Abstract
The mechanism of substrate translocation through the ribosome is central to the rapid and faithful translation of mRNA into proteins. The rate-limiting step in translocation is an unlocking process that includes the formation of an “unlocked” intermediate state, which requires the convergence of large-scale conformational events within the ribosome including tRNA hybrid states formation, closure of the ribosomal L1 stalk domain, and subunit ratcheting. Here, by imaging of the pretranslocation ribosome complex from multiple structural perspectives using two- and three-color single-molecule fluorescence resonance energy transfer, we observe that tRNA hybrid states formation and L1 stalk closure, events central to the unlocking mechanism, are not tightly coupled. These findings reveal that the unlocked state is achieved through a stochastic-multistep process, where the extent of conformational coupling depends on the nature of tRNA substrates. These data suggest that cellular mechanisms affecting the coupling of conformational processes on the ribosome may regulate the process of translation elongation.
Keywords: single-molecule FRET, translation, translocation, tRNA hybrid states, translation regulation
Protein synthesis in the cell is principally regulated through the control of ribosome biogenesis (1) and translation initiation (2). Yet, a growing body of evidence, including recent direct observations of translating ribosomes (3), suggests that the process of translation elongation may also be highly regulated (4). During elongation, polypeptides are synthesized on the two subunit (30 and 50S in bacteria), approximately 2.4 MDa ribosome according to the mRNA codon sequence. Critical to the elongation cycle, tRNA and mRNA substrates bound within the Aminoacyl (A) and peptidyl (P) sites of the pretranslocation complex must be translocated by up to approximately 30–50 Å to the P and exit (E) sites, respectively.
In bacterial cells, rapid, directional, substrate movements with respect to the ribosome are catalyzed by the conserved GTPase, Elongation factor-G (EF-G). EF-G is believed to catalyze translocation by remodeling the global architecture of the pretranslocation ribosome complex and the array of highly-conserved contacts between rRNA, mRNA, and tRNA observed crystallographically (5–9). However, recent evidence, including single-molecule fluorescence resonance energy transfer (smFRET) measurements (10–13), demonstrates that large-scale remodeling of structural elements important to translocation can spontaneously occur within the pretranslocation-ribosome complex. These findings, together with evidence of spontaneous mRNA and tRNA translocation (5, 14–16), suggest that greater knowledge of the nature and timing of conformational processes intrinsic to the pretranslocation ribosome complex may be critical to understanding the translocation mechanism.
The process of translocation is rate limited by an “unlocking” process in the ribosome that remodels the nature of substrate-ribosome interactions in order to facilitate substrate movement (5, 6, 17). Cryoelectron microscopy (cryoEM) studies suggest that the formation of an intermediate “unlocked state” of the pretranslocation complex may play a key role in the unlocking mechanism (5, 18–20). In this state, tRNAs adopt “hybrid state” binding configurations, the L1 stalk domain closes and the 30S subunit undergoes a counterclockwise, ratchet-like rotation with respect to the 50S. Hybrid state tRNA configurations arise from the movement of the 3′-CCA termini of A- and P-site tRNAs to adjacent binding sites in the large subunit prior to codon-anticodon movement in the small subunit [forming A/P and P/E states, respectively (13, 21–26)]. Closure of the L1 stalk domain, comprised of helices 76–78 of 23 S rRNA and the ribosomal protein L1, describes its movement from an extended position, distal to the subunit interface, toward the E-site and the large subunit body (11, 18–20). In a closed position, elements of the L1 stalk can make stabilizing contacts with the P/E hybrid tRNA (13, 19), consistent with earlier observations (27). Subunit ratcheting is a complex set of motions that entails the remodeling of numerous bridging contacts found at the subunit interface that are involved in substrate positioning (11, 18–20, 28). As each of these conformational events can occur spontaneously (10–13, 29, 30), the process of achieving the unlocked state is understood to be intrinsic to the pretranslocation ribosome complex.
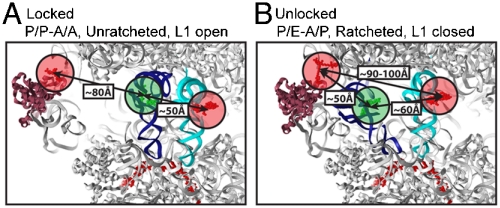
At present, conflicting evidence exists as to the extent to which the conformational events underpinning unlocked state formation are coupled. CryoEM studies (29, 30), and recent smFRET investigations (12), suggest that the pretranslocation complex fluctuates between just two conformational states: A locked state where the P-site tRNA is classically bound, the subunits are unratcheted, and the L1 stalk is open; and an unlocked state in which tRNA hybrid states are formed, subunits are ratcheted, and the L1 stalk is closed (Fig. 1A, B). In this view, conformational transitions between the locked and unlocked states are tightly coupled. By contrast, high-resolution crystallographic data suggest that domain motions in the ribosome related to subunit ratcheting can occur independently (9, 31). Additionally, smFRET studies suggest that intermediate configurations of the L1 stalk exist (11), that the A-and P-site tRNAs can move independently, and that hybrid-state formation and subunit ratcheting occur on distinct time scales (10, 13).
