synthesis of progeny virus and did not block cellular proliferation (Fig. 2), a phenotype of BZLF1 described in established cell lines (36). Such early expression of BZLF1 is surprising because in vivo it could render infected cells susceptible to being killed by already primed effector T cells (37, 38). We investigated this paradox by determining whether the early expression of BZLF1 provided some selective advantage to infected cells.
BZLF1 is a member of the AP-1 family (4), members of which are critically involved in the control of proliferation. We therefore considered that BZLF1 might contribute to this key process. In preliminary experiments we found that a BZLF1-deficient EBV was impaired in its capacity to transform resting B cells purified from blood or adenoids and evaluated this phenotype in detail.
Primary B cells were prepared from adenoids, and three subpopulations of differentiated B cells were obtained by FACS sorting cells for surface markers: naïve B cells (IgD+, CD38–, CD27–), memory B cells (IgD–, CD38–, CD27+), and germinal center (GC) B cells (IgD–, CD38+). Unsorted B cells served as a control. Wild-type EBV (2089) and the BZFL1-knockout mutant EBV 2809 (BZLF1-KO) (3) were used to infect each class of B cells with an MOI of 0.01 for 24 h.
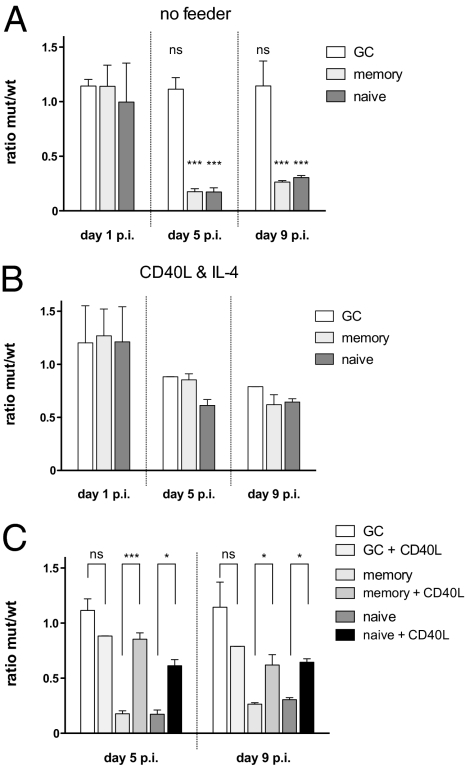
Naïve and memory B cells encompass the majority of quiescent B cells in secondary lymphoid organs, whereas GC B cells are activated, cycling, and proapoptotic cells. Wild-type EBV transformed naïve and memory B cells with equal efficiency (39), whereas BZLF1-KO EBV clearly showed a much reduced capacity to induce these cells to proliferate (Fig. 4A). Both viruses transformed GC B cells inefficiently but equally (39). The observed phenotypes indicated that EBV-induced proliferation of resting B cells was enhanced by the early expression of the BZLF1 gene, whereas that of preactivated, presumably cycling, cells, such as GC B cells, was not. We confirmed this finding by showing that the phenotype of naïve or memory B cells infected with BZLF1-KO EBV could be rescued by stimulating their CD40 receptor in the presence of IL-4 (Fig. 4 B and C). Interestingly, BZLF1’s contribution in inducing proliferation of resting B cells is unique, because a BRLF1-knockout EBV did not show any discernable phenotype in newly infected primary B cells (Fig. S8).
Fig. 4.
Functional role of BZLF1 expression during the early phase of infection. (A) Expression of BZLF1 provides a proliferative advantage to resting naïve and memory B cells but not to GC B cells. Subpopulations of primary B cells from three different donors were sorted and infected with wild-type EBV or BZLF1-KO EBV and were plated on plastic (no feeder). Proliferation of infected cells was measured by FACS as described (42), and the ratios of the number of viable, GFP-positive cells infected with the BZLF-KO EBV (mut) to the number of cells infected with wild-type EBV were calculated as indicated. Asterisks indicate the statistical significance of the means of three independent experiments in a two-way ANOVA analysis with Bonferroni posttests: ***P < 0.001; ns, not significant. (B) Activation of resting B cells by CD40L feeder cells, and IL-4 for 24 h on day 1 p.i. overcomes the need for BZLF1 expression in memory and naïve B-cells. Proliferating B cells were analyzed as in A. (C) Comparison of the results shown in A and B. The confidence values of untreated versus CD40L/IL-4 activated B cells subpopulations are shown. ***P < 0.001; *P < 0.05; ns, not significant.
