Abstract
IL-4 signaling promotes IgE class switching through STAT6 activation and the induction of Ig germ-line ε (GLε) transcription. Previously, we and others identified a transcription factor, Nfil3, as a gene induced by IL-4 stimulation in B cells. However, the precise roles of nuclear factor, IL-3-regulated (NFIL3) in IL-4 signaling are unknown. Here, we report that NFIL3 is important for IgE class switching. NFIL3-deficient mice show impaired IgE class switching, and this defect is B-cell intrinsic. The induction of GLε transcripts after LPS and IL-4 stimulation is significantly reduced in NFIL3-deficient B cells. Expression of NFIL3 in NFIL3-deficient B cells restores the impairment of IgE production, and overexpression of NFIL3 in the presence of cycloheximide induces GLε transcripts. Moreover, NFIL3 binds to Iε promoter in vivo. Together, these results identify NFIL3 as a key regulator of IL-4-induced GLε transcription in response to IL-4 and subsequent IgE class switching.
Keywords: IL-4 signal, immunoglobulin, germ-line transcription
IgE plays a central role in allergic immune response and is essential for host defense against pathogens in mucosal tissues (1). IgE binds to Fc-epsilon receptor (FcεRI) on mast cells and basophils, and cross-linking by antigen causes the release of inflammatory mediators (2). Normally, serum levels of IgE are very low compared with IgG, but elevated levels of IgE are observed in patients with allergic diseases (3). Reducing IgE concentrations alters allergic immune responses, such as those observed in asthma, anaphylaxis, and hyper-IgE disorder (4).
Cytokine stimulation of B cells in the germinal center can induce Ig heavy chain class switch recombination (CSR). IL-4 and CD40 (or LPS) stimulation induces germ-line ε (GLε) transcription from the Iε promoter. The production of GLε is essential before CSR, which is required for the production of IgE (3). Several transcription factors, including STAT6 and NF-κB, positively regulate this promoter activity (3). On the other hand, transcriptional repressors, such as Bcl-6 and Id2, are implicated in negative regulation of this promoter (5, 6). The balance of positive and negative transcriptional regulation is important for the control of IgE production. Although STAT6 has been shown to activate Iε promoter in response to IL-4, the induction of GLε by IL-4 is delayed, occurring after 16–24 h of stimulation (7). De novo protein synthesis is required for this transcription. These observations suggest a cascade of transcription factors, downstream of IL-4 receptor signaling, may be involved in the induction of GLε and IgE class switching.
Previously, cDNA representational difference analysis showed that the Nfil3 gene was induced by LPS plus IL-4 (8). Microarray analysis also identified the Nfil3 gene as the most strongly induced transcription factor by IL-4 that requires the presence of STAT6, thereby implicating that nuclear factor, IL-3-regulated (NFIL3) may have an important role in the regulation of transcription in response to IL-4 (9). NFIL3 (also called E4BP4), a basic leucine zipper (bZIP) type transcription factor, which has similar binding sites to those of CREB/ATF and C/EBP family protein, was initially identified as both a transcriptional repressor and transcriptional activator (10,11–12). Nfil3 is induced by several cytokines and hormones, including IL-3, IL-4, IL-6, IL-10, IL-15, parathyroid hormone (PTH), and insulin (8, 9, 13,14,15,16,17,18–19). Induction of Nfil3 by IL-4 also has been reported in T cells (14). In contrast, TGF-β stimulation suppresses Nfil3 expression induced during T helper (Th)1/Th2 polarization (20). Interestingly, de novo protein synthesis is not required for Nfil3 induction by PTH but is required for its induction by IL-3 (15, 16). IL-4- and IL-10-induced NFIL3 expression is STAT6- and STAT3-dependent, respectively (9, 19). This evidence suggests that NFIL3 is induced via the JAK-STAT pathway at an early time point after cytokine stimulation.
The precise function of Nfil3 in vivo is largely unknown. NFIL3 has been implicated in the regulation of circadian rhythm (10, 21,22–23). In immune cells, overexpression of NFIL3 in an IL-3-dependent B-cell line prevents apoptosis induced by IL-3 depletion, suggesting an antiapoptotic role for NFIL3 (15). Recently, it has been reported that NFIL3 KO mice showed the developmental defect of natural killer (NK) cells (24). NFIL3 is also implicated in malignant transformation that involves STAT3 activation (25). In this study, we generated and analyzed NFIL3 KO mice to understand the in vivo function of NFIL3. We demonstrate that NFIL3 is critical for IgE class switching in response to IL-4.
Results
NFIL3 Is Rapidly Induced by IL-4 Stimulation Independent of de Novo Protein Synthesis.