Fig. 1.
Structural models of locked and unlocked states of the pretranslocation ribosome complex showing estimated distances between fluorescently labeled components. A The locked ribosome configuration consistent with the low-FRET (approximately 0.1) state observed on complexes with labeled L1 and P-site tRNA, and the high-FRET (approximately 0.54) state observed on complexes with labeled A- and P-site tRNAs. B The unlocked ribosome configuration consistent with the high-FRET (approximately 0.65) state observed in complexes with labeled L1 and P-site tRNA, and the intermediate-FRET (approximately 0.4) state observed with both tRNAs labeled. The ribosome is shown in (Gray), A-site tRNA in (Cyan), P-site tRNA in (Blue), mRNA in (Red) and the L1 protein in (Magenta). The estimated center of mass of donor (Green) and acceptor (Red) fluorophores used are shown as semitransparent circles.
Here, to directly assess the order and timing of conformational transitions leading to the unlocked state, high-resolution, two- and three-color smFRET imaging of pretranslocation complexes have been performed from multiple structural perspectives. As previously described (13), complexes containing fluorescently labeled A- and P-site tRNAs were used to monitor the rates of hybrid-state formation. Complexes bearing fluorescently labeled L1 protein and P-site tRNA were used to monitor the rate at which formation of the unlocked state occurred. Complexes containing all three labeled components were used to directly monitor the relative motions of P-site tRNA with respect to A-site tRNA and the L1 stalk. The resulting observations, supported by site-directed mutagenesis of rRNA and structural modeling, reveal that conformational events leading to the unlocked state are not tightly coupled. These data suggest that formation of the unlocked intermediate state of the ribosome preceding translocation likely occurs by way of a multistep process. Similar observations obtained on pretranslocation complexes bearing distinct tRNAs show that a correlation exists between the rates of unlocked-state formation and the rate of translocation. These findings suggest that controlling the extent of conformational coupling between hybrid-state formation, L1 stalk closure, and subunit ratcheting may serve as a regulatory mechanism during elongation that could contribute substantially to the global rate of protein synthesis.
Results
Pretranslocation Ribosome Complexes Bearing Site-Specifically Labeled L1 Protein are Functional in Translation Elongation Reactions.
In order to investigate the order and timing of conformational events underpinning formation of the unlocked state of the ribosome, pretranslocation complexes were generated with 70S particles containing fluorescently labeled L1 protein (Fig. S1), labeled P-site tRNAfMet and A-site fMet-Phe-tRNAPhe. Single-molecule FRET data were obtained using a wide-field, prism-based total internal reflection imaging system at 40 ms time resolution following previously established procedures (13) (Materials and Methods).
Five positions of L1 labeling were considered, each returning FRET values consistent with existing structural data for locked and unlocked ribosome configurations (Fig. S2). Four of the five L1 labeling sites considered yielded evidence indicative of at least one intermediate-FRET state (Fig. S2). Complexes labeled at position S55C of the L1 protein were chosen for in-depth analyses as this site of labeling provided; (i) the greatest overall L1 incorporation efficiency, (ii) the greatest dynamic range in experimentally observed FRET values, and (iii) isotropic-fluorophore-tumbling characteristics amenable to estimating FRET-distance relationships (32). The competency of such complexes in tRNA accommodation into the A-site, peptide-bond formation, hybrid-state formation, and translocation was demonstrated using previously established puromycin-reactivity assays (13, 26), and presteady-state measurements of translocation rates (Fig. S3 and Table S1 (33).
The Unlocked State of the Ribosome is Spontaneously Achieved in the Pretranslocation Complex.
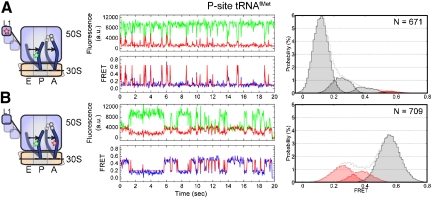
As anticipated from prior smFRET studies (11, 12), pretranslocation complexes containing L1 (Cy5-S55C) and P-site tRNAfMet (Cy3-s4U8) were observed to be intrinsically dynamic, reporting specifically on the spontaneous formation of a high-FRET, unlocked state, where the elbow of the P/E hybrid tRNA and the L1 protein tethered to the closed L1 stalk come into close proximity (ca. 50 Å). The dynamic behavior of these complexes (Fig. 2A) was assessed using hidden Markov modeling and transition density plot analyses (34). After considering Markov chains of increasing complexity, the minimal model adequately describing the smFRET data was shown to be a looped model containing four distinct FRET states (approximately 0.1, 0.25, 0.4, and 0.65; Fig. S4). The existence of these four states were further evidenced by direct inspection of smFRET trajectories and confirmed through experiments performed at fourfold increased time resolution (10 ms) (Fig. S5). Data idealized to this model generated a sequence of dwell times for each FRET trajectory that was used to construct the distributions of data points assigned to each FRET state observed (Fig. 2A, Right). Dwell time information were subsequently used to estimate the rates of interconversion between FRET states (Table S2) (35).