Discussion
The EBV life cycle has been a puzzle, because it must establish itself latently in newly infected cells to avoid killing them but eventually support its productive phase in some infected cells to allow the synthesis of progeny virus resulting in the death of those infected cells. We have learned that EBV achieves these antithetical ends by capitalizing both on the cells’ gradual de novo methylation of the infecting viral DNA and the different activities of its regulatory gene, BZLF1.
EBV DNA is unmethylated upon infecting cells and becomes methylated over time (Fig. S3). Early in infection the BZLF1-encoded protein Zta is expressed (Fig. 1) but fails to induce the viral lytic cycle to yield progeny virus (Fig. 2B). Our findings confirm those of a previous publication (25) but are in conflict with a recent report (40). If Zta supported such a productive cycle, the host cell would be killed, aborting the EBV latent phase and potential long-term survival in the host. Instead, BZLF1 stimulates the ability of the infected resting B cells to proliferate, which has not been observed previously (41) (Fig. 4). This activity of Zta may be similar to that of its cousin, the cellular transcription factor AP-1 (7). The inability of Zta to induce the EBV lytic cycle early after infection probably results from its inability to bind and transactivate unmethylated promoters critical for this induction. As the latently infected B cells proliferate to expand the reservoir of cells harboring EBV, the viral DNA is progressively methylated (Fig. S3). Some of the methylated CpGs are within promoters necessary to support the lytic cycle and within binding sites for Zta. When methylated, these binding sites are bound preferentially by Zta (Fig. S7). EBV thus has evolved an epigenetic switch that depends on the gradual de novo methylation of its viral genome and the differential binding capacity of Zta.
It is likely that no single viral gene but rather a number of these genes critically regulates the onset of the EBV lytic phase as a function of CpG methylation of its DNA. The situation might be even more complex, because some viral promoters seem to be regulated differently in different cells. For example, the BMRF1 gene is readily expressed upon infection of B cells (Fig. 1 and Fig. S1) but fails to be expressed detectably in HEK293 cells upon DNA transfection (Fig. 3). Our observations suggest that Zta can regulate certain promoters in a gradual, quantitative, or absolute (on/off) manner, depending on the status of CpG methylation and the cellular background.
In contrast, certain viral promoters that have CpGs within binding sites for and are transactivated by Zta need not to be methylated to be stimulated by this activator. The BRLF1 promoter, which was the first identified to have CpGs within its Zta binding sites (10, 12), does not require CpGs to support the EBV latent phase (Fig. S6). CpGs also are dispensable in supporting BRLF1 expression during the EBV lytic cycle upon expression of BZLF1 (SI Results). Immunoprecipitation of Zta with unmethylated or methylated EBV DNA identified multiple promoters, including those of the genes BMRF1, BSLF2/BMLF1, BSRF1, and BBLF4 required for BRLF1 expression during the EBV lytic cycle that Zta preferentially binds when the DNA template is methylated (Fig. S7). We favor the notion that, once these promoters are methylated, enhanced Zta binding activates them to support the EBV lytic phase.
Formerly it appeared as if two distinct, well-separated phases of viral gene expression existed in B cells: They either were latently infected or had been induced to support the EBV lytic phase, which eventually would give rise to virus progeny. However, this view is no longer tenable. Previously, two viral genes, classified as lytic viral genes and homologs of cellular anti-apoptotic Bcl-2 proteins, were found to be transiently expressed in cells newly infected by EBV and to be required only initially for cellular transformation (42). Similarly, the viral homolog of the cellular IL-10 gene probably contributes to immune evasion during the early phase of EBV infection and also is expressed early upon infection of primary B cells (43). In a third example, primary B cells also express BZLF1 early after infection (Fig. 1 and ref. 35). The role of these lytic genes has been enigmatic but now is understood to be part of a biphasic control needed to initiate and maintain latent infection in the absence of productive infection.
Our experiments suggest but do not prove that BZLF1 expressed early after infection might transactivate cellular genes to activate quiescent cells to induce their cell-cycle entry. The cellular targets in different subpopulations of B cells might differ according to their status of activation and/or differentiation. So far, only a few viral targets of BZLF1 have been identified (44, 45), and its selective binding to CpG-methylated ZREs might contribute directly to their activation from a chromatin repressed state (46).
