Abstract
In many Gram-negative pathogens, their virulent behavior is regulated by quorum sensing, in which diffusible signals such as N-acyl homoserine lactones (AHLs) act as chemical messaging compounds. Enzymatic degradation of these diffusible signals by, e.g., lactonases or amidohydrolases abolishes AHL regulated virulence, a process known as quorum quenching. Here we report the first crystal structure of an AHL amidohydrolase, the AHL acylase PvdQ from Pseudomonas aeruginosa. PvdQ has a typical α/β heterodimeric Ntn-hydrolase fold, similar to penicillin G acylase and cephalosporin acylase. However, it has a distinct, unusually large, hydrophobic binding pocket, ideally suited to recognize C12 fatty acid-like chains of AHLs. Binding of a C12 fatty acid or a 3-oxo-C12 fatty acid induces subtle conformational changes to accommodate the aliphatic chain. Furthermore, the structure of a covalent ester intermediate identifies Serβ1 as the nucleophile and Asnβ269 and Valβ70 as the oxyanion hole residues in the AHL degradation process. Our structures show the versatility of the Ntn-hydrolase scaffold and can serve as a structural paradigm for Ntn-hydrolases with similar substrate preference. Finally, the quorum-quenching capabilities of PvdQ may be utilized to suppress the quorum-sensing machinery of pathogens.
Keywords: crystal structure, Pseudomonas aeruginosa, quorum sensing, pyoverdine, catalytic mechanism
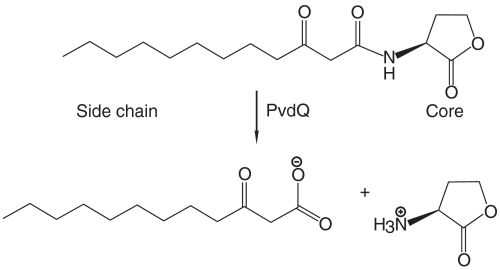
Since the sixties it has been appreciated that bacteria are not just individualistic organisms but have a “social behavior.” This bacterial socialization, also called quorum sensing, is involved in many important processes such as motility and virulence (1). Among the best-characterized quorum sensing signaling molecules, also referred to as autoinducers, are the N-acyl homoserine lactones (AHLs) (2, 3), which consist of an acyl chain linked to a homoserine lactone core (HSL) via an amide bond (Fig. 1). Several varieties of these compounds with different acyl chain lengths and substitutions have been found in Gram-negative bacteria (4).
Fig. 1.
The hydrolysis of 3-oxo-dodecanoic homoserine lactone performed by PvdQ.
In recent years enzymes were discovered that are capable of interrupting bacterial communication by degrading the signaling molecules such as AHLs, a process known as quorum quenching (5). Although the biological relevance of quorum quenching is not fully understood, it has been implicated in several important processes. For example, in Agrobacterium tumefaciens quorum quenching is able to down-regulate the energy-consuming process of conjugation under nutrient-limiting conditions (6). Furthermore, it has been shown that the interruption of bacterial communication by quorum quenching can prevent pathogens from displaying virulent behavior (7). Therefore, quorum quenching could be used in the development of new antimicrobial strategies because many pathogens depend on quorum sensing in regulating virulence (6, 8).
A structurally well-characterized quorum-quenching enzyme is the N-acyl homoserine lactone lactonase of Bacillus thuringiensis (9, 10), which belongs to the metallo-β-lactamase superfamily. This enzyme disrupts quorum sensing by cleaving the ester bond in the homoserine lactone ring, making it inactive in signaling. The lactonase structures show that the AHL acyl chain binds in a solvent-exposed groove along the enzyme surface, allowing the binding of acyl chains of different lengths (11). Huang et al. (12) and Sio et al. (13) have characterized a different quorum-quenching enzyme from the opportunistic pathogen Pseudomonas aeruginosa, which prefers long-chain AHLs such as C10 to C14. This enzyme, called PvdQ, subverts quorum sensing at micromolar concentrations of AHLs by hydrolyzing the peptide bond between the acyl chain and the HSL core (Fig. 1) leading to a significantly reduced virulence (14). Interestingly, the pvdQ gene is located in the pyoverdine gene cluster, suggesting that PvdQ also plays a role in pyoverdine biosynthesis (15). Pyoverdine is a siderophore used as an iron scavenger under iron-limiting conditions (16). Under these conditions PvdQ is up-regulated (17), and it has been hypothesized that the enzyme is able to couple production of the iron scavenger with virulence repression to survive under nutrient-depleted conditions (13).