Fig. 2.
P/E hybrid state formation and L1 stalk closure are not tightly coupled in pretranslocation complexes containing initiator tRNA in the P-site. The dynamics of the pretranslocation complex containing P-site tRNAfMet and A-site fMet-Phe-tRNAPhe are shown from two distinct structural perspectives. A L1- and P-site tRNA-labeled complexes. B A- and P-site tRNA-labeled complexes. Left Cartoon models of the labeling sites in the pretranslocation complex and the putative dynamic elements; Cy5 (Red Star), Cy3 (Green Star). Center Single-molecule fluorescence (Cy3, (Green); Cy5, (Red)) and FRET (Blue) trajectories, where FRET idealization obtained through hidden Markov Modeling methods (Red) is overlaid on the FRET trace. Right Nonzero FRET state occupancies identified through idealization. (Pink) and (Red) shaded distributions correspond to those states representing P/E hybrid tRNA configurations and the unlocked state of the ribosome, respectively.
Atomic-resolution-structural modeling of the pretranslocation complex indicated that the low-FRET state, reflecting an interdye distance of approximately 80 Å (R0 ≈ 56 Å), was consistent with the “locked” configuration. The high-FRET state, reflecting an estimated interdye distance of approximately 50 Å, closely approximated the L1 to P-site tRNA distance observed in structures resembling the “unlocked” state (18–20, 29, 30). The intermediate-FRET states observed (approximately 0.25 and 0.4), while suggestive of uncoupled P-site tRNA and the L1 stalk motions (Fig. S6 and Table S3 and S4), may arise from a multitude of distinct conformational events in the ribosome and could not be unambiguously assigned. Thus, the complete kinetic analysis of the smFRET data shown in Table S2 was reduced to the rates of transitions into, and out of, low-FRET (0.1; k→low,klow→) and high-FRET (0.65; k→high,khigh→) states (Table 1). Such transitions provide estimates of the rates into, and out of, states structurally consistent with locked and unlocked ribosome configurations, respectively. As shown in Figure 2A and Table 1, both histogram and rate analyses suggested that the pretranslocation complex only rarely and transiently fluctuated out of the locked configuration.
Table 1.
Results of hidden Markov modeling analysis
| L1-tRNA FRET | tRNA-tRNA FRET | ||||||||||||||
| P-site tRNA | Occupancies (%) | Rates (sec-1) | Occupancies (%) | Rates (sec-1) | |||||||||||
| 0.1 |
0.25 |
0.4 |
0.65 |
klow→ |
k→low |
khigh→ |
k→high |
0.54 |
0.4 |
0.25 |
kA/A |
kA/P |
kP/P |
kP/E |
|
| tRNAfMet | 76 ± 1 | 18 ± 1 | 4.0 ± 0.4 | 2.0 ± 0.3 | 1.3 ± 0.1 | 7.7 ± 0.4 | 25 ± 2 | 0.12 ± 0.01 | 66 ± 1 | 13 ± 1 | 20 ± 1 | 10.0 ± 0.2 | 1.6 ± 0.2 | 5.7 ± 0.1 | 2.8 ± 0.3 |
| tRNAPhe | 63 ± 2 | 18 ± 1 | 8.1 ± 0.6 | 11 ± 1 | 2.5 ± 0.3 | 9.7 ± 0.7 | 17 ± 1 | 0.44 ± 0.03 | 22 ± 1 | 15 ± 1 | 63 ± 1 | 6.8 ± 0.4 | 1.4 ± 0.1 | 2.2 ± 0.2 | 3.8 ± 0.3 |
As described in SI Text, idealized smFRET trajectories report on the total occupancy of the observed FRET states, and the rates of interconversion between these states. For L1- and P-site tRNA complexes the kinetic data are summarized by the rates of transitions into, and out of, low (approximately 0.1) and high (approximately 0.65) FRET states (k→low,klow→; k→high,khigh→). For A- and P-site tRNA-labeled complexes, kinetics are summarized as the rates forming classical and hybrid configurations (kP/P,kP/E). In all cases the approximate standard errors are given.
L1 Stalk Closure Is Not Tightly Coupled to P/E Hybrid-State Formation.
To probe the coupling of hybrid-state formation and L1 stalk closure, tRNA positions were monitored on similar pretranslocation complexes bearing site specifically labeled A- and P-site tRNAs (13, 26). As previously described, three FRET states were observed reporting on the exchange between classical (approximately 0.55 FRET) and two distinct hybrid tRNA configurations: hybrid state-1 (approximately 0.4 FRET; H1) in which A- and P-site tRNAs occupy A/P and P/E hybrid states; and hybrid state-2 (approximately 0.25 FRET; H2) in which the P-site tRNA resides in a P/E hybrid state, and A-site tRNA remains largely classically bound (13) (Fig. 2B). Comparing P/E hybrid-state occupancy (approximately 33%, H1 + H2, Fig. 2B) to that of the unlocked state (approximately 2%), determined on the corresponding L1- and P-site tRNA-labeled complexes (Fig. 2A), revealed that the unlocked conformation is significantly less stable than the P/E hybrid state (Table 1). Specifically, the estimated rate of P/E hybrid-state formation (kP/E) was approximately two to threefold faster than transitions out of the low-FRET, locked state (klow→) and approximately 20-fold faster than transitions into the high-FRET, unlocked state (k→high) (Table 1). These data suggested that excursions to the P/E hybrid state can occur without formation of the unlocked state and that P/E hybrid-state formation on the L1- and P-site tRNA-labeled complex can occur without any observable departure from the low-FRET state. These observations support a model in which P/E hybrid-state formation and L1 stalk closure are not tightly coupled and that formation of the unlocked state is a multistep process that occurs only via the convergence of these conformational events.
