Abstract
Autophagy is a catabolic pathway that is important for turnover of long-lived proteins and organelles, and has been implicated in cell survival, tumor progression, protection from infection, neurodegeneration, and cell death. Autophagy and caspases are required for type II autophagic cell death of Drosophila larval salivary glands during development, but the mechanisms that regulate these degradation pathways are not understood. We conducted a forward genetic screen for genes that are required for salivary gland cell death, and here we describe the identification of Drosophila dynein light chain 1 (ddlc1) as a gene that is required for type II cell death. Autophagy is attenuated in ddlc1 mutants, but caspases are active in these cells. ddlc1 mutant salivary glands develop large fibrillar protein inclusions that stain positive for amyloid-specific dyes and ubiquitin. Ectopic expression of Atg1 is sufficient to induce autophagy, clear protein inclusions, and rescue degradation of ddlc1 mutant salivary glands. Furthermore, ddlc1 mutant larvae have decreased motility, and mutations in ddlc1 enhance the impairment of motility that is observed in a Drosophila model of neurodegenerative disease. Significantly, this decrease in larval motility is associated with decreased clearance of protein with polyglutamine expansion, the accumulation of p62 in neurons and muscles, and fewer synaptic boutons. These results indicate that DDLC1 is required for protein clearance by autophagy that is associated with autophagic cell death and neurodegeneration.
Keywords: macroautophagy, protein degradation, neurodegeneration
Macroautophagy (autophagy) is a conserved catabolic pathway (1). Autophagy involves the formation of autophagosomes around cytoplasmic components, and fusion with lysosomes enables degradation of autophagosome cargo. Studies of yeast identified Atg genes that are required for autophagy during starvation (2–4). Autophagy is also activated during starvation in animals (5), but the complexity of higher animals, and the association of autophagy with diseases and cell death, raises questions about how autophagy is regulated and cargo is recruited and delivered to lysosomes in multicellular organisms.
The role of autophagy in programmed cell death has been a subject of debate (6). Autophagy has a well established role in cell survival, and decreased Atg gene function can promote cell death (7). There is also evidence that autophagy plays a protective role against neurodegeneration in the setting of disease (8, 9), as well as under basal conditions (10, 11). Autophagosomes have been associated with dying cells during development (12, 13), and recent studies indicate that autophagy can promote the degradation and clearance of cells during cell death (14–17). This apparent paradoxical role of autophagy in both cell survival and death raises questions about how autophagy is regulated and influences different cell fates.
Drosophila larval salivary glands possess type II cell death morphology during development, and contain numerous autophagosomes (14, 18). A rise in the steroid 20-hydroxyecdysone 12 h after puparium formation triggers eversion of the developing adult head and salivary gland cell death, and salivary glands are completely destroyed within 4 h (18). Salivary glands fail to completely degrade in Atg mutants, and induction of autophagy by expression of Atg1 is sufficient to induce caspase-independent cell death (14). Caspases are also required for salivary gland degradation, and function in the fragmentation of DNA, cleavage of nuclear Lamins, and likely many other caspase substrates in these cells (19). Inhibition of both caspases and autophagy has a stronger persistence phenotype than inhibiting either of these pathways alone, indicating that caspases and autophagy work in an additive manner to degrade salivary glands (14).
Dyneins have been implicated in both autophagy and cell death (20, 21). Dynein light chain encodes an 8-kD protein that binds to a variety of cargo and is highly conserved from Chlamydomonas to humans (22). In Drosophila, Ddlc1 is cytoplasmic and ubiquitously expressed, and complete loss of function of ddlc1 causes embryonic lethality and ectopic apoptosis (23). Partial loss of function mutations in Drosophila ddlc1 cause female sterility, alter sensory neuron development, and cause defects in axon guidance (24).
Here we characterize a ddlc1 mutant that was isolated in a genetic screen for genes that are required for salivary gland cell death. We show that salivary glands of ddlc1 animals are defective in autophagy and degradation. These mutant salivary gland cells develop large protein inclusions that can be degraded by ectopic expression of Atg1, a regulator of autophagy. Furthermore, we show that ddlc1 enhances motility defects in a model of the neuromuscular disease spinobulbar muscular atrophy, and suggest that this is because of the accumulation of protein aggregates in neurons and muscles.