Together our combined studies have uncovered a surprising mechanism for the regulation of the EBV biphasic life cycle. The virus has evolved its own epigenetic time-dependent regulation mediated through its host cell’s gradual methylation of viral DNA and the differential, CpG methylation-dependent activities of BZLF1. The early expression of BZLF1 could mimic the function of AP-1 family members, which are instrumental for the activation and the G1-to-S transition of quiescent EBV-infected B cells.
Materials and Methods
Plasmids.
The wild-type EBV plasmid (p2089), the BZLF1-knockout mutant EBV (2809), and the BZLF1 expression plasmid p509 have been described (3, 26, 47). The BRLF1 promoter mutant EBV (4023) and the CD2 reporter EBV (3875) are described in SI Results and SI Material and Methods.
Immunoprecipitation of Methylated DNA.
Primary B cells from adenoids were infected with B95.8 (MOI 1), DNA was prepared, and MeDIP was performed as described previously (48). For microarray analysis, DNA was amplified using the GenomePlex Complete Whole Genome Amplification Kit (Sigma-Aldrich).
Microarray Design.
A custom-made tiling EBV-microarray was established with DNA of the B95.8 EBV strain. Its DNA was PCR-amplified in 285 separate, partially overlapping segments with a length of 500–700 bp. Details of the microarray are given in SI Materials and Methods and are available upon request.
Microarray Hybridization.
Labeling of selectively enriched DNA or input DNA was performed using the BioPrime Total Genomic Labeling System (Invitrogen), and the DNAs were labeled with Alexa Fluor 3 or Alexa Fluor 5. Hybridizations were performed in a Tecan HS 4000 Pro Hybridizing Station. Details on the analysis of the microarray data are included in SI Materials and Methods.
In Vitro Pulldown Assays with GFP:BZLF1.
The chimeric GFP:BZLF1 protein was transiently expressed in HEK293 cells, and nuclear extracts were prepared. Details of their preparation and immunoprecipitation of fragmented EBV DNA are included in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Christine Göbel for her important experimental contributions, Elisabeth Kremmer for her continuous support with monoclonal antibodies and advice, Emmanuel Drouet, Martin Rowe, and Ulrich Rothbauer for their generous gifts of hybridoma cell lines, plasmid constructs, and the GFP binder. We acknowledge New England Biolabs for providing the M.SssI gene. This work was supported by Deutsche Forschungsgemeinschaft Grants SPP1230, SFB455, and SFBTR5 and National Institutes of Health Grant CA70723.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911948107/DCSupplemental.
References
- 1.Kieff E, Rickinson AB. In: Fields Virology. Knipe DM, et al., editors. Philadelphia: Lippincott - Williams & Wilkins; 2007. pp. 2603–2654. [Google Scholar]
- 2.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 3.Feederle R, et al. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 2000;19:3080–3089. doi: 10.1093/emboj/19.12.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farrell PJ, Rowe DT, Rooney CM, Kouzarides T. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 1989;8:127–132. doi: 10.1002/j.1460-2075.1989.tb03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Countryman J, Miller G. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc Natl Acad Sci USA. 1985;82:4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takada K, Shimizu N, Sakuma S, Ono Y. trans activation of the latent Epstein-Barr virus (EBV) genome after transfection of the EBV DNA fragment. J Virol. 1986;57:1016–1022. doi: 10.1128/jvi.57.3.1016-1022.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sinclair AJ. bZIP proteins of human gammaherpesviruses. J Gen Virol. 2003;84:1941–1949. doi: 10.1099/vir.0.19112-0. [DOI] [PubMed] [Google Scholar]
- 8.Yu X, Wang Z, Mertz JE. ZEB1 regulates the latent-lytic switch in infection by Epstein-Barr virus. PLoS Pathog. 2007;3:e194. doi: 10.1371/journal.ppat.0030194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Speck SH, Chatila T, Flemington E. Reactivation of Epstein-Barr virus: Regulation and function of the BZLF1 gene. Trends Microbiol. 1997;5:399–405. doi: 10.1016/S0966-842X(97)01129-3. [DOI] [PubMed] [Google Scholar]
- 10.Bhende PM, Seaman WT, Delecluse HJ, Kenney SC. The EBV lytic switch protein, Z, preferentially binds to and activates the methylated viral genome. Nat Genet. 2004;36:1099–1104. doi: 10.1038/ng1424. [DOI] [PubMed] [Google Scholar]