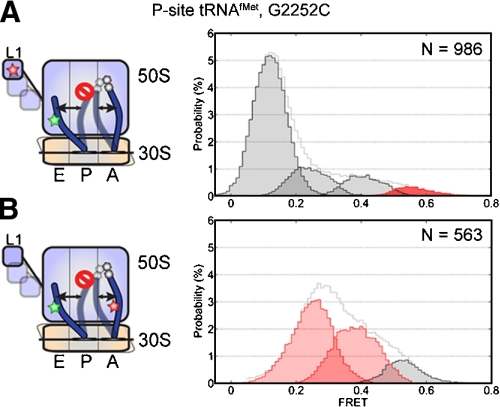
To test this model, identical experiments were performed on pretranslocation complexes bearing a G2252C mutation in the P loop of 23S rRNA, previously shown to promote the P/E hybrid state (13). As anticipated, complexes bearing labeled A- and P-site tRNAs in the context of this mutation showed approximately 86% P/E hybrid-state occupancy. In this mutant background L1- and P-site tRNA-labeled complexes showed approximately 3% high-FRET, unlocked state occupancy while the low FRET, locked configuration remained approximately 68% occupied (Fig. 3). To explain these data, the low-FRET state must represent an aggregate of locked and partially unlocked (P/E hybrid) ribosome configurations, where P/E hybrid-state formation does not necessarily lead to observable changes in FRET or changes in the rate exiting the low-FRET state (klow→). This model specifies that the site of labeling on the L1 protein can maintain an interdye distance of approximately 80 Å with respect to P-site tRNA in the P/P to P/E hybrid transition. Efforts to structurally model this constraint argue that the site of labeling on the L1 protein must move away from the P-site tRNA elbow for this to occur. This may be achieved if; (i) the L1 stalk adopts an extended open position in which it moves away from the 50S subunit body, (ii) the L1 stalk rotates away from the P-site tRNA elbow, or (iii) the labeled domain of the L1 protein moves independently of the L1 stalk. As described in SI Text, a ratcheted, P/E hybrid configuration of the pretranslocation ribosome complex consistent with the 80 Å distance constraint could be modeled by allowing the L1 stalk to adopt an extended open position similar to that found in the high-resolution structure of the 50S subunit from D. radiodurans (36) (Fig. S7).
Fig. 3.
Stabilization of the P/E hybrid state does not substantially increase unlocked state occupancy. Data from pretranslocation complex containing P-site tRNAfMet and A-site fMet-Phe-tRNAPhe, in the context of a G2252C mutation in the P loop of 23S rRNA are shown from two distinct structural perspectives. A L1- and P-site tRNA-labeled complexes. B A- and P-site tRNA-labeled complexes.
Evidence of Uncoupled Motions of P-site tRNA and the L1 stalk is Also Present with Elongator tRNA in the P-site.
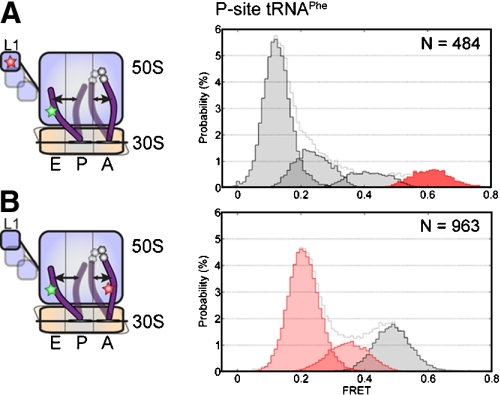
To examine whether uncoupled P/E hybrid-state formation and L1 stalk closure were specific to complexes bearing P-site initiator tRNAfMet, identical interrogations were performed on pretranslocation complexes containing P-site elongator tRNAPhe and A-site NAc-Phe-Lys-tRNALys. Such complexes, shown again to be fully functional in translocation (Table S1), visited analogous states with similar FRET values using both labeling strategies. Consistent with the observation that such complexes translocate approximately 1.5-fold more quickly than those containing initiator tRNAfMet (Table S1) (21, 33), elongator tRNA-programmed pretranslocation complexes showed substantially greater P/E hybrid-state occupancy (approximately 78% vs. 33%), and modestly increased unlocked-state occupancy (approximately 11% vs. approximately 2%) (Figs. 2 and 4 and Table 1). Such distinctions resulted from an approximately 1.4-fold increase in the rate of P/E hybrid-state formation (kP/E ≈ 3.8 sec-1 vs. 2.8 sec-1) and a twofold increase in P/E hybrid-state stability. Simultaneously, two and threefold increases were observed in the rates out of low-FRET (klow→) and into high-FRET states (k→high), respectively. In line with the conclusions obtained from complexes bearing P-site tRNAfMet, the rate of P/E hybrid-state formation was approximately twofold faster than the rate of transition out of the low-FRET state (klow→) and approximatley 10-fold faster than that of the unlocked state (k→high). These results confirmed that L1 stalk closure and P/E hybrid-state formation can occur in an uncoupled fashion on pretranslocation complexes containing elongator tRNAs.