Results
Dynein Light Chain 1 Is Required for Salivary Gland Degradation.
Genes encoding transcription factors, caspases, Atg genes, and regulators of growth have been identified that are required for proper destruction of larval salivary glands (25). In addition, hundreds of genes are induced within 4 h of salivary gland degradation (26), suggesting that additional genes may function in the death of these cells. To identify genes that function in salivary gland cell death, we screened a collection of 1,475 stocks with single lethal P-element-LacZ enhancer trap reporters for expression immediately before cell death, and failure of salivary gland degradation 24 h after puparium formation. From this screen, we identified the P-element line l(1)G0371 inserted into the dynein light chain 1 gene ddlc1, a gene that was detected in our previous DNA microarray studies of salivary glands (26). This P-element insertion was mapped 124 bp downstream of the first exon. This ddlc1 allele is pupal lethal, and head eversion occurs at 15 h after puparium formation.
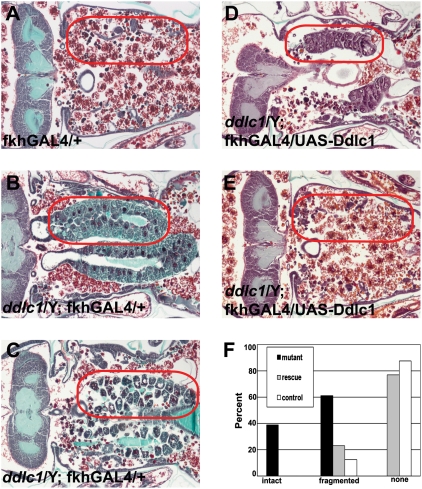
Histological examination of ddlc1 mutant animals 12 h after head eversion (AHE; 8 h after normal salivary gland degradation) revealed a defect in salivary gland cell death. All ddlc1 pupae failed to complete salivary gland degradation, with animals having either intact or partially fragmented glands (Fig. 1 B, C and F). By contrast, most control animals lack salivary glands (Fig. 1 A and F). Significantly, precise excision of the l(1)G0371 P-element reverted the defect in salivary gland degradation (Fig. S1). We confirmed the salivary gland cell death defect of ddlc1 by using the ddlc1e73 allele (24), and 100% of ddlc1e73 animals failed to destroy their salivary glands by 12 h AHE (Fig. S1). To confirm that the failure in salivary gland degradation phenotype was caused by loss of ddlc1 function, we performed a transgenic rescue experiment. We generated transgenic flies and expressed Ddlc1 in the salivary glands of ddlc1 mutant flies. Histological examination indicated that expression of Ddlc1 in salivary glands largely rescued the ddlc1 mutant defect in degradation of this tissue (Fig. 1 D, E, and F). In addition, expression of Ddlc1 in many tissues throughout development using da-GAL4 rescued the viability of 63% of ddlc mutant adult males.
Fig. 1.
ddlc1 is required for salivary gland degradation. (A) Control animals lack salivary glands 12 h AHE (red circles; n = 16). (B and C) ddlc1 mutant animals fail to degrade their salivary glands 12 h AHE, with some being intact (B) and others being fragmented (C) (n = 28). (D and E) Expression of Ddlc1 in salivary glands of ddlc1 mutants rescues the degradation defect phenotype, with a small number having fragmented salivary glands (D) and most having none (E) (n = 22). (F) Percentages of animals with intact, fragmented, and no salivary glands 12 h AHE.
ddlc1 Mutants Have Altered Caspase Levels.