PvdQ is expressed as a proenzyme that is autoproteolytically activated by posttranslational cleavage resulting in the excision of a 23-residue prosegment and the formation of an 18 kDa α-chain and a 60 kDa β-chain (13). The posttranslational modification of PvdQ resembles the maturation of Ntn-hydrolases such as penicillin G acylase (PGA) (18) and glutaryl-7-amino-cephalosporanic acid acylase (CA) (19), which share 24% and 12% sequence identity with PvdQ, respectively. In Ntn-hydrolases this posttranslational modification makes the N terminus of the β-chain available as the active site nucleophile. In various structurally characterized Ntn-hydrolases the N terminus of the newly formed β-chain can be a Ser, Thr, or Cys, which is responsible for both catalysis and autoproteolysis (18, 20, 21). Upon autoproteolysis the α-amino group is unveiled, which is proposed to act as a general base in catalysis (18). Although the substrate preference and biological context of Ntn-hydrolases differ substantially, they all share the highly conserved αββα-fold with two stacked antiparallel β-sheets sandwiched between α-helical bundles (22).
To gain more insights into the substrate preference of PvdQ and the catalytic mechanism behind quorum quenching we solved its crystal structure. The structures presented herein clearly show that PvdQ is an Ntn-hydrolase that belongs to the same subfamily as PGA and CA (as defined in the SCOP database) (23). However, PvdQ has a much more extensive hydrophobic substrate-binding pocket adapted to the long fatty-acid-like tails of P. aeruginosa’s N-acyl homoserine lactones.
Results and Discussion
PvdQ Is a Member of the Ntn-hydrolase Superfamily.
We have solved the 1.8 Å crystal structure of wild type, fully mature PvdQ (Table S1). The structure of PvdQ clearly establishes the enzyme as an Ntn-hydrolase by its typical αββα-fold (Fig. 2B and C) and its heterodimeric organization, with a 60 kDa β-chain and an 18 kDa α-chain. Although posttranslational modification results in the formation of two chains, both chains remain tightly interwoven, forming one single Ntn-hydrolase enzyme. The central stacked antiparallel β-sheets and the protruding A- and B-knobs on either side give PvdQ a heart-shaped structure with a deep, solvent-accessible crevice in the center (Fig. 2A and B). The N-terminal Serβ1 of the β-chain is located at the bottom of this crevice and provides the catalytic nucleophile. No density extends from the N-terminal serine, indicating that the enzyme underwent complete autoproteolysis (Fig. 2C). The fully matured enzyme is composed of 717 amino acids; 171 in the α- and 546 in the β-chain; however, no electron density could be observed for the first five N-terminal residues and the last two C-terminal residues of the α-chain.
Fig. 2.
3D structure of PvdQ. (A) A solvent-accessible surface area representation shows the heart-shape with the α-chain in orange and the β-chain in blue. (B) A secondary structure representation. The α-chain is depicted with magenta β-strands and yellow α-helices while the β-chain has light blue β-strands and orange α-helices. (C) A front view showing the three conserved disulfide bridges. All disulfide bridges (indicated in green) lie on the periphery of the enzyme. The N-terminal nucleophile is located in the center of the enzyme. The σA-weighted electron density, contoured at 1.2σ, shows that no density continues from the newly formed N-terminus after processing, indicating that the enzyme underwent complete autoproteolysis (inset).
A Dali search (24) revealed that the β-chain of PvdQ is structurally similar to the β-chain of CA (PDB entry 1FM2; Z score 46). More distant structural homologues are PGA (PDB entry 1E3A; Z score 27), penicillin V acylase [PDB entry 3PVA (25); 10% identity, Z score 16], and the proteasome subunits [PDB entry 1RYP (26); 7% identity, Z score 9]. While the β-chain of PvdQ shares similarities with representatives of several Ntn-hydrolase subfamilies (as defined in the SCOP database), the typical arrangement of the α-helices in the α-chain can only be found in the α-chains of heterodimeric members of the PGA Ntn-hydrolase subfamily with Z scores between 9.0 and 16.0.
An interesting feature of PvdQ is the presence of six cysteines, which are all involved in disulfide-bridge formation (Fig. 2C). Proteins that reside in the periplasm often contain disulfide bridges due to the oxidizing environment (27). Although PvdQ, PGA, and CA are all localized in the periplasm, PvdQ is the first structurally characterized Ntn-hydrolase within this family to have any disulfide bridges. The disulfides are located in both the α- and β-chains on the periphery of the protein. In the β-chain they connect structural elements that are close together in the sequence (Cys217-Cys237 and Cys339-Cys352), while they are further apart in the α-chain (Cys44-Cys125). A BLAST search (28) against the UniProt database (29) revealed that the presence and location of these disulfide bridges is highly conserved in the PvdQ AHL acylases from Pseudomonas species (Fig. S1) as well as in aculeacin acylase from Actinoplanes utahensis and penicillin V acylase from Streptomyces mobaraensis.
PvdQ Has a Hydrophobic Pocket Near the N-terminal Nucleophile.