Fig. 4.
P/E hybrid state formation and L1 stalk closure are not tightly coupled in pretranslocation complexes containing initiator tRNA in the P-site. Data from pretranslocation complex containing P-site tRNAPhe and A-site NAc-Phe-Lys-tRNALys are shown from two distinct structural perspectives. A L1- and P-site tRNA-labeled complexes. B A- and P-site tRNA-labeled complexes.
Three-Color FRET Experiments Support the Notion of Uncoupled tRNA and L1 Stalk Motions.
To directly test whether hybrid-state formation and L1 stalk closure can occur independently, pretranslocation ribosome complexes containing Cy3-labeled P-site tRNA, Cy5-labeled A-site tRNA, and Cy5.5-labeled L1 protein were examined using a three-color smFRET imaging strategy (37). In such experiments, the Cy3 fluorophore linked to P-site tRNA is capable of transferring energy to both Cy5 and Cy5.5. FRET between Cy5 and Cy5.5 is expected to be negligible except upon formation of the unlocked configuration when the dyes linked to the L1 protein and A-site tRNA should come within approximately 90–100 Å of one another. In a simple two-state model, where P/E hybrid-state formation and L1 stalk closure are perfectly coupled, decreased FRET efficiency to the Cy5-labeled A-site tRNA, resulting from P/E hybrid-state formation, should be concomitant with increased FRET efficiency to Cy5.5 as a result of L1 stalk closure. If formation of the unlocked state is a multistep process, where P/E hybrid-state formation and L1 stalk closure can occur independently, more complex FRET trajectories are anticipated.
As observed in the two-color smFRET experiments, pretranslocation complexes bearing Cy3-labeled P-site tRNAfMet, displayed anticorrelated changes in Cy3/Cy5 fluorescence (Fig. 5A, Fluorescence) and Cy5-FRET fluctuations (Fig. 5A, FRET) consistent with the hybrid-classical exchange. However, due to differences in the optical treatment of fluorescence in the three-color experiment (Supporting Information Text), the signal-to-noise ratio of these measurements and the observed FRET values were lower. Thus, the dominant high-FRET state observed, corresponding to the classical tRNA positions, had a FRET value of approximately 0.5. Transitions to lower-FRET states (approximately 0.16–0.35), corresponding to H1 and H2 transitions, were observed but could not be differentiated quantitatively.
Fig. 5.
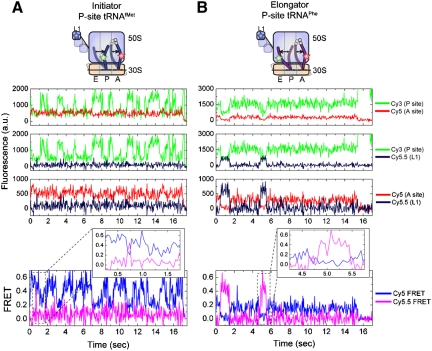
Uncoupled motions of P-site tRNA and the L1 stalk observed through multicolor smFRET. smFRET trajectories acquired from pretranslocation complexes bearing Cy5.5-labeled L1, Cy5-labeled A-site tRNA, and Cy3-labeled P-site tRNA. Data is shown for complexes containing A P-site tRNAfMet and A-site fMet-Phe-tRNAPhe, or B P-site tRNAPhe and A-site NAc-Phe-Lys-tRNALys. From Top to Bottom, the first three panels show Cy3/Cy5, Cy3/Cy5.5, Cy5/Cy5.5 fluorescence trajectories from a single complex. Bottom panels, and insets, show anticorrelated changes in Cy5- and Cy5.5 FRET channels indicative of FRET between Cy5 and Cy5.5 fluorophores in the unlocked state.
Consistent with the two-color smFRET data and a multistep model of unlocked-state formation, Cy5.5 fluorescence and Cy5.5-FRET trajectories within the same complex displayed a dominant low-FRET (approximately 0.03) state (Fig. 5A, FRET). Transitions to higher-FRET (approximately 0.4–0.5), closed L1-stalk states, although clearly observed, were rare. Evidence of anticorrelated changes in Cy5 and Cy5.5 fluorescence concomitant with these transitions, reflected most clearly as anticorrelated changes in Cy5- and Cy5.5-FRET channels (Figure 5A, FRET Inset), confirmed the assignment of such events to transitions to the unlocked state. Only in this configuration do the L1 stalk and A-site tRNA achieve a proximity close enough for FRET (ca. 90–100 Å) (Fig. 1). These data are consistent with the model that tRNAfMet transitions to the P/E hybrid state substantially faster than L1 stalk closure and unlocked-state formation.