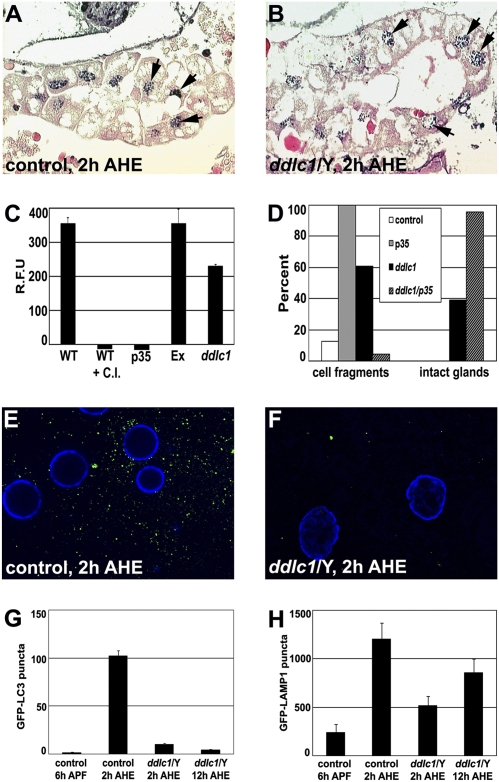
Ectopic caspase activity was previously shown to be present in Drosophila ddlc1 mutants (23), and Ddlc1 inhibits the proapoptotic activity of Bim in mammalian cells (21). As caspases are required to complete salivary gland cell degradation (14, 18, 19), we hypothesized that ddlc1 may regulate caspases in salivary glands. Caspase-dependent DNA fragmentation was detected in both the control and the ddlc1 mutant salivary glands by the TUNEL assay 2 h AHE (Fig. 2 A and B). Caspase activity levels were quantified using the florigenic caspase substrate DEVD-AMC. Both wild type (WT) and control animals lacking the P-element following precise excision had comparable levels of caspase activity, and this activity could be inhibited by either a chemical inhibitor (Ac-DEVD-CHO) or genetically by expression of the caspase inhibitor p35 (Fig. 2C). Although ddlc1 mutant animals have reduced caspase activity, the presence of caspase activity and fragmented DNA suggests that residual cysteine protease activity may be the reason that salivary gland degradation is not completely inhibited in ddlc1 mutants.
Fig. 2.
ddlc1 mutants have altered caspase activity and autophagy. (A and B) TUNEL assay to detect DNA fragmentation (arrows) in control (A) and ddlc1 mutant (B) salivary glands 2 h AHE (n = 5 animals/genotype). (C) Average caspase activity (±SEM) of pupal lysates collected 4 h after puparium formation and measured by cleavage of fluorogenic caspase-3 substrate Z-DEVD-AMC (n = 3). WT, wild-type Canton S; WT + C.I., Canton S + caspase inhibitor DEVD-CHO; p35, daGAL4 × uas-p35: Ex, precise excision of the P element; ddlc1, ddlc1/Y. (D) Percent animals with salivary gland cell fragments and intact glands in paraffin sections of pupae 12 h AHE. Genotypes: Control (fkhGAL4/+) (n = 16), p35 (fkhGAL4/uas-p35) (n = 13), ddlc1 mutant (ddlc1/Y) (n = 28), and p35 expression in ddlc1 mutant (ddlc1/Y; fkhGAL4/uas-p35) (n = 24). (E and F) Representative fluorescence microscopy images of salivary glands expressing GFP-LC3 (green) and DNA (blue). (E) Control (P-excision; +/+; fkhGAL4/uas-GFP-LC3) salivary glands have large numbers of puncta 2 h AHE (n = 20 glands). (F) ddlc1 mutant (ddlc1/Y; +/+; fkhGAL4/uas-GFP-LC3) salivary glands have few puncta 2 h AHE (n = 17 glands). (G) Average number of GFP-LC3 puncta (±SEM) in the genotypes shown in E and F and in ddlc1 mutant salivary glands 12 h AHE (n = 23 glands). (H) Average number of GFP-LAMP1 puncta (±SEM) in salivary glands of control (P-excision; +/+; +/tub-GFP-LAMP1) 6 h after puparium formation, 2 h AHE, and in ddlc1 mutant (ddlc1/Y; +/+; +/tub-GFP-LAMP1) 2 h AHE, and 12 h AHE (n = 21 glands).