A surface representation of PvdQ shows a pocket in the vicinity of the N-terminal nucleophile in the interior of the enzyme (Fig. 3A), which is closed off from the solvent by Pheβ24 serving as a gate. The pocket lies on top of the large central β-sheets, and amino acid residues from both the α- and β-chain contribute to its build-up. The lining of the pocket is formed by a constellation of mainly bulky hydrophobic residues originating from α7, α9, β4, β5, and the loops that connect β5–β6, α10-β17, β7–β8, and β9–β10. These loops fold away from the large central β-sheet, providing space for the pocket. This gives the pocket a hydrophobic character. The residues that form the lining of the substrate-binding site are highly conserved among PvdQ homologues from different Pseudomonads (Fig. S1).
Fig. 3.
Comparison of liganded and unliganded PvdQ. Surface slice-throughs of PvdQ showing the conformational changes upon substrate binding. (A) Apo-enzyme and (B) Dodecanoic acid bound. The substrate-binding site of PvdQ is built up from mainly bulky hydrophobic residues. Upon binding of 3-oxo-dodecanoic acid (C) (light green model) or dodecanoic acid (D) (light green model) residues move with respect to the apo-enzyme (dark blue). The weaker σA-weighted density corresponding to 3-oxo-C12 is contoured at 1σ while C12 is contoured at 1.2σ. The carboxylates of the ligands form hydrogen bonding interactions with the N-terminal nucleophile and the oxyanion hole residues. Wat1 bridges the Serβ1 Oγ and the free α-amino group, which in turn is coordinated by Asnβ269 and Hisβ23.
The Hydrophobic Pocket of PvdQ Shows Induced Fit upon Ligand Binding.
The volume of the hydrophobic pocket near the catalytic center is 144 Å3, which would be too small to accommodate the C12-acyl chains that PvdQ prefers as a substrate (13). To gain more insights into substrate binding, soaking experiments were performed with 3-oxo-C12-HSL, an auto-inducer of P. aeruginosa, and C12-HSL, an auto-inducer analogue.
The structure of the complex immediately shows that the acyl chain of the compound binds in the hydrophobic pocket, which has opened up to the solvent to provide space for the ligand (Fig. 3B). The residue that is responsible for the transition from closed to open state is Pheβ24 (Fig. 3C and D), which forms a gate between the binding pocket and solvent (Fig. 3A). Other residues that move upon ligand binding are, for example, Ileα146 and Trpβ186. A σA-weighted (30) electron density map clearly showed a stretch of continuous density, in which an acyl chain could be fitted (Fig. 3C and D). However, from the density it is clear that not the substrate AHL, but the hydrolysis product dodecanoic- or 3-oxo-dodecanoic acid is bound in the hydrophobic pocket, which means that crystallized PvdQ can still hydrolyze AHLs. Both compounds have almost exclusively Van der Waals interactions with the enzyme, except for the carboxylate moiety, which makes hydrogen bonds with the N-terminal nucleophile and the backbone amide of Valβ70. Dodecanoic acid binds very close to the N-terminal nucleophile and induces a second conformation of the Serβ1 side chain; in each conformation the Serβ1 hydroxyl group makes a hydrogen bond with either of the C12-carboxylate oxygen atoms. While the electron density of dodecanoic acid is very well defined, the density corresponding to the polar part of 3-oxo-dodecanoic acid is weaker. The 3-oxo moiety of 3-oxo-C12 does not make any hydrogen-bonding interactions with the enzyme, and the polar 3-oxo-group may facilitate easy diffusion of the 3-oxo-C12 chain out of the hydrophobic binding pocket.
The product-bound structures can be superimposed onto apo-PvdQ with an rmsd of 0.14 Å, indicating that binding of substrate does not introduce any major main-chain rearrangements. However, upon ligand binding PvdQ undergoes subtle conformational changes in the side chains that line the substrate-binding pocket. The displacement of the side chains increases the pocket volume from 144 Å3 to about 260 Å3. Furthermore, the substrate-binding residues become more rigid as reflected by a decrease in B-factors from 21 Å2 to 17 Å2, compared to average protein B-factors of 28 Å2 and 27 Å2, in the apo- and dodecanoic acid bound enzyme, respectively. In summary, the adaptation of the ligand-binding pocket explains the preference of PvdQ for long fatty-acid-like side chains of AHLs.
The Catalytic Center of PvdQ.
The binding of the hydrolysis product gives valuable insights into the catalytic mechanism of PvdQ. In the dodecanoic acid bound structure the carboxylate carbon is only 2.9 Å away from the Serβ1 Oγ, indicating the serine as the putative nucleophile. Furthermore, one of the carboxylate oxygens of dodecanoic acid is in close proximity to the backbone amide of Valβ70 and the Nδ atom of Asnβ269, at 2.8 Å and 4.2 Å, respectively, in a configuration suitable for stabilizing the transient oxyanion transition state during the reaction (Fig. 3C and D). The buildup of this oxyanion hole and the interactions with the reaction product are very similar to the oxyanion hole and substrate interactions observed in penicillin G acylase (18, 31).
