Abstract
Timely elimination of damaged mitochondria is essential to protect cells from the potential harm of disordered mitochondrial metabolism and release of proapoptotic proteins. In mammalian red blood cells, the expulsion of the nucleus followed by the removal of other organelles, such as mitochondria, are necessary differentiation steps. Mitochondrial sequestration by autophagosomes, followed by delivery to the lysosomal compartment for degradation (mitophagy), is a major mechanism of mitochondrial turnover. Here we show that mice lacking the essential autophagy gene Atg7 in the hematopoietic system develop severe anemia. Atg7−/− erythrocytes accumulate damaged mitochondria with altered membrane potential leading to cell death. We find that mitochondrial loss is initiated in the bone marrow at the Ter119+/CD71High stage. Proteomic analysis of erythrocyte ghosts suggests that in the absence of autophagy other cellular degradation mechanisms are induced. Importantly, neither the removal of endoplasmic reticulum nor ribosomes is affected by the lack of Atg7. Atg7 deficiency also led to severe lymphopenia as a result of mitochondrial damage followed by apoptosis in mature T lymphocytes. Ex vivo short-lived hematopoietic cells such as monocytes and dendritic cells were not affected by the loss of Atg7. In summary, we show that the selective removal of mitochondria by autophagy, but not other organelles, during erythropoeisis is essential and that this is a necessary developmental step in erythroid cells.
Keywords: Atg7, mitophagy, lymphopenia, cell death, reactive oxygen species
Autophagy is responsible for the continuous turnover of cytoplasmic constituents, from long-lived proteins to organelles, such as mitochondria (1), peroxisomes (2), endoplasmic reticulum (ER) (3), and even select areas of the nucleus (4). It was originally described as an alternative cell death pathway, where complete digestion of the cells would be achieved from within (5). This role is now controversial, and evidence is accumulating to suggest that autophagy is also a cell survival mechanism (6).
The mitochondrion is the site of aerobic energy production in eukaryotic cells, of which reactive oxygen species (ROS) are an inevitable by-product. ROS can lead to mitochondrial DNA mutations, dysfunction, and therefore apoptosis (7). Mitochondrial quality control is exerted through an intraorganelle proteolytic system and fission and fusion steps and, last, severely damaged mitochondria are removed by mitophagy (8). In this role, autophagy could indeed be perceived as a cytoprotective mechanism preventing cell death.
During definitive mammalian erythropoiesis, late-stage erythroblasts enucleate and change from larger motile irregular-shaped cells to small uniform biconcave cells by degrading organelles and proteins. The nucleus is known to be expelled from developing erythrocytes (9). On the other hand, the accumulation of autophagosomes observed in early electron micrographs of erythroblasts suggested that organelles may be removed through autophagy (10, 11). Furthermore, two recent studies have implicated mitophagy in red blood cell (RBC) maturation. The Bcl-2 family member Nix, which is up-regulated during terminal erythroid differentiation, was found to target mitochondria to autophagosome-like structures, and its absence in mice caused a mild, nonlethal anemia (12). Moreover, a mild anemia was also found in the absence of the Atg1-related serine-threonine kinase Ulk1 (13) caused by a delay in the removal of mitochondria and ribosomes in erythroid cells. However, as neither Ulk1 nor Nix are essential autophagy genes, the role of autophagy in this process remains to be confirmed in vivo.
The autophagy pathway involves the engulfment of cytoplasmic material by a double-membraned vesicle, the autophagosome, which then fuses with lysosomes for degradation of its cargo. Two ubiquitin-like conjugation systems are required for the elongation phase of autophagosome formation, and Atg7 encodes the E1-like enzyme required in both of these conjugation systems (14, 15). Because autophagy-gene knockout mice display either embryonic (16) or neonatal lethality (17, 18), we generated a mouse knocked out for Atg7 in the hematopoietic system only. This model allowed us to investigate the role of this essential autophagy gene in erythroid as well as other hematopoietic cells.
Results
A Conditional Autophagy Knockout in the Hematopoietic System.
Atg7Flox/Flox mice (18) were crossed to Vav-iCre mice (19), expressing the codon-improved Cre recombinase (iCre) in hematopoietic stem cells and fetal and adult hematopoietic lineages (19, 20), resulting in a hematological Atg7 knockout, hereafter called Vav-Atg7−/−.
The iCre recombinase efficiently excised the loxP-flanked region at the genomic level at both alleles, resulting in any Atg7Flox allele becoming Atg7Null (Fig. S1A). The Atg7 protein was absent in Vav-Atg7−/− hematopoietic cells (splenocytes and bone marrow-derived dendritic cells) yet still present in the nonhematopoietic kidney cells (Fig. S1B). The absence of autophagy has previously been demonstrated in Atg7Null/Null cells (18, 21). Here we found by electron microscopy (EM) that, whereas wild-type (WT) bone marrow (BM) cells show some autophagic vacuoles, in Atg7−/− BM these were rare, mostly immature, and open (Fig. S1C). In addition, using endogenous staining of LC3, we determined the constitutive levels of autophagy by immunofluorescence. Whereas all WT BM cells had some punctate staining, very few Atg7−/− cells had any puncta at all (Fig. S1D).