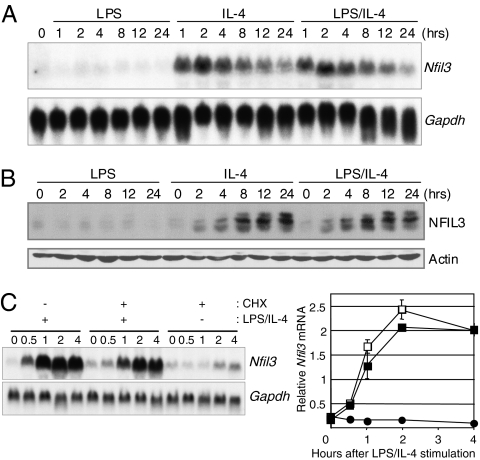
In a previous study, we identified genes that are regulated by STAT6 in response to IL-4 in B cells through microarray experiments (9). Among these genes, a transcription factor Nfil3 was identified as the transcription factor most strongly induced by IL-4. Nfil3 also was induced by IL-4 in T cells (14). To confirm the microarray studies, we examined the induction of Nfil3 mRNA and NFIL3 protein by LPS alone, IL-4 alone, or LPS plus IL-4 stimulation in M12.4.1 B-cell line. Nfil3 mRNA was induced within 1 h and NFIL3 protein within 2 h of IL-4 alone or LPS plus IL-4 stimulation (Fig. 1 A and B). Multiple forms of NFIL3 proteins were detected, and the amount of higher molecular weight species increased with the time. Only the smallest band was detected after phosphatase treatment in vitro, suggesting that NFIL3 can be phosphorylated by IL-4 stimulation or without IL-4 stimulation in B cells (Fig. S1). To determine if the rapid induction of NFIL3 requires protein synthesis, M12.4.1 cells were treated with the protein synthesis inhibitor cycloheximide (CHX) before stimulation. Northern blot analysis showed that CHX treatment did not block the rapid induction of NFIL3-encoding mRNA by IL-4 (Fig. 1C, Left). Quantification by real-time RT-PCR also indicates that CHX did not affect Nfil3 gene induction by IL-4 stimulation (Fig. 1C, Right), demonstrating that its induction is independent of de novo protein synthesis and suggesting that the Nfil3 gene is a direct target of STAT6. Taken together, the rapid induction of NFIL3 by IL-4 suggests that NFIL3 could play a role in the modulation of gene regulation downstream of IL-4.
Fig. 1.
NFIL3 expression is rapidly induced by IL-4 stimulation and is CHX-resistant. Rapid induction of Nfil3 mRNA (A) and NFIL3 protein (B) expression in M12.4.1 cells stimulated with IL-4 or LPS/IL-4. Expression of Nfil3 mRNA and NFIL3 protein was determined by Northern blot analysis and Western blot analysis, respectively. Data are representative of two independent experiments. (C) Induction of Nfil3 mRNA does not require de novo protein synthesis. RNA prepared from M12.4.1 cells stimulated as indicated was subjected to Northern blot analysis (Left) and real-time RT-PCR (Right). Relative expression of Nfil3 was normalized by the expression of Hprt1 mRNA. CHX(−) IL-4 (+) cells (▪), CHX(+) IL-4 (+) cells (□), and CHX(+) IL-4 (−) cells (•), respectively. Data are representative of two independent experiments.
Normal B-Cell and T-Cell Development in NFIL3-Deficient Mice.
We generated NFIL3-deficient mice to examine the role of NFIL3 in vivo. The Nfil3 gene consists of two exons, and the entire coding region is located in the second exon. ES cells were generated by homologous recombination in which the second exon was replaced with the neomycin-resistant gene by gene targeting (Fig. S2A). Homozygous NFIL3 KO mice generated from these ES cells were born at the expected Mendelian ratio (117:201:119 for WT, heterozygous, and KO mice, respectively), appeared grossly normal, and were fertile. To confirm the absence of NFIL3 protein in NFIL3 KO mice, cell lysates from IL-4-stimulated splenocytes for 24 h were prepared and loss of NFIL3 expression was confirmed by Western blot analysis. NFIL3 protein was detected in WT splenocytes but not in NFIL3 KO splenocytes (Fig. S2E), indicating that the Nfil3 gene is successfully disrupted in NFIL3 KO mice. We determined whether NFIL3 deficiency affects lymphocyte and myeloid cell development by flow cytometry. In bone marrow, spleen, and peritoneal cavity, the numbers of B cells in NFIL3 KO mice were comparable to those in WT mice (Fig. S3 A–E and Table S1). Similarly, the numbers of T cells in NFIL3 KO mice were normal in the thymus and spleen (Fig. S3 F–J and Table S1). The numbers of the other lineages, including myeloid and erythroid, were also normal, but NK-cell population (CD3−pan-NK+NKp46+) cells were significantly reduced consistent with a recent report (24) (Fig. S3 K–N and Table S1). Taken together, lack of NFIL3 does not have a pronounced effect on the development of hematopoietic cells, with the exception of NK cells.
Impaired IgE Class Switching in NFIL3-Deficient Mice.
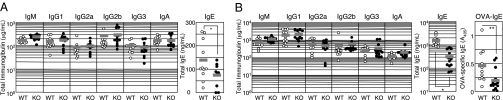
IL-4 signaling is a major regulator of Ig heavy chain class switching to the IgG1 and IgE isotypes, which occurs via rearrangement of the Ig heavy chain locus (3, 26). To explore the role of NFIL3 in class switching, we examined baseline serum Ig concentration in sera from NFIL3 KO and WT mice by ELISA (Fig. 2A). The concentration of all Ig subclasses except IgE was similar when NFIL3 KO and WT mice were compared. However, the IgE level in NFIL3 KO mice was reduced, on average, to 50% of that of WT mice, suggesting that NFIL3 is important for the normal production of IgE in vivo. To determine whether NFIL3 deficiency affects the ability to induce IgE production in response to antigen, mice were immunized with ovalbumin (OVA) with alum (Fig. 2B). Total Ig concentration of all subclasses except IgE was comparable in sera between OVA-immunized NFIL3 KO and WT mice. Total IgE levels in NFIL3 KO mice were ~5-fold lower, on average, compared with those in sera from WT mice. Further, OVA-specific IgE in NFIL3 KO mice was significantly lower than that seen in WT mice. These data suggest that NFIL3 is specifically required for efficient production of IgE.