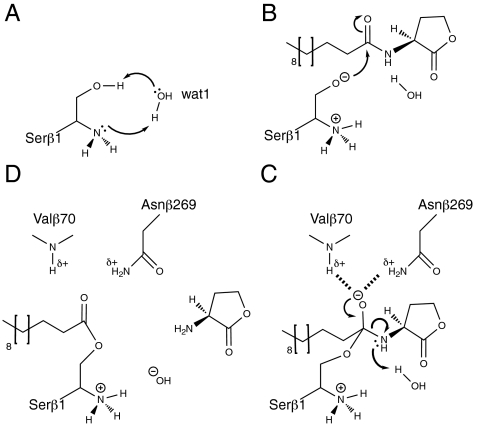
Because Ntn-hydrolases lack a classic catalytic base to activate the N-terminal nucleophile, it has been proposed that the α-amino group of Serβ1 deprotonates its own Oγ via a bridging water molecule that acts as a “virtual base” (18). Apo- and liganded PvdQ do indeed show such an organization of the active site with a water molecule (Wat1; Fig. 3C and D) at hydrogen bonding distance from both the Serβ1 Oγ (3.0 Å) and the Serβ1 α-amino group (2.9 Å). After transfer of the Oγ proton to the α-amino group, but prior to nucleophilic attack, the reactive charge-separated state can be stabilized by the Nδ lone pair of Hisβ23, which is at hydrogen bonding distance from the α-amino-group of Serβ1 (2.9 Å; Fig. 3C and D). The backbone amide of Hisβ23 has a hydrogen bonding interaction with the seryl Oγ (2.5 Å).
In addition to Hisβ23, the structure indicates several other residues that may partake in substrate recognition and catalysis. The Oδ atom of Asnβ269 is at 2.6 Å from the α-amino group of Serβ1, orienting the lone pair of the α-amino group in line with Wat1 and the Serβ1 Oγ to facilitate proton transfer. The side chain of Asnβ269 is kept in position by a hydrogen bond with the Nη atom of the highly conserved Argβ297 (2.9 Å), which also forms hydrogen bonds with the backbone carbonyl of the N-terminal nucleophilic Serβ1 (2.7 Å). An additional hydrogen bond of 2.9 Å is observed between the alternate conformation of Serβ1 Oγ and the Nδ atom of Asnβ269 in the dodecanoic acid bound PvdQ structure. Except for the dual conformation of Serβ1, this arrangement and the type of amino acids in the catalytic center is typical for Ntn-hydrolases.
The Catalytic Mechanism of PvdQ Proceeds Via a Covalently Bound Intermediate.
In order to obtain more detailed information on the catalytic mechanism of PvdQ, crystals were soaked shortly at pH 5.5 to capture a reaction intermediate. At low pH the catalytic base activity of the α-amino group is reduced, and the deacylation of the acyl-enzyme intermediate by a hydroxylate ion is also lowered. A σA-weighted difference omit map clearly showed the presence of a covalent ester link between the Serβ1 Oγ and the carbonyl carbon of the dodecanoic acid (Fig. 4). No density was present anymore for the homoserine lactone group. A similar covalent intermediate in an Ntn-hydrolase has been observed in γ-glutamyl transpeptidase (32). The ester carbonyl oxygen forms 2.9 Å and 3.5 Å hydrogen bonds with the Valβ70 backbone amide and the Asnβ269 side chain Nδ2 (Fig. 4), establishing that these groups form the oxyanion hole. Wat1, the virtual base, is still present in the ester intermediate structure, located 2.8 Å from the carbonyl carbon of the ester bond. However, to prevent a clash with dodecanoic acid, it has shifted 2 Å with respect to the situation in the apo-enzyme.
Fig. 4.
Covalent acyl-enzyme intermediate between PvdQ and dodecanoic acid when soaked shortly at pH 5.5. This ester intermediate is stabilized by hydrogen bonds with the oxyanion hole at the bottom of the figure. Wat1 is bound closely to nucleophile to hydrolyze the acyl-enzyme intermediate. The σA-weighted omit density is contoured at 1σ.
Our data allows us to present a catalytic mechanism of N-acyl homoserine lactone hydrolysis by PvdQ. After activation of the N-terminal nucleophilic serine hydroxyl group by the virtual base Wat1 as explained above, the Serβ1 hydroxylate attacks the carbonyl carbon of the scissile bond of the substrate (Fig. 5A and B). The attack results in the formation of a tetrahedral transition state, which is stabilized by the oxyanion hole residues (Fig. 5C). Wat1, which makes hydrogen bonds with both the α-amino group and the side chain Oγ of Serβ1, donates its proton to the amine of the HSL-leaving group, which results in the collapse of the transition state into the ester intermediate (Fig. 5D). Once the covalent acyl-enzyme intermediate has formed, the Wat1 hydroxylate can attack the newly formed ester intermediate, resulting in a similar transition state stabilized by the oxyanion hole and concomitant release of the dodecanoic acid (see Fig. S2). This catalytic mechanism is similar to the mechanism proposed for penicillin G acylase (18, 31).
Fig. 5.