The Absence of Atg7 in the Hematopoietic System Is Lethal and Leads to Severe Anemia.
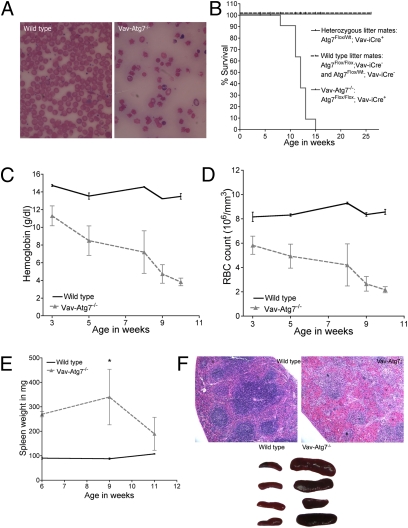
Vav-Atg7−/− mice were born healthy, fertile, and to Mendelian frequencies, with the Atg7 genotypes in iCre-expressing tissues being 47% Flox/Flox; 24% Flox/Null; 29% Null/Null. Blood smears and counts revealed that Vav-Atg7−/− mice developed a progressively worsening anemia (Fig. 1 A, C, and D). All Vav-Atg7−/− mice died between the age of 8 and 14 weeks (Fig. 1B). Approximately 1 week before death, mice became symptomatic: weight loss, withdrawal from the group, and lethargy became apparent. Nine-week-old asymptomatic Vav-Atg7−/− mice displayed significant splenomegaly (Fig. 1E) that disappeared as the anemia progressed. The structural organization of the Vav-Atg7−/− spleen into white (purple) and red (pink) pulps had disappeared (Fig. 1F). The splenomegaly is a typical sign of stress erythropoiesis observed during anemia.
Fig. 1.
The absence of Atg7 in the hematopoietic system leads to lethal anemia. (A) Representative blood smears from anemic 11-week-old mice. (B) Survival curves. (C) Hemoglobin levels. (D) Red blood cell counts. (E) Spleen weight in mg (*P = 0.0159). (F) Upper: H&E-stained paraffin-embedded spleen sections. Lower: representative spleen size, 10-week-old mice.
The Distribution of Erythroid Developmental Stages Is Altered in Vav-Atg7−/− Mice.
The ability of BM progenitor cells to give rise to erythroid colonies was investigated by performing a colony-forming cell assay. This revealed that Vav-Atg7−/− and WT BM cells gave rise to similar numbers of erythroid colonies (BFU-E), suggesting that the anemia is not caused by a defect at the stage of production of erythroid progenitors (Fig. S2).
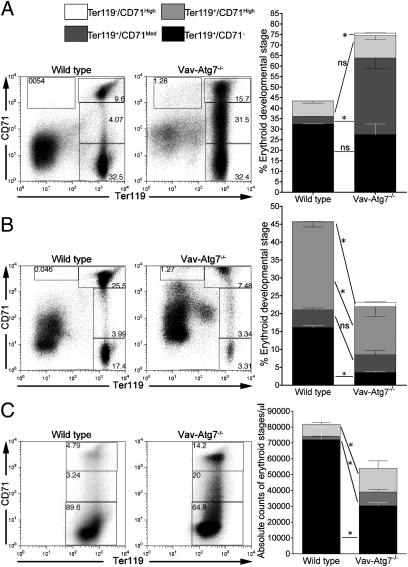
Stages of erythroid development were then analyzed further. Using the surface markers CD71 (transferrin receptor) and Ter119 (glycophorin A), which distinguish four nucleated erythroid developmental stages, Ter119−/CD71High (proerythroblasts), Ter119+/CD71High (basophilic erythroblasts), Ter119+/CD71Med (chromatophilic erythroblasts), and Ter119+/CD71− (orthochromatophilic erythroblasts) (22), we found significantly increased numbers of immature erythroid stages in the spleen, BM, and blood of Vav-Atg7−/− mice (Fig. 2). Blood erythroid cells, though enucleated, still express Ter119, and only reticulocytes retain CD71 expression. The ability of Atg7−/− RBCs to enucleate was normal, as shown by the blood smears and confirmed by flow cytometry using the cell-permeant nucleic acid dye LDS751 (Fig. S3).
Fig. 2.
Altered erythroid developmental stages in Vav-Atg7−/− mice. Distribution of erythroid developmental stages in (A) the spleen of 6-week-old mice (n = 4), (B) the BM of 6-week-old mice (n = 4), and (C) the blood of 9-week-old mice (n = 4). ns: P > 0.05; *P < 0.05. Stages as gated on dot plots, and individual stage frequencies are plotted in bar graphs. One of three representative experiments is shown.
Interestingly, up-regulation of the transferrin receptor was observed in Vav-Atg7−/− BM Ter119+/CD71High cells. Such up-regulation of the transferrin receptor may reflect increased erythropoietic activity to compensate for defective red blood cell development.