- 11.Karlsson QH, Schelcher C, Verrall E, Petosa C, Sinclair AJ. Methylated DNA recognition during the reversal of epigenetic silencing is regulated by cysteine and serine residues in the Epstein-Barr virus lytic switch protein. PLoS Pathog. 2008;4:e1000005. doi: 10.1371/journal.ppat.1000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhende PM, Seaman WT, Delecluse HJ, Kenney SC. BZLF1 activation of the methylated form of the BRLF1 immediate-early promoter is regulated by BZLF1 residue 186. J Virol. 2005;79:7338–7348. doi: 10.1128/JVI.79.12.7338-7348.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holley-Guthrie EA, Quinlivan EB, Mar EC, Kenney S. The Epstein-Barr virus (EBV) BMRF1 promoter for early antigen (EA-D) is regulated by the EBV transactivators, BRLF1 and BZLF1, in a cell-specific manner. J Virol. 1990;64:3753–3759. doi: 10.1128/jvi.64.8.3753-3759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janz A, et al. Infectious Epstein-Barr virus lacking major glycoprotein BLLF1 (gp350/220) demonstrates the existence of additional viral ligands. J Virol. 2000;74:10142–10152. doi: 10.1128/jvi.74.21.10142-10152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neuhierl B, Feederle R, Hammerschmidt W, Delecluse HJ. Glycoprotein gp110 of Epstein-Barr virus determines viral tropism and efficiency of infection. Proc Natl Acad Sci USA. 2002;99:15036–15041. doi: 10.1073/pnas.232381299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrold RE, Marchini A, Fruehling S, Longnecker R. Glycoprotein 110, the Epstein-Barr virus homolog of herpes simplex virus glycoprotein B, is essential for Epstein-Barr virus replication in vivo. J Virol. 1996;70:2049–2054. doi: 10.1128/jvi.70.3.2049-2054.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiesner M, et al. Conditional immortalization of human B cells by CD40 ligation. PLoS One. 2008;3:e1464. doi: 10.1371/journal.pone.0001464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaefer BC, Strominger JL, Speck SH. Host-cell-determined methylation of specific Epstein-Barr virus promoters regulates the choice between distinct viral latency programs. Mol Cell Biol. 1997;17:364–377. doi: 10.1128/mcb.17.1.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minarovits J. Epigenotypes of latent herpesvirus genomes. Curr Top Microbiol Immunol. 2006;310:61–80. doi: 10.1007/3-540-31181-5_5. [DOI] [PubMed] [Google Scholar]
- 20.Low M, Hay J, Keir HM. DNA of herpes simplex virus is not a substrate for methylation in vivo. J Mol Biol. 1969;46:205–207. doi: 10.1016/0022-2836(69)90068-0. [DOI] [PubMed] [Google Scholar]
- 21.Kintner C, Sugden B. Conservation and progressive methylation of Epstein-Barr viral DNA sequences in transformed cells. J Virol. 1981;38:305–316. doi: 10.1128/jvi.38.1.305-316.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohn F, Weber M, Schübeler D, Roloff TC. Methylated DNA immunoprecipitation (MeDIP) Methods Mol Biol. 2009;507:55–64. doi: 10.1007/978-1-59745-522-0_5. [DOI] [PubMed] [Google Scholar]
- 23.Paulson EJ, Speck SH. Differential methylation of Epstein-Barr virus latency promoters facilitates viral persistence in healthy seropositive individuals. J Virol. 1999;73:9959–9968. doi: 10.1128/jvi.73.12.9959-9968.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tierney RJ, et al. Methylation of transcription factor binding sites in the Epstein-Barr virus latent cycle promoter Wp coincides with promoter down-regulation during virus-induced B-cell transformation. J Virol. 2000;74:10468–10479. doi: 10.1128/jvi.74.22.10468-10479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugden B. In: Immune Deficiency and Cancer: Epstein-Barr Virus and Lymphoproliferative Malignancies. Purtilo DT, editor. New York: Plenum Press; 1984. pp. 165–177. [Google Scholar]