Evidence corroborating the two-color smFRET data and the multistep model of unlocked-state formation were also observed for triple-labeled pretranslocation complexes containing Cy3-labeled P-site tRNAPhe. In line with the increased P/E hybrid-state stability of this complex observed in two-color smFRET experiments, the dominant Cy5-FRET state evidenced had a FRET value between approximately 0.16 and 0.35 (Fig. 5B). Within the same complex, Cy5.5-FRET trajectories showed a dominant low-FRET (approximately 0.03) state punctuated by transitions to relatively long-lived higher-FRET states. Again, such transitions were accompanied by anticorrelated changes in the Cy5-FRET channel indicative of unlocked-state formation (Figure 5B, FRET Inset).
Discussion
Spontaneous Formation of the Unlocked State of the Ribosome Is a Multistep Process.
Here, using both two- and three-color smFRET imaging it was shown that P/E hybrid-state formation and L1 stalk closure can occur through uncoupled processes in the pretranslocation complex. Although the L1- and P-site tRNA labeling strategy yields smFRET signals that report on the motions of two dynamic components, the advantage of this approach is that coincident motions of these elements towards each other generates a high-FRET state consistent with the unlocked configuration of the pretranslocation ribosome complex. In line with this view, structural models and experiments performed on the EF-G-bound ribosome (32), suggest that the unlocked state can only be achieved when both L1 stalk closure and P/E hybrid-state formation are accompanied by subunit ratcheting. As we and others (12, 29, 30) show, this key intermediate in the translocation reaction coordinate can be spontaneously achieved, perhaps helping to explain translocation in the absence of EF-G or nucleotide hydrolysis (5, 14–16). While the site-specific labeling of ribosomal components may lead to subtle perturbations of the metastable ribosome energy landscape (28), the correlation between unlocked-state occupancy and translocation rates for the distinct pretranslocation complexes investigated, as well as the nearly wild-type activities of dye-labeled complexes, argue that the present observations describe general features of pretranslocation ribosome complex dynamics.
The present study supports a model in which formation of the unlocked state requires the convergence of forward transitions in at least three distinct, large-scale structural exchange processes. Taken together with previous observations (11), these data suggest that P/E hybrid-state formation (kP/E) occurs relatively rapidly, while L1 stalk closure (k0.1→) and subunit ratcheting may occur more slowly and at comparable rates. While the stochastic nature of these three processes inherently specifies that they can occur in a concerted fashion with finite probability, their distinct time scales of motion suggest that unlocked-state formation principally follows a multistep, multipathway mechanism. The requirement for at least three distinct mobile elements in the ribosome to converge implies that achieving this state is a low-probability event that likely follows a conformational capture- or induced fit-like process (38). Given that the unlocked state, once achieved, is also highly transient in nature, we conclude that formation of this intermediate state may contribute substantially to the rate-limiting unlocking process preceding substrate movement.
L1 Stalk Motions May Determine the Effective Rate That the Unlocked State Is Achieved and Its Lifetime.
Key insights into the dynamics of the pretranslocation complex and the mechanism of translocation are afforded by the present observation that the P/E hybrid state is achieved approximately 10–20-times more rapidly than unlocked-state formation (Table 1). The first, stated above, is that P/E hybrid state formation, L1 stalk closure and subunit ratcheting are not tightly coupled and that multiple kinetic and structural pathways can lead to unlocked-state formation. Additional insights pertain to the predominant low-FRET state observed on L1- and P-site tRNA-labeled pretranslocation complexes containing either a G2252C mutation (Fig. 3) or P-site tRNAPhe (Fig. 4). In such complexes the P/E hybrid state was greater than 75% occupied while the unlocked state was only weakly populated (3–11%). The striking conclusion that can be drawn from this observation is that the low-FRET state represents ribosome conformations containing both classical and hybrid P-site tRNA. Given the suggested correlation between P/E hybrid-state occupancy and subunit ratcheting (18–20, 29, 30, 39, 40), these data suggest that the persistent low-FRET state observed represents a P/E hybrid, ratcheted ribosome configuration where the site of L1 labeling occupies a position approximately 80Å distal to the P/E tRNA elbow. Intermediate-FRET states (approximately 0.25–0.4) may therefore be predominantly attributed to conformational events in the particle that reflect isolated motions of the L1 stalk (Figs. S4 and Fig. S5). In line with the distinct time scales of L1 stalk opening and subunit unratcheting noted previously (11), formation and stability of the unlocked state may be limited by L1 stalk motions into and out of the closed configurations, respectively.
Structural Modeling of L1 Stalk Motions.