Fig. 2.
IgE production is impaired in NFIL3 KO mice. (A) Serum Ig levels in NFIL3 KO and WT mice as determined by ELISA (n =9–13; *P < 0.069 for IgE). (B) Total Ig and OVA-specific IgE levels in sera from WT and NFIL3 KO mice injected twice i.p. with OVA. Sera were collected 14 days after the last immunization, and total Ig levels were determined by ELISA (n = 9–11). Total and OVA-specific IgE levels in sera from immunized WT and NFIL3 KO mice were also analyzed (n = 10–12; *P < 0.0001 for total IgE and **P < 0.028 for OVA-specific IgE).
We also examined the Nfil3 level in the B cells from OVA-immunized mice by real-time RT-PCR. After OVA immunization, expression of Nfil3 in splenic B cells was increased compared with that in B cells from unimmunized mice. This induction was not observed in B cells from OVA-immunized STAT6 KO mice (Fig. S4). These data indicate that IL-4/STAT6 signaling is involved in in vivo induction of NFIL3 expression in B cells and that induced NFIL3 can be involved in IgE class switching.
B-Cell Intrinsic Defect in IgE Production in the Absence of NFIL3 Expression.
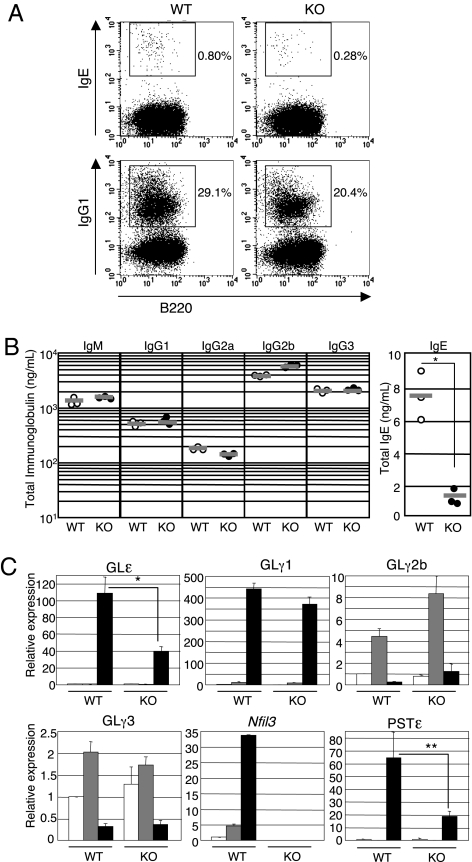
The defects of IgE production in NFIL3 could be secondary to B-cell intrinsic defects or attributable to defects in other cells, such as T cells, that are required for stimulation of IgE production by B cells. To determine directly whether the defect in IgE production by NFIL3 KO mice is B-cell intrinsic, we assessed production of IgE and IgG1 by cultured B cells in vitro. Splenic B cells stimulated with LPS/IL-4 for 5 days were stained and analyzed for intracellular IgE and IgG1 by flow cytometry. To prevent the detection of IgE bound to CD23, staining buffer containing EGTA was used. The levels of intracellular IgE expression of NFIL3 KO B cells were lower than those of WT B cells, although the expression levels were extremely low (Fig. 3A). Interestingly, although the serum levels of IgG1 in NFIL3 KO mice were normal (Fig. 2), the level of intracellular IgG expression of NFIL3 KO B cells was slightly lower than that of WT B cells. An equal percentage of B cells expressed sIgG3 and sIgG2b in response to LPS when NFIL3 KO and WT B cells were compared (Fig. S5). To quantify the impairment of IgE production by NFIL3 KO B cells further, secreted Ig levels in the culture supernatants from B cells stimulated were determined by ELISA (Fig. 3B). The production of IgM, IgG2a, IgG2b, and G3 by LPS-stimulated NFIL3 KO B cells was similar to that of WT cells. Secreted IgG1 produced by LPS/IL-4-stimulated NFIL3 KO B cells was also comparable to that of WT cells. However, IgE secretion by LPS/IL-4-stimulated NFIL3 KO B cells was significantly decreased relative to WT cells. Taken together, these results demonstrate that NFIL3 deficiency in B cells impairs their ability to class switch to IgE.
Fig. 3.