Autophagy Genes Are Transcriptionally Regulated in Wild-Type Bone Marrow Erythroid Developmental Stages.
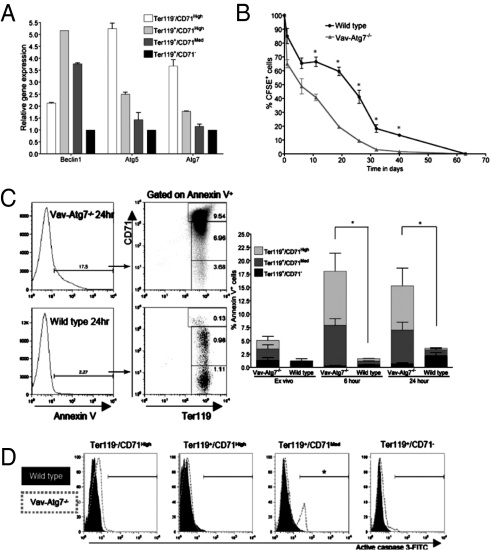
The expression of autophagy genes was investigated in WT nucleated erythroid developmental stages purified from the BM by flow sorting based on their CD71 and Ter119 expression as in Fig. 2B. Gene expression was normalized to GAPDH, and the most mature stage (Ter119+/CD71−), with the lowest overall transcription levels, was used as reference. All genes tested were expressed in BM erythroid cells (Fig. 3A). The relative abundance of autophagy gene transcripts was highest at the proerythroblast and basophilic stages, stages found to accumulate in the spleen and BM of Vav-Atg7−/− mice (Fig. 2 A and B).
Fig. 3.
Atg7−/− erythroid cells have a decreased lifespan and a significantly increased susceptibility to cell death in both blood and spleen. (A) qPCR amplification of Beclin1, Atg5, and Atg7 cDNA in the indicated BM erythroid developmental stages from 8-week-old WT mice. (B) Survival of CFSE-labeled WT and Vav-Atg7−/− red blood cells in WT hosts (*P < 0.05). This experiment was repeated three times with similar results. (C) Annexin V levels in peripheral red blood cells ex vivo, after 6 and 24 h of culture. The percentage of Annexin V+ cells was determined by gating on live cells and is shown in the left panel. Bar heights indicate total percentage of Annexin V+ cells (interaction: *P < 0.05, two-way ANOVA, n = 3). (D) Representative histograms of active caspase 3 levels on spleen erythroid stages from 9-week-old mice, gated as in Fig. 2A (*P = 0.0159, n = 4).
Atg7−/− Erythroid Cells Have a Decreased Lifespan and a Significantly Increased Susceptibility to Cell Death in Blood and Spleen.
Next, as the absence of Atg7 in Vav-Atg7−/− mice is not restricted to the erythroid lineage, we investigated whether the anemic phenotype might be caused by lack of autophagy in other hematopoietic lineages. For that, we injected equal numbers of carboxyfluorescein succinimidyl ester (CFSE)-labeled Atg7−/− or WT RBCs into WT hosts. The CFSE-labeled cells were tracked after 24 h and then weekly in peripheral blood. Within the first 24 h, we observed a substantial drop in Atg7−/− CFSE+ cell number, and subsequently the overall RBC disappearance rate was comparable between knockout and WT (Fig. 3B). This finding led us to investigate whether increased cell death was responsible for the accelerated disappearance of Atg7−/− RBCs. Experiments were systematically performed using Annexin V and intracellular staining of active caspase 3, except on mature blood RBCs, which lyse during permeabilization. Atg7−/− peripheral blood erythroid cells showed significantly higher levels of Annexin V staining ex vivo and after culture in a nutrient-rich and Epo-containing medium (Fig. 3C). The percentage of Annexin V+ among Atg7−/− cells correlated with the percent loss of Atg7−/− CFSE+ RBCs in vivo (Fig. 3B). Interestingly, in Vav-Atg7−/−, CD71+ cells had the highest levels of Annexin V staining, whereas in WT cells it was the mature CD71− erythrocytes. Higher levels of Annexin V were also found on spleen and BM Atg7−/− erythroid cells ex vivo. In the spleen, levels of active caspase 3 were highest in Ter119+/CD71Med cells from Vav-Atg7−/− (Fig. 3D). Taken together, these results suggest that the absence of Atg7 causes an erythroid-intrinsic defect leading to cell death in both nucleated and enucleated stages of development.
ErGFP-Atg7−/− Mice Develop Anemia and Their RBCs Show Increased Levels of Death.
To formally exclude that the observed anemia was due to a defect in other hematopoietic cells, we generated conditional erythroid Atg7 knockout mice (ErGFP-Atg7−/−). This was achieved by breeding Atg7Flox/Flox mice to ErGFP-Cre mice, which express GFP and iCre under the control of their endogenous erythropoietin receptor (EpoR).