To test if caspases contribute to ddlc1 mutant salivary gland degradation, we expressed p35 in ddlc1 mutant salivary glands and examined them using paraffin histology. As previously shown (14, 18), all control animals that express p35 in salivary glands possess partly degraded condensed salivary gland cell fragments with neither lumen nor basement being membrane present at 12 h AHE (Fig. 2D). In control ddlc1 mutants, 39% of animals have intact salivary glands, whereas 61% are partly degraded (Fig. 2D). By contrast, p35 expression in the salivary glands of ddlc1 mutant animals results in nearly complete inhibition of degradation, with 96% of these animals having intact salivary glands (Fig. 2D). In addition, expression of the caspase Dronc in salivary glands of ddlc1 mutants rescued the degradation of this tissue 12 h AHE, but also caused premature degradation of salivary glands (Fig. S2). Combined, these data indicate that caspases are active and contribute to the partial salivary gland degradation phenotype in ddlc1 mutant animals, and suggest that Ddlc1 influences both caspases and noncaspase factor(s) that participate in the death of this tissue.
ddlc1 Mutant Glands Have Impaired Autophagy.
Autophagosomes and Atg genes are induced just before salivary gland cell death, and autophagy is required for complete degradation of this tissue (14, 18, 26). We tested whether autophagy is defective in ddlc1 mutant salivary glands using the autophagosome marker GFP-LC3 (27). As expected, the number of GFP-LC3 puncta increased in control salivary glands following the rise in steroid that triggers salivary gland cell death 2 h AHE (Fig. 2 E and G). By contrast, the number of GFP-LC3 puncta was significantly lower in ddlc1 mutant salivary glands at the same stage (Fig. 2 F and G; P < 0.0001). In addition, the number of GFP-LC3 puncta did not increase in ddlc1 mutant salivary glands 10 h later (Fig. 2G; P < 0.0001), suggesting that the decrease in the number of puncta is not a result of a developmental delay. In addition, ectopic expression of a dominant-negative form of Atg1 in ddlc1 mutant salivary glands did not alter the ddlc1 mutant persistent salivary gland phenotype (Fig. S3), suggesting that autophagy is largely impaired in ddlc1 mutant salivary glands.
As lysosomes are required for autophagy, we tested if lysosome numbers are altered in ddlc1 mutant salivary glands by expressing a GFP-Lysosome–associated membrane protein 1 (LAMP1) reporter transgene (27). In controls, the number of GFP-LAMP1 puncta increases following the rise in steroid that triggers salivary gland cell death 2 h AHE (Fig. 2H and Fig S4). Although the number of GFP-LAMP1 puncta was lower in ddlc1 mutant salivary glands at the same stage (Fig. 2H; P = 0.0008; also see Fig. S4), the number of GFP-LAMP1 puncta was not significantly different 10 h later (Fig. 2H; P = 0.14). These data indicate that, although lysosome numbers are reduced in ddlc1 mutant salivary glands, lysosomes appear to be produced in this tissue.
ddlc1 Mutants Contain Protein Inclusions.
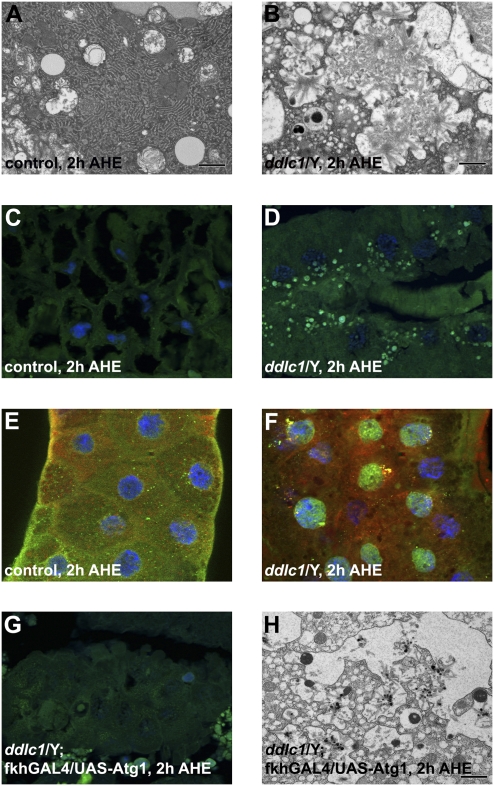
The observation of fewer autophagosomes in ddlc1 mutant salivary gland cells prompted us to investigate their morphology by transmission EM (TEM). The cytoplasmic morphology of WT salivary gland cells includes numerous mitochondria, extensive rough endoplasmic reticulum, and autolysosomes that are typically observed in these cells 2 h AHE (Fig. 3A). Although ddlc1 mutant salivary glands contained some autophagic structures, numerous fibrillar inclusions were present. These inclusions appear to be dispersed throughout the cytoplasm and were not surrounded by membranes (Fig. 3B). Glue protein is produced and secreted by salivary glands (28), and our observation of amyloid-like inclusions prompted us to consider if protein secretion is defective in ddlc1 mutants; salivary gland glue secretion is normal (Fig. S5).