Impaired IgE production in NFIL3 KO B cells. (A) B cells from WT and NFIL3 KO mice were stimulated with LPS/IL-4 for 5 days, and cell intracellular expression of IgE and IgG1 was examined by flow cytometry. Two experiments were performed with similar results. (B) Ig levels secreted by B cells stimulated for 5 days with LPS (IgM, IgG2a, IgG2b, and IgG3) or LPS/IL-4 (IgE and IgG1) were determined by ELISA (n = 3; *P = 0.0024). (C) Reduced GLε and PSTε in NFIL3 KO B cells. Cells were unstimulated (open bars) or stimulated with LPS alone (gray bars) or LPS/IL-4 (closed bars) for 3 and 5 days, after which RNA was prepared. Germ-line transcript (GLT) and PST were analyzed by real-time RT-PCR. Relative expression of GLT and PST was normalized by the expression of Hprt1 mRNA. The expression of RNA in the unstimulated WT cells was set as 1. The graphs represent means and SD from the three WT mice and three NFIL3 KO mice (*P < 0.005, GLε expression between WT and KO B cells stimulated with LPS/IL-4 for 3 days; **P < 0.02, PSTε expression between WT and KO B cells stimulated with LPS/IL-4 for 5 days).
Reduced GLε Transcription in the Absence of NFIL3 Expression.
Extensive studies indicate that GL transcription through switch regions is required for CSR, likely secondary to altered accessibility of the switch region to the enzymes involved in CSR (27). Therefore, we determined whether the impaired IgE production in NFIL3 KO B cells is attributable to the impaired transcription of GLε RNA in response to LPS/IL-4. We examined GLε and postswitch ε transcripts (PSTε) by real-time RT-PCR in the splenic B cells cultured for 3 and 5 days with LPS or LPS/IL-4. The levels of GLγ1, GLγ2b, and GLγ3 are not largely affected by the loss of NFIL3 expression after 3 days of stimulation by LPS or LPS/IL-4. However, GLε was strongly induced in WT B cells but was reduced at lower levels in NFIL3 KO B cells by LPS/IL-4 stimulation (Fig. 3C). The PSTεs were also significantly decreased in NFIL3 KO B cells after 5 days of stimulation. This reduction was also confirmed by Northern blot analysis (Fig. S6). These data suggest that NFIL3 regulates GLε transcription in response to LPS/IL-4.
Normal IL-4 Signaling and Proliferation of NFIL3-Deficient B Cells.
Impaired GLε transcription could be secondary to an alteration in IL-4 signaling. To determine if IL-4 signaling is normal in NFIL3 KO B cells, splenic B cells cultured with LPS were stimulated with IL-4 and the activation of STAT6 and Akt was evaluated. Normal activation of STAT6 and Akt was observed in NFIL3 KO B cells when compared with WT cells, suggesting that the IL-4 signaling pathway is intact in the absence of NFIL3 expression (Fig. S7). Because cellular proliferation is required for the class switching to IgG1 and IgE (28,29–30), we determined whether the impaired class switching to IgE in NFIL3 KO B cells was attributable to the defect of B-cell proliferation. Thymidine incorporation of NFIL3 KO B cells in response to all stimuli indicated was comparable to that of WT B cells (Fig. S8A). Cell division numbers of NFIL3 KO B cells and WT B cells in response to LPS/IL-4 were comparable (Fig. S8B). These results indicate that the IgE class switching defect of NFIL3 KO B cells is not related to a proliferative defect.
NFIL3 Expression Is Required for and Promotes IgE Class Switching.
To determine whether the reconstitution of NFIL3 KO B cells with NFIL3 can restore the impairment of IgE production, NFIL3 KO B cells were retrovirally transduced with NFIL3 using pMiT vector (31), which coexpresses Thy1.1 and NFIL3. Transduced NFIL3 KO B cells were cultured in the presence of LPS/IL-4 for 4 days, and the secreted IgE levels were determined by ELISA. Consistently, NFIL3-transduced NFIL3 KO B cells secreted IgE at the same level as WT B cells, but control virus-transduced NFIL3 KO B cells did not secrete IgE (Fig. 4A). The effect of reintroduction of NFIL3 on IgG1 expression was not pronounced. NFIL3-transduced WT B cells showed elevated IgE secretion and similar IgG1 secretion compared with those of control virus-transduced WT B cells in the presence of LPS/IL-4 (Fig. 4B). These results indicate that NFIL3 expression controls IgE production.
Fig. 4.
Expression of NFIL3 is required for and promotes IgE class switching. NFIL3 KO B cells (A) and WT B cells (B) were infected with either NFIL3 or control retrovirus and cultured in the presence of LPS/IL-4 for 4 days. Over 95% of cells were infected with each virus according to the expression of Thy1.1 by flow cytometry. The supernatants of cell culture were analyzed by ELISA (n = 3; *P < 0.001, **P < 0.001). Three separate infections and cultures were performed, and data are representative of three independent cultures. (C) Overexpression of NFIL3 promotes early expression of GLε transcription. M12NFIL3 and M12Vector cells were stimulated with LPS/IL-4 for the indicated time. RNAs and cell lysates were subjected to Northern blot and Western blot analysis, respectively. The relative expression levels of GLε transcripts were quantified and are shown in the graph. Two experiments were performed with similar results. (D) Inhibition of NFIL3 induction by CHX results in the reduction of GLε transcription. M12NFIL3 and M12Vector cells pretreated with CHX were stimulated with LPS/IL-4 for the indicated time. RNAs and cell lysates were subjected to Northern blot and Western blot analysis, respectively. Two experiments were performed with similar results.