We found that ErGFP-Atg7−/− mice develop a mild anemia, characterized by significantly increased absolute numbers of Ter119+/CD71Med cells in peripheral blood at 7 weeks of age (Fig. S4A). The distribution of spleen erythroid developmental stages was also significantly altered in ErGFP-Atg7−/− mice (Fig. S4B). Similar to Vav-Atg7−/−, ErGFP-Cre mice had splenomegaly (Fig. S4C). In addition, a significantly higher number of peripheral blood RBCs from ErGFP-Atg7−/− showed signs of cell death (Fig. S4D). Finally, as expected, no other hematopoietic lineage was affected in ErGFP-Atg7−/− mice.
Overall, analysis of this erythroid-specific conditional Atg7 knockout model shows that the absence of Atg7 in erythroid cells is sufficient to reproduce the anemic phenotype of Vav-Atg7−/− mice; however, it is not as severe.
To understand this difference in severity, we tested whether this could be due to lower excision levels in this model. In the original study it was demonstrated that the ErGFP-Cre-mediated recombination efficiency among Ter119+ cells in adult spleen and bone marrow was only 61.7% and 76.3%, respectively (23). To compare this with Vav-iCre-driven excision, we performed a qPCR analysis of Atg7 mRNA levels in sorted Ter119+ cells from fetal liver and adult bone marrow from both mouse models. The relative Atg7 transcript abundance was dramatically decreased in Ter119+ cells from both models when compared to wild-type fetal Ter119+ cells (100%). Whereas Atg7 transcript levels from ErGFP-Atg7−/− fetal and adult Ter119+ cells were decreased to 15% and 25%, respectively, thereby confirming findings in ref. 23, the decrease was significantly higher in Vav-Atg7−/− cells, with remaining levels of only 5% in fetal and 1% in adult Ter119+ cells (Fig. S4E).
Atg7−/− Erythroid Cells Accumulate Selectively Damaged Mitochondria.
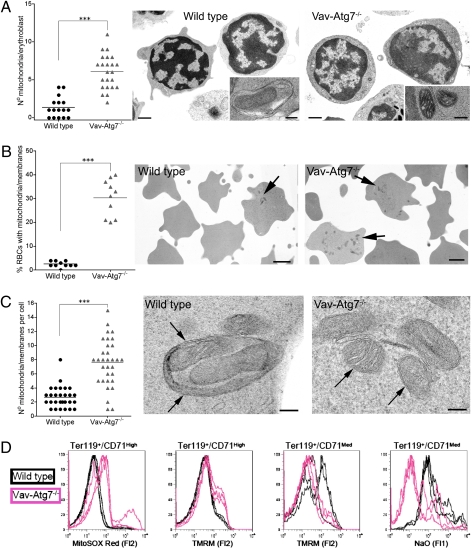
As autophagy has been implicated in the removal of mitochondria in erythroid development (12, 13), we investigated the ability of Atg7−/− cells to lose their mitochondria. We found a significantly higher number of mitochondria but no other organelles in Atg7−/− BM erythroblasts by EM (Fig. 4A). Autophagosomal structures were only rarely observed in WT cells, but on the rare occasion these carried mitochondria. On the other hand, in Atg7−/− cells, mitochondria were associated with isolation membrane-like structures (Fig. 4A, insets). Enucleated RBCs are usually devoid of organelles; however, in Vav-Atg7−/− mice, a significant proportion of peripheral blood cells contained mitochondrial remnants, only very rarely observed in WT cells (Fig. 4B). There were also more mitochondria per cell in Vav-Atg7−/− (Fig. 4C). Strikingly, when mitochondrial remnants were found in WT peripheral blood cells, these were often within autophagosomes, whereas those seen in Vav-Atg7−/− blood cells were in the proximity of isolation vesicle-like structures (Fig. 4C).
Fig. 4.
Atg7−/− erythroid cells accumulate damaged mitochondria. (A) Left: total mitochondrial count/BM erythroblast counted by EM. Right: representative EM. (Scale bars, 1 mm in lower power and 100 nm in inset.) (B) Left: percent of peripheral RBCs containing remnants of mitochondria or other membranes. Right: representative EM. (Scale bars, 1 μm.) Arrows: mitochondrial remnants. (C) Left: number of mitochondria and membranes per cell. Right: representative EM. (Scale bars, 100 nm.) In WT, arrows point at autophagosome; in Atg7−/−, arrows point at mitochondria. (D) Overlays of three WT and three Vav-Atg7−/− BM erythroid developmental stages stained with MitoSOX Red, TMRM, and NaO (gated on the indicated erythroid stages as shown in Fig. 2B) (***P < 0.0001).
We excluded that these results were due to increased numbers of reticulocytes in the blood of Vav-Atg7−/− mice (Fig. S5A) by sorting blood cells based on staining for the RNA-specific dye thiazole orange (TO) to distinguish reticulocytes from mature RBCs (Fig. S6). We thus confirmed that Atg7−/− reticulocytes are defective in clearing their mitochondria, leading to a significant number of mature RBCs with remaining mitochondria.