- 26.Delecluse HJ, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc Natl Acad Sci USA. 1998;95:8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dickerson SJ, et al. Methylation-dependent binding of the Epstein-Barr virus BZLF1 protein to viral promoters. PLoS Pathog. 2009;5:e1000356. doi: 10.1371/journal.ppat.1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinclair AJ. Unexpected structure of Epstein-Barr virus lytic cycle activator Zta. Trends Microbiol. 2006;14:289–291. doi: 10.1016/j.tim.2006.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothbauer U, et al. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Fixman ED, Hayward GS, Hayward SD. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J Virol. 1992;66:5030–5039. doi: 10.1128/jvi.66.8.5030-5039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gruffat H, et al. Epstein-Barr virus mRNA export factor EB2 is essential for production of infectious virus. J Virol. 2002;76:9635–9644. doi: 10.1128/JVI.76.19.9635-9644.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quinlivan EB, et al. Direct BRLF1 binding is required for cooperative BZLF1/BRLF1 activation of the Epstein-Barr virus early promoter, BMRF1. Nucleic Acids Res. 1993;21:1999–2007. doi: 10.1093/nar/21.8.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Q, Summers WC. Identification of the 12-O-tetradecanoylphorbol-13-acetate-responsive enhancer of the MS gene of the Epstein-Barr virus. J Biol Chem. 1992;267:12049–12054. [PubMed] [Google Scholar]
- 34.Urier G, Buisson M, Chambard P, Sergeant A. The Epstein-Barr virus early protein EB1 activates transcription from different responsive elements including AP-1 binding sites. EMBO J. 1989;8:1447–1453. doi: 10.1002/j.1460-2075.1989.tb03527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wen W, et al. Epstein-Barr virus BZLF1 gene, a switch from latency to lytic infection, is expressed as an immediate-early gene after primary infection of B lymphocytes. J Virol. 2007;81:1037–1042. doi: 10.1128/JVI.01416-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cayrol C, Flemington EK. The Epstein-Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 1996;15:2748–2759. [PMC free article] [PubMed] [Google Scholar]
- 37.Steven NM, et al. Immediate early and early lytic cycle proteins are frequent targets of the Epstein-Barr virus-induced cytotoxic T cell response. J Exp Med. 1997;185:1605–1617. doi: 10.1084/jem.185.9.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pudney VA, Leese AM, Rickinson AB, Hislop AD. CD8+ immunodominance among Epstein-Barr virus lytic cycle antigens directly reflects the efficiency of antigen presentation in lytically infected cells. J Exp Med. 2005;201:349–360. doi: 10.1084/jem.20041542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ehlin-Henriksson B, Gordon J, Klein G. B-lymphocyte subpopulations are equally susceptible to Epstein-Barr virus infection, irrespective of immunoglobulin isotype expression. Immunology. 2003;108:427–430. doi: 10.1046/j.1365-2567.2003.01601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halder S, et al. Early events associated with infection of Epstein-Barr virus infection of primary B-cells. PLoS One. 2009;4:e7214. doi: 10.1371/journal.pone.0007214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katsumura KR, Maruo S, Wu Y, Kanda T, Takada K. Quantitative evaluation of the role of Epstein-Barr virus immediate-early protein BZLF1 in B-cell transformation. J Gen Virol. 2009;90:2331–2341. doi: 10.1099/vir.0.012831-0. [DOI] [PubMed] [Google Scholar]
- 42.Altmann M, Hammerschmidt W. Epstein-Barr virus provides a new paradigm: A requirement for the immediate inhibition of apoptosis. PLoS Biol. 2005;3:e404. doi: 10.1371/journal.pbio.0030404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeidler R, et al. Downregulation of TAP1 in B lymphocytes by cellular and Epstein-Barr virus-encoded interleukin-10. Blood. 1997;90:2390–2397. [PubMed] [Google Scholar]
- 44.Mauser A, et al. The Epstein-Barr virus immediate-early protein BZLF1 induces expression of E2F-1 and other proteins involved in cell cycle progression in primary keratinocytes and gastric carcinoma cells. J Virol. 2002;76:12543–12552. doi: 10.1128/JVI.76.24.12543-12552.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones RJ, et al. Roles of lytic viral infection and IL-6 in early versus late passage lymphoblastoid cell lines and EBV-associated lymphoproliferative disease. Int J Cancer. 2007;121:1274–1281. doi: 10.1002/ijc.22839. [DOI] [PubMed] [Google Scholar]
- 46.Heather J, Flower K, Isaac S, Sinclair AJ. The Epstein-Barr virus lytic cycle activator Zta interacts with methylated ZRE in the promoter of host target gene egr1. J Gen Virol. 2009;90:1450–1454. doi: 10.1099/vir.0.007922-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hammerschmidt W, Sugden B. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell. 1988;55:427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- 48.Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.