To elucidate the mechanism by which NFIL3 regulates IgE class switching, we determined whether overexpression of NFIL3 enhances GLε expression in response to LPS/IL-4. Northern blot analysis using NFIL3-overexpressing M12.4.1 cells (M12NFIL3 cells) and vector-infected M12.4.1 cells (M12Vector cells) treated with LPS/IL-4 demonstrated that GLε transcripts were detected as early as 1 h after stimulation in M12NFIL3 cells. In contrast, GLε transcripts were detected after 4 h in M12Vector cells (Fig. 4C). The expression levels of GLε transcripts in M12NFIL3 cells were higher than in M12Vector cells over the 8 h of stimulation. These results suggest that NFIL3 expression induces early expression of GLε transcription.
Next, we examined whether NFIL3 expression promotes the induction of GLε transcription. M12NFIL3 and M12Vector cells pretreated with CHX were stimulated with LPS/IL-4 for up to 8 h, and GLε transcription was monitored (Fig. 4D). CHX treatment blocked NFIL3 protein synthesis but not Nfil3 mRNA in both cells. Exogenous NFIL3 expression in M12NFIL3 cells was present in the presence of CHX. Interestingly, GLε transcripts were rapidly induced by LPS/IL-4 in the presence of CHX in M12NFIL3 cells but were not detected in M12Vector cells. These results suggest that NFIL3 expression promotes IL-4 induction of GL transcription in the absence of protein synthesis.
NFIL3 Binds to Iε Promoter in Vivo.
To elucidate the mechanism by which NFIL3 regulates GLε transcription, we determined whether NFIL3 binds to the Iε promoter region in vivo by ChIP assay. NFIL3-containing chromatin complexes from fixed M12NFIL3 cells unstimulated or stimulated with LPS/IL-4 were subjected for ChIP-PCR analysis. NFIL3 binding to Iε promoter was detected and increased by LPS/IL-4 stimulation (Fig. 5). However, apparent binding of NFIL3 was not detected in Iγ1, Iγ2a, and β-globin genes as a negative control. These results indicate NFIL3 binds to Iε promoter region in vivo and may account for the specificity of NFIL3’s effect on IgE class switching.
Fig. 5.
NFIL3 binds Iε promoter in vivo. M12NFIL3 cells were unstimulated or stimulated with LPS/IL-4, and soluble chromatin complex was applied for ChIP by anti-NFIL3 and control antibodies. NFIL3 binding to the indicated genes in the coprecipitated DNA was assessed by real-time PCR. The relative enrichment by control IgG in unstimulated cells was set as 1. The averages of relative NFIL3 binding activity from three experiments are indicated (*P = 0.06, **P =0.03, ***P < 0.01).
Discussion
In this study, we have generated and analyzed NFIL3-deficient mice. It has been shown that the Nfil3 gene is expressed in a variety of tissues, including spleen, bone marrow, testis, placenta, skeletal muscle, ovary, lung, and heart (12, 32,33–34). However, NFIL3 KO mice demonstrated normal fertility, growth, lymphocyte development, and myeloid cell development, except for NK cells, as Gascoyne et al. (24) recently reported. These observations indicate that NFIL3 is not a critical transcription factor for the development of hematopoietic cells, except for NK cells. Notably, despite normal B-cell development, NFIL3 deficiency caused impaired class switching to IgE in vivo and in vitro. These observations are consistent with the low level of NFIL3 expression in resting B cells in mice and with the rapid and strong induction of NFIL3 by IL-4 stimulation. The cell activation and differentiation induced by cytokine stimulation may require NFIL3 induction in some situations. Indeed, NFIL3 is induced by several cytokines and hormones, including IL-3, IL-4, IL-6, insulin, and PTH (8, 9, 13,14,15,16,17,18–19). This suggests that NFIL3 is the critical mediator to induce cytokine-dependent cellular activation and differentiation.
We clearly demonstrated that B cells from NFIL3 KO mice are greatly impaired in their ability to class switch to IgE in vitro (Fig. 3). However, we also observed a slight reduction of IL-4 production by CD4+ T cells after OVA immunization in NFIL3 KO mice compared with WT mice in vivo (Fig. S9). In addition, NK cell development is altered in these mice. Thus, the reduced levels of IgE in NFIL3 KO mice may be multifactorial.
How does NFIL3 regulate IgE class switching? We hypothesize that NFIL3 regulates GLε transcription. ChIP experiments demonstrated that NFIL3 could bind to Iε promoter region in response to LPS/IL-4, although we could not find the consensus binding sequence for NFIL3 in Iε promoter region. It has been reported that the interaction of STAT6 with NF-κB promotes Iε transcription (35, 36). Therefore, it is possible that NFIL3 can interact with these transcription factors, and thereby regulate GLε transcription. NFIL3 interacts with TATA-binding protein (TBP)-binding protein Dr1 through its repression domain of NFIL3, and, interestingly, non-DNA binding forms of NFIL3 can activate transcription (37). Therefore, NFIL3 may positively regulate GLε transcription with indirect binding to the sequence in Iε promoter, and NFIL3 may interact with the other transcriptional factors or coactivators. The experiments using CHX treatment and overexpression of NFIL3 in M12.4.1 B cells indicate that NFIL3 induction is required for the initiation of GLε transcription. Reintroduction of NFIL3 into NFIL3 KO B cells can restore impaired IgE class switching. This evidence also supports the hypothesis that NFIL3 controls IgE class switching by regulating GLε transcription. It has been suggested that STAT6 activation by IL-4 is required for the direct regulation of Iε and Aid promoter activity to induce IgE CSR (3). However, our results clearly demonstrate that STAT6 is also required for rapid NFIL3 induction to induce GLε transcription maximally in response to LPS/IL-4, because a low level of induction of GLε transcripts could be detected in spite of obvious activation of STAT6 in the absence of NFIL3. Thus, our study provides an additional mechanism by which IL-4-induced STAT6 activation regulates GLε transcription.