Previous structural data (41) and smFRET experiments (11) suggest that the L1 stalk can adopt at least four distinct L1 stalk positions on the 70S ribosome. In the present study, experiments on both wild-type (Figs. 2A and 4A) and P/E-stabilized L1- and P-site tRNA-labeled pretranslocation complexes (Fig. 3), in which a low-FRET state is observed to predominate, suggest additional complexities may exist in the motions of the L1 stalk. While L1 stalk motions consistent with this FRET state may in principle arise through stalk bending, rotation and/or uncoupled motions of the L1 protein and the L1 stalk, an extended-open L1 stalk position may be more likely given that it is consistent with existing structural data (18, 19). Our modeling efforts suggest that such an extended-open L1 stalk configuration may be achieved by a hinge-like remodeling event centered on the highly conserved, three-way junction of helices 75, 76, and 79 of 23S rRNA (Fig. S8). Further experiments, will be necessary to examine this notion. While the mechanistic significance of such L1 stalk mobility is presently unclear, opening and closing cycles of the L1 stalk on the otherwise unlocked pretranslocation complex may contribute to substrate movement (19). An extended-open configuration of the L1 stalk may facilitate E-site tRNA release following translocation.
Implications for the Regulation of Translation Elongation.
The existance of independent conformational degrees of freedom in the ribosome predicts that regulating the extent of conformational coupling in the pretranslocation complex may provide a potential mechanism for controlling the rate of translation elongation. The coupling efficiencies underpinning unlocked-state formation may be regulated by environmental factors, posttranscriptional or translational modifications of the ribosome, ribosome interactions with mRNA, and/or ligand binding events. Indeed, the estimated rates of spontaneous unlocked state formation (k→high) are approximately 5–10-fold slower than EF-G-catalyzed translocation rates measured under saturating conditions (10 μM EF-G) (Tables 1 and Table S1). Thus, a key role of EF-G may be to accelerate translocation by altering the rate of unlocked-state formation (17). Thus, EF-G promoted unlocked-state formation may occur via distinct and/or concerted structural mechanisms.
A deeper understanding of how EF-G influences the rate and extent of conformational events on the ribosome will most certainly benefit from additional labeling strategies and further advances in three-color smFRET imaging. Such efforts may ultimately facilitate the simultaneous monitoring of conformational degrees of freedom in the ribosome during transit of the mRNA open reading frame and detailed investigations into how such events may be regulated in the cell.
Materials and Methods
Preparation of Dye-Labeled Ribosome Complexes.
Ribosomal protein L1 was cloned, purified, and labeled with either the Cy5 or Cy5.5 fluorophore as described in SI Text. Translation factors were prepared as described in ref. 42. All tRNA were purchased (Sigma), fluorescently labeled with either Cy3 or Cy5, and purified by hydrophobic interaction chromatography (13). Aminoacylation of tRNAs, except tRNAPhe (Cy3-s4U8) (SI Text), was performed as previously described in ref. 13. 70S ribosomes were isolated and, where appropriate, reconstituted with dye-labeled L1 as described in ref. 32. Initiation complexes were formed with fMet-tRNAfMet and 5′-biotinylated mRNA in the presence of initiation factors (13). Ribosomes were nonenzymatically initiated with NAc-Phe-tRNAPhe by incubation with excess tRNA for 10 min at 37 °C. Pretranslocation complexes were formed by incubating the surface-immobilized ribosome complexes with EF-Tu(GTP)-aa-tRNA for 2 min at room temperature (13).
Acquisition and Analysis of smFRET Data.
All smFRET experiments were performed at room temperature in Tris Polymix buffer (50 mM Tris-OAc pH 7.5, 100 mM KCl, 15 mM Mg(OAc)2, 5 mM NH4OAc, 0.5 mM Ca(OAc)2, 0.1 mM EDTA, 5 mM putrescine, 1 mM spermidine, and 5 mM BME), in the presence of an oxygen scavenging and triplet-state quenching system (43), on a prism-based total internal reflection fluorescence microscope (13). Microscope specifications and the details of data collection for two- and three-color FRET experiments can be found in SI Text. All data were acquired with Metamorph (Molecular Devices), and analyzed according to an automated protocol implemented in Matlab (Mathworks) and QuB (www.qub.buffalo.edu), as described in SI Text.
Supplementary Material
Acknowledgments.
The authors thank members of the Blanchard lab and Dr. Joachim Frank’s lab (Columbia University, Howard Hughes Medical Institute) for insightful comments on the manuscript. This work was supported by National Institutes of Health Grant 1R01GM079238-01, the Alice Bohmfalk Charitable Trust, and NYSTAR.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908597107/DCSupplemental.