Depolarization of the mitochondrial membrane measured by tetramethylrhodamine (TMRM) is a very early marker of apoptosis and possibly precedes their autophagic engulfment in erythroid cells (12). If damaged mitochondria are not disposed of in time, it results in oxidation of the cardiolipins in the inner mitochondrial membrane, detected by N-nonyl-acridine orange (NaO) (24, 25). Mitochondrial superoxides (detected by MitoSOX) generated during normal oxygen phosphorylation constitute the major ROS in mitochondria and are responsible for cardiolipin oxidation (25). This is followed by cell death due to leakage of apoptotic factors from the mitochondrial intermembranal space.
Mitochondrial damage was therefore next investigated by flow cytometry. First, we observed that the levels of intact cardiolipins as well as mitochondrial mass (MitoTracker Green) decreased over erythroid development in WT BM and that mitochondrial membrane potential went from highly polarized, then depolarized, to again repolarized, and finally completely hypopolarized (Fig. S7), possibly thereby triggering mitochondrial degradation during these stages. In Vav-Atg7−/− BM cells, on the other hand, an accumulation of mitochondrial superoxide was found in Ter119+/CD71High (Fig. 4D). Using TMRM, a proportion of BM Atg7−/− erythroblasts were found to contain mitochondria with altered membrane potential, hyperpolarizing first then depolarizing (Fig. 4D). Atg7−/− BM Ter119+/CD71Med cells were also found to have lower levels of intact cardiolipins than their WT counterparts.
To investigate whether autophagy may be involved in the removal of other organelles, ribosomes were stained with TO in RBCs. Interestingly, not only were Vav-Atg7−/− RBCs devoid of RNA at the Ter119+/CD71− stage like WT cells but Ter119+/CD71Med cells had lower levels of RNA than their WT counterparts (Fig. S5B), suggesting that Atg7-deficient RBCs were able to degrade their ribosomes. Finally, we found that the degradation of ER during erythroid development was not affected by the loss of Atg7 (Fig. S5C).
Mitochondrial Proteins Were Preferentially Detected in Atg7−/− RBC Membrane Preparations by Mass Spectrometry.
A quantitative mass spectrometry approach was used to compare the relative abundance of proteins contained by Atg7−/− and WT RBCs. Membrane preparations were used to eliminate the most abundant cytoplasmic proteins in RBCs, hemoglobins, which would otherwise make the identification of other less abundant proteins impossible. Among the proteins detected preferentially in Atg7−/− samples were the transferrin receptor (CD71) and its precursor (serotransferrin precursor) (Table 1; Table S1). This finding is consistent with the accumulation of CD71-expressing cells as measured by flow cytometry (see Fig. 2C). An increase in a number of proteins of the endomembrane system was also identified in the absence of Atg7, such as clathrin heavy chain (26), two lysosomal proteins, and Atg12, an autophagy-related protein which when conjugated to Atg5 by Atg7 forms an integral part of autophagosomal membranes (27). Most importantly, this analysis revealed that mitochondrial proteins were consistently and preferentially detected in Atg7−/− samples only, further confirming that in the absence of autophagy, peripheral blood cells have impaired removal of mitochondria.
Table 1.
Unique proteins identified in Atg7-/- RBC membrane preparations by mass spec.
| Biological Function | Protein Name | Swiss-Prot Accession No. |
| RBC developmental receptors | Transferrin receptor protein 1 | Q62351 |
| Serotransferrin precursor | Q921I1 | |
| Endo-lysosomal and autophagosomal proteins | Clathrin heavy chain 1 | Q68FD5 |
| Lysosomal α glucosidase precursor | P70699 | |
| Lysosomal associated transmembrane protein 4A | Q60961 | |
| Autophagy related protein 12 | Q9CQY1 | |
| Mitochondrial proteins | 60 kDa heat-shock protein mitochondrial precursor | P63038 |
| ATP synthase subunit α mitochondrial precursor | Q03265 | |
| Acylglycerol kinase mitochondrial precursor | Q9ESW4 | |
| Malate dehydrogenase mitochondrial precursor | P08249 | |
| Coproporphyrinogen III oxidase mitochondrial precursor | P36552 |
Lymphocytes but Not Myeloid Cells Are Affected by the Loss of Atg7.
Absolute numbers of T and B lymphocytes in peripheral blood were significantly reduced in Vav-Atg7−/− mice (Fig. S8 A and B). This lymphopenia was observed in mice as young as 3 weeks old and, unlike the anemia, did not progress. Significantly more ex vivo active caspase 3 levels were detected in both CD4+ and CD8+ lymphocytes in the absence of Atg7. More apoptosis was also observed after in vitro culture (Fig. S8C). Staining with MitoSOX and MTG revealed that CD4+ and CD8+ cells from Vav-Atg7−/− mice had higher levels of mitochondrial superoxide, increased mitochondrial mass (Fig. S9A), and a higher number of mitochondria as counted by EM (Fig. S9B).
The myeloid lineage, in contrast, was not affected by the absence of Atg7. Ex vivo absolute numbers of myeloid (CD11b+) and dendritic cells (CD11c+ DCs) were found unaltered in the blood and spleen of Vav-Atg7−/− mice.