The exposure of LPS (or CD40) and IL-4 induces IgE and IgG1 class switching in murine B cells. The regulatory elements in Iε and Iγ1 promoter regions, which contain STAT6, NF-κB, AP1, and C/EBP sites, are very similar. However, NFIL3 deficiency did not affect IgG1 class switching in vivo and had a small effect in vitro. How are IgE and IgG1 class switching differently regulated? Several reports demonstrate differential regulation of Iε and Iγ1 promoter activities. The arrangement and affinity of the transcription factor binding sites in the IL-4-responsive region are different when the Iε and Iγ1 promoters are compared (38). Interestingly, unique bZip proteins bind to these promoter regions (38). The binding sequence of NFIL3 is very similar to that of ATF, AP1, and C/EBP sites (10). Therefore, it is possible that NFIL3 may interact with these regulatory elements differently in the IgE and IgG1 loci. Moreover, Iε promoter activity is specifically regulated by 3′ enhancer region (39, 40). Indeed, our analysis by ChIP experiments clearly showed that NFIL3 bound to Iε promoter region but not to Iγ1 or Iγ2a promoter region, indicating a specific contribution of NFIL3 to the regulation of Iε promoter activity in response to IL-4 (Fig. 5). Moreover, we observed that IL-4 production by CD4+ T cells after OVA immunization was slightly decreased in NFIL3 KO mice compared with WT mice (Fig. S9). This reduction may have some effect on NFIL3-mediated IgE but not on IgG1 class switching in part in vivo. IL-4 regulation of NFIL3 may account for the mechanism by which the serum IgE levels are kept at low levels, and IL-4 regulates IgE and IgG1 class switching differentially.
NFIL3 is well studied in the regulation of circadian oscillation of gene expression (23, 41, 42). Recently, the release of hematopoietic stem cells has been shown to be regulated by circadian oscillation, and a chemokine CXCL12 has been shown to play a role in the regulation of this fluctuation (43). It is of interest to examine the relation between circadian rhythm and immune response. In the experimental asthma model, IgE-mediated activation of mast cells and other cells has been implicated in airway hyperresponsiveness and cellular infiltration (44). Therefore, it might be valuable to examine if NFIL3 deficiency alters asthmatic phenotypes in the experimental asthma model. Clinically, the relation between diurnal variations in lung physiology and nocturnal asthma has been well documented (45, 46). Thus, NFIL3 has multiple functions not only in B cells but in the other cell types in the regulation of allergic responses. Therefore, NFIL3 expression in the local tissues may be a mechanism to regulate allergic diseases.
Materials and Methods
Mice.
All mice were bred and maintained under pathogen-free conditions. Mice aged 8–10 weeks with a 129× C57BL/6 mixed background were used for all experiments, except for STAT6 KO mice (BALB/c), which were obtained from the Jackson Laboratory. All experimental mouse protocols adhered to Institutional Animal Care and Use Committee (IACUC) guidelines and were approved by the IACUC of the University of Iowa. The generation of NFIL3 KO mice is described in SI Materials and Methods.
Other Materials and Methods.
Detailed methods, including cell culture, Western blotting, flow cytometry, immunization, ELISA, retroviral infection, quantitative RT-PCR, and ChIP experiments, are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the Rothman Laboratory and Colgan Laboratory at the University of Iowa for helpful suggestions on the manuscript. The retroviral plasmid pMiT was a kind gift from Dr. Thomas Mitchell (University of Louisville, Louisville, KY). We thank Dr. Kazuo Kinoshita (Shiga Medical Center Research Institute, Shiga, Japan) for providing the primer information for RT-PCR. This study was supported by National Institutes of Health Grant R01 AI54821 (to P.B.R) and a grant from the Arthritis Foundation (to D.M.L).
Footnotes
The authors declare no conflict of intrest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909235107/DCSupplemental.