The mechanism of acyl-enzyme intermediate formation. (A) Wat1 is involved in the activation of the Serβ1 nucleophile by relaying the proton from the Oγ to the α-amino group. (B) Upon nucleophile activation the N-terminal nucleophile attacks the carbonyl carbon of the scissile bond in the substrate. (C) The transition state is stabilized by the oxyanion hole formed by a backbone amide and a side-chain amide. Upon protonation of the α-amino group by wat1 the transition state collapses into an ester intermediate (D).
Generality of the Hydrophobic Substrate-Binding Site.
Ever since their first characterization (33) Ntn-hydrolases have been found to be a highly versatile class of enzymes. While their catalytic mechanism and the amino acid residues surrounding the N-terminal nucleophile are strictly conserved, their substrate specificity has been shown to be quite broad, ranging from small hydrophilic or hydrophobic compounds to large proteins. Consequently, Ntn-hydrolases are utilized in a wide variety of cellular processes from protein degradation (26) to nucleotide biosynthesis (34). The discovery of PvdQ introduces yet another important role for Ntn-hydrolases, the hydrolysis of long acyl amides involved in quorum sensing. Our crystal structures provide the molecular details of the distinct substrate specificity of PvdQ for the fatty-acid acyl moiety of long-chain homoserine lactone autoinducers. However, no data is available yet on the interaction between the enzyme and the homoserine lactone ring, though it is conceivable that the homoserine lactone moiety binds in the deep solvent-accessible crevice between the A- and B-knobs (Figure 2A and B).
Comparison of PvdQ with two well-characterized Ntn-hydrolases, PGA and CA, shows that in PvdQ the substrate-binding pocket is formed by the same secondary structure elements as in PGA and CA but that its size is much larger to allow binding of C12-acyl chains. In PGA and CA the key residues for substrate recognition are Metα142 and Argβ57, respectively. In PvdQ these residues are replaced by smaller residues (Leuα146 and Asnβ57). Furthermore, α7, on which residue α142 is located, is pushed upward by Trpβ186, which introduces more space for the substrate. These differences explain the increased size of the hydrophobic pocket of PvdQ and its deeper protrusion into the interior of the enzyme, explaining the unique substrate preference of PvdQ for long acyl chains.
A BLAST search revealed other Ntn-hydrolases that are homologous to PvdQ. Most notable are aculeacin acylase from Actinoplanes utahensis (37% identity to PvdQ) (35) and penicillin V acylase from Streptomyces mobaraensis (36% identity) (36). Interestingly, these enzymes have a substrate preference similar to PvdQ. Aculeacin acylase cleaves off long acyl chains, such as linoleic, myristic, or palmitic acid, from the cyclic hexapeptide core of the antifungal echinocandin (35). Penicillin V acylase from Streptomyces mobaraensis has a preference for capsaicin, which consists of an 8-methyl-6-nonene side chain connected to a vanillyl core via a peptide bond. Multiple sequence alignment (Fig. S1) indicates that residue β57 and α146 are a glutamine and a leucine or a glutamate and a serine in penicillin V acylase and aculeacin acylase, respectively, and Trpβ186 is also present in these enzymes. These observations suggest that the buildup of the substrate-binding pocket in these enzymes is similar to PvdQ. Thus, since aculeacin acylase and penicillin V acylase are more closely related to PvdQ than to PGA and CA (with which they share on average only 15% sequence identity), these long acyl-chain-recognizing Ntn-hydrolases may form a separate group for which the structure of PvdQ can serve as a paradigm. QuiP, a second Ntn-hydrolase present in the genome of P. aeruginosa, also acts on long-chain AHLs (37). Although QuiP belongs to the Ntn-hydrolase superfamily and exhibits a similar substrate specificity for AHLs, the low sequence identity to PvdQ (16%) and the presence of several insertions/deletions make it difficult to draw conclusions on the buildup of its substrate-binding pocket. Residues that are conserved are the N-terminal nucleophilic serine, Hisβ23, Argβ297, and several hydrophobic residues that contribute to the lining of the hydrophobic cavity in PvdQ. In contrast, the disulfide bond forming cysteines are not conserved.
The 3D-structure of PvdQ also gives valuable information on how to alter its substrate specificity toward shorter homoserine lactones, such as C6- and C8-HSL quorum sensing molecules produced by Burkholderia and Yersinia, on which PvdQ shows weak activity (13). In that way PvdQ could disturb quorum sensing regulated virulence in pathogenic organisms that rely on AHLs other than the ones from Pseudomonas.
The role of PvdQ in quorum quenching has been well established (12–14). In addition, pvdQ-gene deletion abrogates pyoverdine biosynthesis (38). Although the precise role of PvdQ in this latter pathway is not known, Visca et al. (15) have proposed the need for an enzyme that removes a long acyl chain from a pyoverdine precursor. The crystal structures of PvdQ show that the enzyme has evolved as an Ntn-hydrolase with a distinct binding pocket for long acyl chains and indeed might be able to accommodate the acyl chain of a pyoverdine precursor. The pyoverdine core can easily be accommodated in the crevice between the A- and B-knobs (Fig 2A and B). Whether this specialized acyl-chain binding pocket was initially acquired for AHL hydrolysis or pyoverdine maturation requires further investigation.