Discussion
Autophagy was initially described to function as an alternative cell death mechanism. In contrast, most in vivo studies of autophagy, including the present one, support its role as a survival mechanism (28): the ablation of Atg5 (29) or Atg7 (21) caused cell death in neurons, and Atg5−/− BM chimeras showed increased susceptibility to death in T cells (30). Although direct crosstalk between autophagy and apoptosis is likely to occur at the molecular level (31), here we show evidence of an indirect link between autophagy and death. Our results indicate that the role of autophagy is in mitochondrial clearance preventing cell death in hematopoietic cells.
The absence of Atg7-mediated mitophagy in erythroid cells leads to an accumulation of mitochondria from the erythroblast stage in the bone marrow to the reticulocyte stage in peripheral blood, resulting in cell death. As the presence of mitochondria in reticulocytes is not, per se, toxic, considering that wild-type reticulocytes contain mitochondria, the presence of mitochondria as seen in Atg7−/− leading to their death may seem puzzling. The presence of mitochondria being toxic in Atg7−/− cells could mean that damage to mitochondria is induced to signal for their clearance at the basophilic erythroblast stage. Whereas in wild-type cells these are soon cleared, in Atg7−/− cells damaged mitochondria remain and the pathways downstream of mitochondrial damage are allowed to continue, leading to apoptosis. A recent study using Nix−/− mice supports this, as it shows that mitochondria are targeted for autophagosomal degradation by a Nix-dependent loss of mitochondrial membrane potential (12).
The accumulation of mitochondria in Atg7−/− erythroid cells was evident already in the BM and autophagy genes were transcriptionally regulated in WT BM, altogether suggesting an active role for autophagy in BM erythroid cells. Increased death in both nucleated (spleen) and enucleated (blood) erythroid cells was also observed. We therefore propose that mitophagy starts in nucleated erythroblasts and is ongoing until the mature erythrocyte stage is reached.
Some redundancy for mitochondrial removal may exist as, similar to Ulk1-deficient RBCs (13), a limited percentage of Atg7−/− RBCs were found to contain mitochondria by EM. The fact that membranous structures remain in autophagy-deficient RBCs might be due to the defective removal of other organelles; however, this seems unlikely, as we show that both ribosomes and ER are efficiently cleared from Atg7−/− erythrocytes. An alternative explanation for these remaining membranous structures is that these might represent an alternative organelle digestion vesicular pathway which accumulates only in the absence of autophagy. On the other hand, Atg12 was found preferentially in the knockout samples by proteomics analysis of RBC membrane preparations. As the Atg12/Atg5 conjugate was recently reported to have an E3-like activity enhancing Atg8 lipidation in yeast (32), it can therefore be speculated that its up-regulation in the absence of Atg7 might be a way to attempt autophagosome formation and may result in an increase in the formation of isolation membranes accumulating in Atg7−/− erythrocytes. Interestingly, a recent paper shows that there is indeed an Atg7-independent pathway of mitochondrial removal using degradative vacuoles that form in the absence of autophagy (33).
In particular, regarding the removal of ribosomes from developing erythroid cells, these were found increased in Ulk1−/− reticulocytes by EM, and inhibition of autophagy in WT cells resulted in accumulation of RNA bound to ribosomes. This suggested that autophagy is involved in the removal of ribosomes from reticulocytes (13). This was not confirmed in Atg7−/− RBCs. On the contrary, Ter119+/CD71Med cells have a lower RNA content than WT (Fig. S5B), despite Vav-Atg7−/− mice having an increased number of reticulocytes (Fig. S5B). Our results also clearly indicate that ER is degraded normally in the absence of Atg7.
The anemia was reproduced in another in vivo model lacking Atg7 in the erythroid compartment only (ErGFP-Atg7−/− mice); however, the phenotype in these was much milder and the anemia was not progressing in severity as seen in Vav-Atg7−/−.
The significantly higher excision rate of Atg7 in both fetal and adult erythroid cells in VavCre-Atg7−/− as compared to ErGFP-Atg7−/− mice could explain this difference, with more Atg7+ erythroid cells surviving in the latter, giving rise to a mature pool of healthy RBCs. Interestingly, we found that the excision rate during erythropoiesis is comparatively efficient before and after birth in both models, excluding the possibility that the progressive anemia in Vav-Atg7−/− mice is due to better excision in adult cells than in fetal cells. The most likely explanation for the progression of the disease in Vav-Atg7−/− is therefore the fact that vav is active much earlier (20) than the EpoR in hematopoiesis, which may lead to a cumulative problem in the hematopoietic stem cell compartment, therefore affecting the overall levels of erythropoiesis. However, the colony-forming cell assay showed that erythroid progenitors are produced to equal numbers in Vav-Atg7−/− and wild-type mice. As this assay is not informative about their functional capacity, more stringent functional assays of erythroid and other progenitors from Vav-Atg7−/− are needed to exclude that a more severe and progressive anemia in Vav-Atg7−/− is due to a role for autophagy in hematopoietic progenitor cells.