References
- 1.Gould HJ, Sutton BJ. IgE in allergy and asthma today. Nat Rev Immunol. 2008;8:205–217. doi: 10.1038/nri2273. [DOI] [PubMed] [Google Scholar]
- 2.Owen CE. Immunoglobulin E: Role in asthma and allergic disease: Lessons from the clinic. Pharmacol Ther. 2007;113:121–133. doi: 10.1016/j.pharmthera.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Geha RS, Jabara HH, Brodeur SR. The regulation of immunoglobulin E class-switch recombination. Nat Rev Immunol. 2003;3:721–732. doi: 10.1038/nri1181. [DOI] [PubMed] [Google Scholar]
- 4.Corry DB, Kheradmand F. Induction and regulation of the IgE response. Nature. 1999;402(6760, Suppl):B18–B23. doi: 10.1038/35037014. [DOI] [PubMed] [Google Scholar]
- 5.Harris MB, et al. Transcriptional repression of Stat6-dependent interleukin-4-induced genes by BCL-6: Specific regulation of iepsilon transcription and immunoglobulin E switching. Mol Cell Biol. 1999;19:7264–7275. doi: 10.1128/mcb.19.10.7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugai M, et al. Essential role of Id2 in negative regulation of IgE class switching. Nat Immunol. 2003;4:25–30. doi: 10.1038/ni874. [DOI] [PubMed] [Google Scholar]
- 7.Rothman P, Lutzker S, Cook W, Coffman R, Alt FW. Mitogen plus interleukin 4 induction of C epsilon transcripts in B lymphoid cells. J Exp Med. 1988;168:2385–2389. doi: 10.1084/jem.168.6.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu CC, Paul WE. Expressed genes in interleukin-4 treated B cells identified by cDNA representational difference analysis. Mol Immunol. 1998;35:487–502. doi: 10.1016/s0161-5890(98)00031-5. [DOI] [PubMed] [Google Scholar]
- 9.Schroder AJ, Pavlidis P, Arimura A, Capece D, Rothman PB. Cutting edge: STAT6 serves as a positive and negative regulator of gene expression in IL-4-stimulated B lymphocytes. J Immunol. 2002;168:996–1000. doi: 10.4049/jimmunol.168.3.996. [DOI] [PubMed] [Google Scholar]
- 10.Cowell IG. E4BP4/NFIL3, a PAR-related bZIP factor with many roles. BioEssays. 2002;24:1023–1029. doi: 10.1002/bies.10176. [DOI] [PubMed] [Google Scholar]
- 11.Cowell IG, Skinner A, Hurst HC. Transcriptional repression by a novel member of the bZIP family of transcription factors. Mol Cell Biol. 1992;12:3070–3077. doi: 10.1128/mcb.12.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang W, et al. Molecular cloning and characterization of NF-IL3A, a transcriptional activator of the human interleukin-3 promoter. Mol Cell Biol. 1995;15:6055–6063. doi: 10.1128/mcb.15.11.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altura RA, et al. The chimeric E2A-HLF transcription factor abrogates p53-induced apoptosis in myeloid leukemia cells. Blood. 1998;92:1397–1405. [PubMed] [Google Scholar]
- 14.Chen Z, et al. Identification of novel IL-4/Stat6-regulated genes in T lymphocytes. J Immunol. 2003;171:3627–3635. doi: 10.4049/jimmunol.171.7.3627. [DOI] [PubMed] [Google Scholar]
- 15.Ikushima S, et al. Pivotal role for the NFIL3/E4BP4 transcription factor in interleukin 3-mediated survival of pro-B lymphocytes. Proc Natl Acad Sci USA. 1997;94:2609–2614. doi: 10.1073/pnas.94.6.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ozkurt IC, Tetradis S. Parathyroid hormone-induced E4BP4/NFIL3 down-regulates transcription in osteoblasts. J Biol Chem. 2003;278:26803–26809. doi: 10.1074/jbc.M212652200. [DOI] [PubMed] [Google Scholar]
- 17.Ramsborg CG, Papoutsakis ET. Global transcriptional analysis delineates the differential inflammatory response interleukin-15 elicits from cultured human T cells. Exp Hematol (Charlottesville, Va) 2007;35:454–464. doi: 10.1016/j.exphem.2006.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sartipy P, Loskutoff DJ. Expression profiling identifies genes that continue to respond to insulin in adipocytes made insulin-resistant by treatment with tumor necrosis factor-alpha. J Biol Chem. 2003;278:52298–52306. doi: 10.1074/jbc.M306922200. [DOI] [PubMed] [Google Scholar]
- 19.Lang R, Patel D, Morris JJ, Rutschman RL, Murray PJ. Shaping gene expression in activated and resting primary macrophages by IL-10. J Immunol. 2002;169:2253–2263. doi: 10.4049/jimmunol.169.5.2253. [DOI] [PubMed] [Google Scholar]
- 20.Lund R, Aittokallio T, Nevalainen O, Lahesmaa R. Identification of novel genes regulated by IL-12, IL-4, or TGF-beta during the early polarization of CD4+ lymphocytes. J Immunol. 2003;171:5328–5336. doi: 10.4049/jimmunol.171.10.5328. [DOI] [PubMed] [Google Scholar]
- 21.Akashi M, Ichise T, Mamine T, Takumi T. Molecular mechanism of cell-autonomous circadian gene expression of Period2, a crucial regulator of the mammalian circadian clock. Mol Biol Cell. 2006;17:555–565. doi: 10.1091/mbc.E05-05-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitsui S, Yamaguchi S, Matsuo T, Ishida Y, Okamura H. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev. 2001;15:995–1006. doi: 10.1101/gad.873501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ueda HR, et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 24.Gascoyne DM, et al. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol. 2009;10:1118–1124. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- 25.Alvarez JV, et al. Identification of a genetic signature of activated signal transducer and activator of transcription 3 in human tumors. Cancer Res. 2005;65:5054–5062. doi: 10.1158/0008-5472.CAN-04-4281. [DOI] [PubMed] [Google Scholar]