References
- 1.Wilson DN, Nierhaus KH. The weird and wonderful world of bacterial ribosome regulation. Crit Rev Biochem Mol Biol. 2007;42:187–219. doi: 10.1080/10409230701360843. [DOI] [PubMed] [Google Scholar]
- 2.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wen JD, et al. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herbert TP, Proud CG. In: Translational Control in Biology and Medicine. Mathews MB, Sonenberg N, Hershey JW, editors. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2007. pp. 601–624. [Google Scholar]
- 5.Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. The process of mRNA-tRNA translocation. Proc Natl Acad Sci USA. 2007;104:19671–19678. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shoji S, Walker SE, Fredrick K. Ribosomal translocation: One step closer to the molecular mechanism. ACS Chem Biol. 2009;4:93–107. doi: 10.1021/cb8002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selmer M, et al. Structure of the 70 S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 8.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70 S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 9.Berk V, Zhang W, Pai RD, Cate JHD. Structural basis for mRNA and tRNA positioning on the ribosome. Proc Natl Acad Sci USA. 2006;103:15830–15834. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornish PV, et al. Following movement of the L1 stalk between three functional states in single ribosomes. Proc Natl Acad Sci USA. 2009;106:2571–2576. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 13.Munro JB, Altman RB, O’Connor N, Blanchard SC. Identification of two distinct hybrid-state intermediates on the ribosome. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gavrilova LP, Kostiashkina OE, Koteliansky VE, Rutkevitch NM, Spirin AS. Factor-free (“non-enzymic”) and factor-dependent systems of translation of polyuridylic acid by Escherichia coli ribosomes. J Mol Biol. 1976;101:537–552. doi: 10.1016/0022-2836(76)90243-6. [DOI] [PubMed] [Google Scholar]
- 15.Cukras AR, Southworth DR, Brunelle JL, Culver GM, Green R. Ribosomal proteins S12 and S13 function as control elements for translocation of the mRNA:tRNA complex. Mol Cell. 2003;12:321–328. doi: 10.1016/s1097-2765(03)00275-2. [DOI] [PubMed] [Google Scholar]
- 16.Fredrick K, Noller H. Catalysis of ribosomal translocation by sparsomycin. Science. 2003;300:1159–1162. doi: 10.1126/science.1084571. [DOI] [PubMed] [Google Scholar]
- 17.Savelsbergh A, et al. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 18.Connell SR, et al. Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol Cell. 2007;25:751–764. doi: 10.1016/j.molcel.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 19.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 20.Taylor DJ, et al. Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO J. 2007;26:2421–2431. doi: 10.1038/sj.emboj.7601677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dorner S, Brunelle JL, Sharma D, Green R. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat Struct Mol Biol. 2006;13:234–241. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma D, Southworth DR, Green R. EF-G-independent reactivity of a pre-translocation-state ribosome complex with the aminoacyl tRNA substrate puromycin supports an intermediate (hybrid) state of tRNA binding. RNA. 2004;10:102–113. doi: 10.1261/rna.5148704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker SE, Shoji S, Pan D, Cooperman BS, Fredrick K. Role of hybrid tRNA-binding states in ribosomal translocation. Proc Natl Acad Sci USA. 2008;105:9192–9197. doi: 10.1073/pnas.0710146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 25.Pan D, Kirillov S, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian AR, Dabbs ER. Functional studies on ribosomes lacking protein L1 from mutant Escherichia coli. Euro J Biochem. 1980;112:425–430. doi: 10.1111/j.1432-1033.1980.tb07222.x. [DOI] [PubMed] [Google Scholar]
- 28.Munro JB, Sanbonmatsu KY, Spahn CM, Blanchard SC. Navigating the ribosome’s metastable energy landscape. Trends Biochem Sci. 2009;34:390–400. doi: 10.1016/j.tibs.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agirrezabala X, et al. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Julian P, et al. Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc Natl Acad Sci USA. 2008;105:16924–16927. doi: 10.1073/pnas.0809587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 32.Munro JB, Altman RB, Tung CS, Sanbonmatsu KY, Blanchard SC. A fast dynamic mode of the EF-G-bound ribosome. EMBO J. 2005 doi: 10.1038/emboj.2009.384. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Studer SM, Feinberg JS, Joseph S. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J Mol Biol. 2003;327:369–381. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- 34.Qin F. Restoration of single-channel currents using the segmental k-means method based on hidden Markov modeling. Biophys J. 2004;86:1488–1501. doi: 10.1016/S0006-3495(04)74217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin F, Auerbach A, Sachs F. Estimating single-channel kinetic parameters from idealized patch-clamp data containing missed events. Biophys J. 1996;70:264–280. doi: 10.1016/S0006-3495(96)79568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harms J, et al. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 37.Hohng S, Joo C, Ha T. Single-molecule three-color FRET. Biophys J. 2004;87:1328–1337. doi: 10.1529/biophysj.104.043935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leulliot N, Varani G. Current topics in RNA-protein recognition: Control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 39.Ermolenko DN, et al. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol. 2007;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 40.Spiegel PC, Ermolenko DN, Noller HF. Elongation factor G stabilizes the hybrid-state conformation of the 70 S ribosome. RNA. 2007;13:1473–1482. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korostelev A, Ermolenko DN, Noller HF. Structural dynamics of the ribosome. Curr Opin Chem Biol. 2008;12:674–683. doi: 10.1016/j.cbpa.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 43.Dave R, Terry DS, Munro JB, Blanchard SC. Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys J. 2009;96:2371–2381. doi: 10.1016/j.bpj.2008.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.