Not all hematopoietic lineages were equally affected by the loss of Atg7, as absolute numbers of myeloid cells were normal in Vav-Atg7−/− mice. We therefore hypothesize that the removal of damaged mitochondria may play a more important role in long-lived cells. The lifespan of some lymphocytes, particularly T cells, can be years, whereas myeloid cells live only hours to weeks.
Overall, these results may shed light on human anemia of unknown etiology. Two possible diseases would resemble the symptoms found in this mouse model most. First, congenital dyserythropoietic anemia type II (CDAII) is a rare anemia, in which double membranes are found in a large proportion of red cell precursors (34). However, as the molecular basis of CDAII has just been elucidated (35, 36), an important role for autophagy in the pathogenesis of this disease is now unlikely. Second, myelodysplastic syndrome (MDS) comprises a group of anemic disorders characterized by abnormal cell morphology in the bone marrow accompanied by cytopenias in peripheral blood. The hallmark of MDS is ineffective hematopoiesis with anemia, neutropenia, thrombocytopenia, splenomegaly, and leukopenia. Apoptosis is observed in erythroid progenitors as in other bone marrow-derived cells (37). It is associated with mitochondrial release of cytochrome c with subsequent activation of caspase 3 and 9 (38). Whether certain subsets of MDS have defects in autophagy is a question that merits investigation.
In conclusion, we have in the present study demonstrated that the core autophagy gene Atg7 is a key molecular player in mitochondrial autophagy in mammalian hematopoietic cells.
Materials and Methods
Animals.
Mice were bred and housed in Biomedical Services, Oxford in individually ventilated cages. Atg7Flox/Flox were crossed to Vav-iCre+/− mice (from Dimitris Kioussis, London) to obtain Vav-iCre+/−; Atg7Flox/Flox. Genotyping was performed on ear genomic DNA as described before in refs. 18 and 19. Male and female mice were used equally in all experiments and littermates were always used as controls.
Flow Cytometry.
Flow cytometry experiments were performed with a DAKO CyAn instrument unless otherwise stated, and FACS data were analyzed with FlowJo 8.7.3 for Mac (Treestar). SI Materials and Methods provide more staining details.
See further details in SI Materials and Methods.
Real-Time PCR.
RNA from sorted cells was extracted using MicroRNeasy columns (Qiagen), reverse-transcribed (High Capacity RNA-to-cDNA kit), and amplified using gene-specific TaqMan gene expression assays and gene expression master mix (all from Applied Biosystems, Foster City, CA) and run on an ABI 7500 instrument. Samples were run in duplicate.
Red Blood Cell Lifespan.
RBCs, counted on a hemocytometer, were stained with 10 μM CFSE in PBS at 37°C for 15 min and washed twice. RBCs (2 × 109) in PBS were injected into the tail vein of C57BL/6 females [representing about 12% of total blood, and shown as 100% (Fig. 3B)].
Electron Microscopy.
Tissue for routine EM was processed as described previously (39).
Bone Marrow-Derived Dendritic Cells.
Hind limb bones were obtained from 9-week-old mice. BM cells were plated in RPMI1640 with 10% FBS (Sigma, St Louis, Missouri), glutamine, penicillin, and streptomycin. Recombinant murine GM-CSF and IL-4 (PeproTech EC, London) were added on days 0 and 3 at 20 ng/mL each. On day 5, 10 ng/mL of LPS (Sigma) was added overnight for maturation.
Mass Spectrometry.
For RBC membrane preparation, see SI Materials and Methods. Sample analysis by tandem mass spectrometry and assessment of the relative quantitation of identified proteins were performed as described previously in ref. 40.
Statistics.
Statistical analyses were performed using Prism 4 for Mac (GraphPad Software). Error bars represent SEM and P values calculated with a two-tailed Mann–Whitney test unless stated otherwise.
Supplementary Material
Acknowledgments
We thank Dimitris Kioussis (Imperial College, London) for providing the Vav-iCre mice; Bill Wood and Jacqueline Sharpe for the ErGFP-Cre mice; Catherine Porcher (Oxford) for her invaluable advice on erythropoiesis; Joanna Poulton (Oxford) for her input on mitochondrial biology; and Eric Freundt, who inspired us to initiate this project. Funding was provided by the National Institute for Health Research Biomedical Research Centre and the Association for International Cancer Research. M.M. holds a studentship from the Medical Research Council and obtained funds from the Andrew McMichael Fund and the Biotechnology and Biological Sciences Research Council.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913170107/DCSupplemental.