- 26.Rothman P. Interleukin 4 targeting of immunoglobulin heavy chain class-switch recombination. Res Immunol. 1993;144:579–583. doi: 10.1016/s0923-2494(05)80006-9. [DOI] [PubMed] [Google Scholar]
- 27.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 28.Hasbold J, Lyons AB, Kehry MR, Hodgkin PD. Cell division number regulates IgG1 and IgE switching of B cells following stimulation by CD40 ligand and IL-4. Eur J Immunol. 1998;28:1040–1051. doi: 10.1002/(SICI)1521-4141(199803)28:03<1040::AID-IMMU1040>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 29.Hodgkin PD, Lee JH, Lyons AB. B cell differentiation and isotype switching is related to division cycle number. J Exp Med. 1996;184:277–281. doi: 10.1084/jem.184.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rush JS, Liu M, Odegard VH, Unniraman S, Schatz DG. Expression of activation-induced cytidine deaminase is regulated by cell division, providing a mechanistic basis for division-linked class switch recombination. Proc Natl Acad Sci USA. 2005;102:13242–13247. doi: 10.1073/pnas.0502779102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitchell TC, et al. Immunological adjuvants promote activated T cell survival via induction of Bcl-3. Nat Immunol. 2001;2:397–402. doi: 10.1038/87692. [DOI] [PubMed] [Google Scholar]
- 32.Hulme DJ, Blair IP, Dawkins JL, Nicholson GA. Exclusion of NFIL3 as the gene causing hereditary sensory neuropathy type I by mutation analysis. Hum Genet. 2000;106:594–596. doi: 10.1007/s004390000306. [DOI] [PubMed] [Google Scholar]
- 33.Lai CK, Ting LP. Transcriptional repression of human hepatitis B virus genes by a bZIP family member, E4BP4. J Virol. 1999;73:3197–3209. doi: 10.1128/jvi.73.4.3197-3209.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishimura Y, Tanaka T. Calcium-dependent activation of nuclear factor regulated by interleukin 3/adenovirus E4 promoter-binding protein gene expression by calcineurin/nuclear factor of activated T cells and calcium/calmodulin-dependent protein kinase signaling. J Biol Chem. 2001;276:19921–19928. doi: 10.1074/jbc.M010332200. [DOI] [PubMed] [Google Scholar]
- 35.Messner B, Stütz AM, Albrecht B, Peiritsch S, Woisetschläger M. Cooperation of binding sites for STAT6 and NF kappa B/rel in the IL-4-induced up-regulation of the human IgE germline promoter. J Immunol. 1997;159:3330–3337. [PubMed] [Google Scholar]
- 36.Shen CH, Stavnezer J. Interaction of stat6 and NF-kappaB: Direct association and synergistic activation of interleukin-4-induced transcription. Mol Cell Biol. 1998;18:3395–3404. doi: 10.1128/mcb.18.6.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cowell IG, Hurst HC. Protein-protein interaction between the transcriptional repressor E4BP4 and the TBP-binding protein Dr1. Nucleic Acids Res. 1996;24:3607–3613. doi: 10.1093/nar/24.18.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mao CS, Stavnezer J. Differential regulation of mouse germline Ig gamma 1 and epsilon promoters by IL-4 and CD40. J Immunol. 2001;167:1522–1534. doi: 10.4049/jimmunol.167.3.1522. [DOI] [PubMed] [Google Scholar]
- 39.Laurencikiene J, Deveikaite V, Severinson E. HS1,2 enhancer regulation of germline epsilon and gamma2b promoters in murine B lymphocytes: Evidence for specific promoter-enhancer interactions. J Immunol. 2001;167:3257–3265. doi: 10.4049/jimmunol.167.6.3257. [DOI] [PubMed] [Google Scholar]
- 40.Laurencikiene J, Tamosiunas V, Severinson E. Regulation of epsilon germline transcription and switch region mutations by IgH locus 3′ enhancers in transgenic mice. Blood. 2007;109:159–167. doi: 10.1182/blood-2006-02-005355. [DOI] [PubMed] [Google Scholar]
- 41.Okano T, Fukada Y. Chicktacking pineal clock. J Biochem (Tokyo) 2003;134:791–797. doi: 10.1093/jb/mvg221. [DOI] [PubMed] [Google Scholar]
- 42.Ueda HR. Systems biology of mammalian circadian clocks. Cold Spring Harbor Symp Quant Biol. 2007;72:365–380. doi: 10.1101/sqb.2007.72.047. [DOI] [PubMed] [Google Scholar]
- 43.Méndez-Ferrer S, Lucas D, Battista M, Frenette PS. Haematopoietic stem cell release is regulated by circadian oscillations. Nature. 2008;452:442–447. doi: 10.1038/nature06685. [DOI] [PubMed] [Google Scholar]
- 44.Hamelmann E, Tadeda K, Oshiba A, Gelfand EW. Role of IgE in the development of allergic airway inflammation and airway hyperresponsiveness—A murine model. Allergy. 1999;54:297–305. doi: 10.1034/j.1398-9995.1999.00085.x. [DOI] [PubMed] [Google Scholar]
- 45.Jarjour NN. Circadian variation in allergen and nonspecific bronchial responsiveness in asthma. Chronobiol Int. 1999;16:631–639. doi: 10.3109/07420529908998732. [DOI] [PubMed] [Google Scholar]
- 46.Martin RJ. Location of airway inflammation in asthma and the relationship to circadian change in lung function. Chronobiol Int. 1999;16:623–630. doi: 10.3109/07420529908998731. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.