Materials and Methods
Purification of PvdQ.
PvdQ was cloned and overexpressed according to Sio et al. (13). The cell-pellet was resuspended in 3 vol of lysis buffer (50 mM Tris-HCl, pH 8.8, 2 mM EDTA), lysed by sonication and centrifuged at 30,000 g to remove cell debris. The supernatant was applied to a HiTrap Q-Sepharose column (GE Healthcare Life Sciences). PvdQ appeared in the flow-through containing 50 mM Tris-HCl, pH 8.8, and 2 mM EDTA. After bringing the buffer to 700 mM ammonium sulfate, the protein solution was applied to a HiTrap phenyl sepharose column (GE Healthcare Life Sciences). PvdQ eluted at the end of a 700-0 mM ammonium sulfate gradient. Finally, PvdQ was concentrated to 4 mg/ml and applied to a Hiload Superdex 75 16/160 gel filtration column (GE Healthcare Life Sciences), and the major peak was collected. PvdQ shows 2 bands on SDS-PAGE gels corresponding to the α- and β-chains. Dynamic Light Scattering experiments indicated that PvdQ is monodisperse in solution with a particle size corresponding to a molecular mass of approximately 80 kDa.
Thermofluor Assay and Buffer Exchange.
Because PvdQ was purified without any additives during hydrophobic interaction and ion exchange chromatography, the thermal shift assay (39) was used to establish a protein buffer capable of stabilizing PvdQ during storage and crystallization. The thermal shift assay indicated 100 mM Tris-HCl, pH 7.5, containing 5% glycerol, as an adequate buffer for the stabilization of PvdQ. Buffer exchange and concentration of PvdQ to 7 mg/mL was performed with an Amicon filtration device with a 50 kDa MW cutoff filter, and the concentrated protein solution was used in crystallization trials.
Crystallization and Soaks.
PvdQ could be crystallized in a condition of the Wizard Screen (Emerald Biosystems) consisting of 100 mM 2-(N-cyclohexylamino)ethanesulfonic acid, pH 10.0, 20% PEG 8000. Optimized crystallization conditions were obtained using 100 mM N,N-bis(2-hydroxyethyl)glycine (Bicine), pH 9.1, and 23% PEG 6000. The substrate 3-oxo-C12-HSL kindly provided by Miguel Cámara and Paul Williams from the University of Nottingham, while C12-HSL was obtained from Fluka. The substrates were stored in DMSO or ethylacetate; for soaking studies a 100 nl aliquot was added to 10 μl of mother liquor; after which the crystal was added and allowed to soak for 20 min. To capture a reaction intermediate, crystals were serially transferred from mother liquor solutions containing Bicine, pH 9.1, Tris-HCl, pH 8.0, Tris-HCl, pH 7.0, MES, pH 6.0, and finally MES, pH 5.5. Subsequently, the crystals were transferred to mother liquor supplied with MES, pH 5.5, and 30% glycerol for cryoprotection. After that the crystals were allowed to soak in a C12-HSL solution for 1 min, made as described above, and immediately flash-frozen in liquid nitrogen.
Data Collection and Processing.
Cryocrystallographic diffraction data were collected at the European Synchrotron Radiation Facility in Grenoble, France. For cryoprotection crystals were immersed in mother liquor supplemented with 25% glycerol and flash frozen in liquid nitrogen. Crystals were of space group C2221. Data were integrated and scaled with XDS (40) and merged with SCALA (41). Data collection statistics can be found in Table S1.
Structure Solution and Refinement.
Phases were obtained by molecular replacement with the PHASER program (42) from the CCP4 package (43). An ensemble of two models was used as input for PHASER, penicillin G acylase (PGA; 13% idenity; PDB entry 1E3A; (44)) and cephalosporin acylase (CA; 22% identity; PDB entry 1KEH; (45)), which were found using the fold & function assignment system (FFAS; (46)). The conserved residues were kept while variable residues were replaced with serines using the SCWRL modeler (47).
Initial building was done in alternate rounds of statistical density modification/automated building with RESOLVE (48) and manual building in COOT (49). RESOLVE managed to build 75% of the main-chain and 50% of the side-chain atoms; final automated building was performed with ARP/wARP (50), which built 95% of the amino acids. Refinement was done using COOT and Refmac5 with TLS refinement using 3 separate domains (51). Ligand coordinates and dictionaries were created using JLigand v.0.1b (http://www.ysbl.york.ac.uk/~pyoung/JLigand/JLigand.html). The structures were validated with Molprobity (52). Refinement statistics can be found in Table S1. Detection and analysis of active site pockets in PvdQ structures was performed with VOIDOO (53) using the Connolly rolling probe algorithm (54), in which a central pocket coordinate was used as a seed point. Figures were created using the PyMol molecular viewer www.pymol.org (55).