References
- 1.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monastyrska I, Klionsky DJ. Autophagy in organelle homeostasis: Peroxisome turnover. Mol Aspects Med. 2006;27:483–494. doi: 10.1016/j.mam.2006.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krick R, et al. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol Biol Cell. 2008;19:4492–4505. doi: 10.1091/mbc.E08-04-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 6.Scarlatti F, Granata R, Meijer AJ, Codogno P. Does autophagy have a license to kill mammalian cells? Cell Death Differ. 2009;16:12–20. doi: 10.1038/cdd.2008.101. [DOI] [PubMed] [Google Scholar]
- 7.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Twig G, Hyde B, Shirihai OS. Mitochondrial fusion, fission and autophagy as a quality control axis: The bioenergetic view. Biochim Biophys Acta. 2008;1777:1092–1097. doi: 10.1016/j.bbabio.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bessis M. Living Blood Cells and Their Ultrastructure. Berlin: Springer; 1973. pp. 110–140. [Google Scholar]
- 10.Kent G, Minick OT, Volini FI, Orfei E. Autophagic vacuoles in human red cells. Am J Pathol. 1966;48:831–857. [PMC free article] [PubMed] [Google Scholar]
- 11.Heynen MJ, Tricot G, Verwilghen RL. Autophagy of mitochondria in rat bone marrow erythroid cells. Relation to nuclear extrusion. Cell Tissue Res. 1985;239:235–239. doi: 10.1007/BF00214924. [DOI] [PubMed] [Google Scholar]
- 12.Sandoval H, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kundu M, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanida I, et al. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol Biol Cell. 1999;10:1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanida I, Tanida-Miyake E, Ueno T, Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem. 2001;276:1701–1706. doi: 10.1074/jbc.C000752200. [DOI] [PubMed] [Google Scholar]
- 16.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 18.Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Boer J, et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 20.Ogilvy S, et al. Promoter elements of vav drive transgene expression in vivo throughout the hematopoietic compartment. Blood. 1999;94:1855–1863. [PubMed] [Google Scholar]
- 21.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 22.Socolovsky M, et al. Ineffective erythropoiesis in Stat5a(−/−)5b(−/−) mice due to decreased survival of early erythroblasts. Blood. 2001;98:3261–3273. doi: 10.1182/blood.v98.12.3261. [DOI] [PubMed] [Google Scholar]
- 23.Heinrich AC, Pelanda R, Klingmüller U. A mouse model for visualization and conditional mutations in the erythroid lineage. Blood. 2004;104:659–666. doi: 10.1182/blood-2003-05-1442. [DOI] [PubMed] [Google Scholar]
- 24.Garcia Fernandez MI, Ceccarelli D, Muscatello U. Use of the fluorescent dye 10-N-nonyl acridine orange in quantitative and location assays of cardiolipin: A study on different experimental models. Anal Biochem. 2004;328:174–180. doi: 10.1016/j.ab.2004.01.020. [DOI] [PubMed] [Google Scholar]
- 25.Ferlini C, Scambia G. Assay for apoptosis using the mitochondrial probes, Rhodamine123 and 10-N-nonyl acridine orange. Nat Protoc. 2007;2:3111–3114. doi: 10.1038/nprot.2007.397. [DOI] [PubMed] [Google Scholar]
- 26.Poupon V, et al. Clathrin light chains function in mannose phosphate receptor trafficking via regulation of actin assembly. Proc Natl Acad Sci USA. 2008;105:168–173. doi: 10.1073/pnas.0707269105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizushima N, et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 28.Cecconi F, Levine B. The role of autophagy in mammalian development: Cell makeover rather than cell death. Dev Cell. 2008;15:344–357. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 30.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He Y-W. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yousefi S, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 32.Hanada T, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, et al. Mitochondrial clearance is regulated by Atg7-dependent and -independent mechanisms during reticulocyte maturation. Blood. 2009;114:157–164. doi: 10.1182/blood-2008-04-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong KY, Hug G, Lampkin BC. Congenital dyserythropoietic anemia type II: Ultrastructural and radioautographic studies of blood and bone marrow. Blood. 1972;39:23–30. [PubMed] [Google Scholar]
- 35.Bianchi P, et al. Congenital dyserythropoietic anemia type II (CDAII) is caused by mutations in the SEC23B gene. Hum Mutat. 2009;30:1292–1298. doi: 10.1002/humu.21077. [DOI] [PubMed] [Google Scholar]
- 36.Schwarz K, et al. Mutations affecting the secretory COPII coat component SEC23B cause congenital dyserythropoietic anemia type II. Nat Genet. 2009;41:936–940. doi: 10.1038/ng.405. [DOI] [PubMed] [Google Scholar]
- 37.Raza A, et al. Apoptosis in bone marrow biopsy samples involving stromal and hematopoietic cells in 50 patients with myelodysplastic syndromes. Blood. 1995;86:268–276. [PubMed] [Google Scholar]
- 38.Tehranchi R, et al. Granulocyte colony-stimulating factor inhibits spontaneous cytochrome c release and mitochondria-dependent apoptosis of myelodysplastic syndrome hematopoietic progenitors. Blood. 2003;101:1080–1086. doi: 10.1182/blood-2002-06-1774. [DOI] [PubMed] [Google Scholar]
- 39.Ling YM, et al. Vacuolar and plasma membrane stripping and autophagic elimination of Toxoplasma gondii in primed effector macrophages. J Exp Med. 2006;203:2063–2071. doi: 10.1084/jem.20061318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu D, et al. Novel MMP-9 substrates in cancer cells revealed by a label-free quantitative proteomics approach. Mol Cell Proteomics. 2008;7:2215–2228. doi: 10.1074/mcp.M800095-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.