Supplementary Material
Acknowledgments.
M.B. was partly funded by Stichting Technische Wetenschappen under project 790.35.630 of the Nederlandse Organisatie voor Wetenschappelijk Onderzoek. P.N.J. was partly funded by EU grant Antibiotarget MEST-CT-2005-020278.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2wye, 2wyd, 2wyc and 2wyb)
This article contains supporting information online at www.pnas.org/cgi/content/full/0911839107/DCSupplemental.
References
- 1.Bassler BL, Losick R. Bacterially speaking. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Eberhard A, et al. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry. 1981;20:2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- 3.Pearson JP, Passador L, Iglewski BH, Greenberg EP. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitehead NA, et al. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol Rev. 2001;25:365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x. [DOI] [PubMed] [Google Scholar]
- 5.Dong YH, et al. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature. 2001;411:813–817. doi: 10.1038/35081101. [DOI] [PubMed] [Google Scholar]
- 6.Dong YH, Zhang LH. Quorum sensing and quorum-quenching enzymes. J Microbiol. 2005;43:101–109. [PubMed] [Google Scholar]
- 7.Molina L, et al. Degradation of pathogen quorum-sensing molecules by soil bacteria: A preventive and curative biological control mechanism. FEMS Microbiol Ecol. 2003;45:71–81. doi: 10.1016/S0168-6496(03)00125-9. [DOI] [PubMed] [Google Scholar]
- 8.Martin CA, Hoven AD, Cook AM. Therapeutic frontiers: Preventing and treating infectious diseases by inhibiting bacterial quorum sensing. Eur J Clin Microbiol. 2008;27:635–642. doi: 10.1007/s10096-008-0489-3. [DOI] [PubMed] [Google Scholar]
- 9.Liu D, et al. Three-dimensional structure of the quorum-quenching N-acyl homoserine lactone hydrolase from Bacillus thuringiensis. Proc Natl Acad Sci USA. 2005;102:11882–11887. doi: 10.1073/pnas.0505255102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim MH, et al. The molecular structure and catalytic mechanism of a quorum-quenching N-acyl-L-homoserine lactone hydrolase. Proc Natl Acad Sci USA. 2005;102:17606–17611. doi: 10.1073/pnas.0504996102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu D, et al. Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures. Biochemistry. 2008;47:7706–7714. doi: 10.1021/bi800368y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang JJ, Han JI, Zhang LH, Leadbetter JR. Utilization of acyl-homoserine lactone quorum signals for growth by a soil pseudomonad and Pseudomonas aeruginosa PAO1. Appl Environ Microbiol. 2003;69:5941–5949. doi: 10.1128/AEM.69.10.5941-5949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sio CF, et al. Quorum quenching by an N-acyl-homoserine lactone acylase from Pseudomonas aeruginosa PAO1. Infect Immun. 2006;74:1673–1682. doi: 10.1128/IAI.74.3.1673-1682.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papaioannou E, et al. Quorum quenching acylase reduces the virulence of Pseudomonas aeruginosa in a Caenorhabditis elegans infection model. Antimicrob Agents Chemother. 2009;53:4891–4897. doi: 10.1128/AAC.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Visca P, Imperi F, Lamont IL. Pyoverdine siderophores: From biogenesis to biosignificance. Trends Microbiol. 2007;15:22–30. doi: 10.1016/j.tim.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 16.Meyer JM, Hornsperger JM. Role of Pyoverdinepf, the iron-binding fluorescent pigment of Pseudomonas fluorescens, in iron transport. J Gen Microbiol. 1978;107:329–331. [Google Scholar]
- 17.Ochsner UA, Wilderman PJ, Vasil AI, Vasil ML. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol Microbiol. 2002;45:1277–1287. doi: 10.1046/j.1365-2958.2002.03084.x. [DOI] [PubMed] [Google Scholar]
- 18.Duggleby HJ, et al. Penicillin acylase has a single-amino-acid catalytic center. Nature. 1995;373:264–268. doi: 10.1038/373264a0. [DOI] [PubMed] [Google Scholar]
- 19.Kim Y, et al. The 2.0 A crystal structure of cephalosporin acylase. Structure. 2000;8:1059–1068. doi: 10.1016/s0969-2126(00)00505-0. [DOI] [PubMed] [Google Scholar]
- 20.Seemuller E, et al. Proteasome from Thermoplasma acidophilum: A threonine protease. Science. 1995;268:579–582. doi: 10.1126/science.7725107. [DOI] [PubMed] [Google Scholar]
- 21.Kim JH, et al. Structure and function of the glutamine phosphoribosylpyrophosphate amidotransferase glutamine site and communication with the phosphoribosylpyrophosphate site. J Biol Chem. 1996;271:15549–15557. doi: 10.1074/jbc.271.26.15549. [DOI] [PubMed] [Google Scholar]
- 22.Oinonen C, Rouvinen J. Structural comparison of Ntn-hydrolases. Protein Sci. 2000;9:2329–2337. doi: 10.1110/ps.9.12.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andreeva A, et al. Data growth and its impact on the SCOP database: New developments. Nucleic Acids Res. 2007;36:D419–425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suresh CG, et al. Penicillin V acylase crystal structure reveals new Ntn-hydrolase family members. Nat Struct Biol. 1999;6:414–416. doi: 10.1038/8213. [DOI] [PubMed] [Google Scholar]
- 26.Groll M, et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 27.Kadokura H, Katzen F, Beckwith J. Protein disulfide bond formation in prokaryotes. Annu Rev Biochem. 2003;72:111–135. doi: 10.1146/annurev.biochem.72.121801.161459. [DOI] [PubMed] [Google Scholar]
- 28.Altschul SF, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 29.The UniProt Consortium. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2008;36:D190–195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Read R. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr A. 1986;42:140–149. [Google Scholar]
- 31.Alkema WB, et al. Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and site-directed mutagenesis studies. Protein Eng. 2000;13:857–863. doi: 10.1093/protein/13.12.857. [DOI] [PubMed] [Google Scholar]
- 32.Okada T, et al. Crystal structures of gamma-glutamyltranspeptidase from Escherichia coli, a key enzyme in glutathione metabolism, and its reaction intermediate. Proc Natl Acad Sci USA. 2006;103:6471–6476. doi: 10.1073/pnas.0511020103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brannigan JA, et al. A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature. 1995;378:416–419. doi: 10.1038/378416a0. [DOI] [PubMed] [Google Scholar]
- 34.Smith JL, et al. Structure of the allosteric regulatory enzyme of purine biosynthesis. Science. 1994;264:1427–1433. doi: 10.1126/science.8197456. [DOI] [PubMed] [Google Scholar]
- 35.Torres-Bacete J, et al. Newly discovered penicillin acylase activity of aculeacin A acylase from Actinoplanes utahensis. Appl Environ Microbiol. 2007;73:5378–5381. doi: 10.1128/AEM.00452-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang D, et al. Cloning and characterization of penicillin V acylase from Streptomyces mobaraensis. J Biotechnol. 2007;128:788–800. doi: 10.1016/j.jbiotec.2006.12.017. [DOI] [PubMed] [Google Scholar]
- 37.Huang JJ, Petersen A, Whiteley M, Leadbetter JR. Identification of QuiP, the product of gene PA1032, as the second acyl-homoserine lactone acylase of Pseudomonas aeruginosa PAO1. Appl Environ Microbiol. 2006;72:1190–1197. doi: 10.1128/AEM.72.2.1190-1197.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nadal Jimenez P, et al. The role of PvdQ in Pseudomonas aeruginosa virulence under iron limiting conditions. Microbiology+ 2009 doi: 10.1099/mic.0.030973-0. in press. doi: 10.1099/mic.0.030973-0. [DOI] [PubMed] [Google Scholar]
- 39.Ericsson UB, et al. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal Biochem. 2006;357:289–298. doi: 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 40.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 41.Evans P. Scaling and assessment of data quality. Acta Crystallogr D. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 42.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 43.Collaborative Computational Project N, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 44.Hewitt L, et al. Structure of a slow processing precursor penicillin acylase from Escherichia coli reveals the linker peptide blocking the active-site cleft. J Mol Biol. 2000;302:887–898. doi: 10.1006/jmbi.2000.4105. [DOI] [PubMed] [Google Scholar]
- 45.Kim Y, Kim S, Earnest TN, Hol WGJ. Precursor structure of cephalosporin acylase. Insights into autoproteolytic activation in a new N-terminal hydrolase family. J Biol Chem. 2002;277:2823–2829. doi: 10.1074/jbc.M108888200. [DOI] [PubMed] [Google Scholar]
- 46.Jaroszewski L, et al. FFAS03: a server for profile–profile sequence alignments. Nucleic Acids Res. 2005;33:W284–288. doi: 10.1093/nar/gki418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Canutescu AA, Shelenkov AA, Dunbrack RL., Jr A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 2003;12:2001–2014. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Terwilliger T. SOLVE and RESOLVE: Automated structure solution, density modification and model building. J Synchrotron Radiat. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 49.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 50.Morris RJ, Perrakis A, Lamzin VS. ARP/wARP and automatic interpretation of protein electron density maps. Method Enzymol. 2003;374:229–244. doi: 10.1016/S0076-6879(03)74011-7. [DOI] [PubMed] [Google Scholar]
- 51.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 52.Davis IW, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kleywegt GJ, Jones TA. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr D. 1994;50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- 54.Connolly ML. The molecular surface package. J Mol Graphics. 1993;11:139–141. doi: 10.1016/0263-7855(93)87010-3. [DOI] [PubMed] [Google Scholar]
- 55.Delano WL. The PyMOL Molecular Graphics System. Palo Alto, CA, USA: DeLano Scientific LLC; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.