Fig. 3.
ddlc1 mutant salivary glands contain protein inclusions that are reduced when autophagy is induced. (A) WT salivary glands 2 h AHE lack inclusions by TEM, whereas (B) ddlc1 mutant salivary glands possess large cytoplasmic inclusions 2 h AHE. (C) WT salivary glands have no thioflavin-S foci 2 h AHE (n = 10 animals), whereas (D) ddlc1 mutant salivary glands possess numerous green thioflavin-S–stained foci 2 h AHE (n = 10 animals). (E) WT salivary glands exhibit no colocalization of Ref(2)P (green) and ubiquitin (red) 2 h AHE (n = 4 glands), whereas (F) Ref(2)P and Ubiquitin are colocalized in ddlc1 mutant salivary glands 2 h AHE (n = 5 glands) (DNA, blue). (G) Expression of Atg1 in ddlc1 mutant salivary glands eliminates thioflavin-S–stained foci (compare with D) (n = 10), and (H) attenuates fibrillar inclusions by TEM. (Scale bars, 1 μm in A, B, and H.)
The accumulation of protein inclusions is a feature that is common to many age-related neurodegenerative diseases (29), and has been associated with proteasome inhibition (9). Therefore, we tested if the proteasome was inhibited in the ddlc1 mutant salivary glands by expressing the GFP-CL1 reporter (9). Although genetic impairment of the proteasome resulted in accumulation of CL1-GFP, this reporter was not present in ddlc1 mutant salivary glands (Fig. S6), indicating that proteasome function is not impaired in ddlc1 mutants. The fibrillar nature of the ddlc1 mutant salivary gland inclusions are similar to structures that are observed in amyloids associated with many neurodegenerative diseases (29). Therefore, we tested if the protein inclusions in ddlc1 mutant salivary glands have amyloid properties by staining with thioflavin-S (30). Paraffin sections of ddlc1 mutant salivary glands contained numerous thioflavin-S–positive foci that are indicative of amyloid deposition (Fig. 3D), whereas control glands did not possess these structures (Fig. 3C).
Protein inclusions that occur during neurodegeneration typically contain ubiquitin and p62, proteins that are associated with the proteasome and autophagy (31, 32). Therefore, we tested whether the protein inclusions that are observed in ddlc1 mutant salivary glands contain ubiquitin and Drosophila p62 orthologue Ref(2)P (33). Although both ubiquitin and Ref(2)P were detected in WT salivary gland cells, they were not colocalized (Fig. 3E). By contrast, ubiquitin and Ref(2)P were colocalized in large puncta in ddlc1 mutant salivary gland cells (Fig. 3F). Furthermore, Ref(2)P levels were elevated and ubiquitin-positive species of multiple molecular weights were elevated in ddlc1 mutant salivary glands (Fig. S7).
Protein inclusions accumulate when catabolic pathways, including autophagy, are defective during neurodegeneration (9, 34). The reduced number of GFP-LC3 autophagosomes and accumulation of protein aggregates in ddlc1 mutant salivary glands prompted us to test if expression of the Atg1 kinase, a protein that is sufficient to induce autophagy (14, 35), is sufficient to inhibit the accumulation of protein aggregates in these mutants. Significantly, the thioflavin-S–positive inclusions in ddlc1 mutant salivary gland cells were absent when we expressed a moderate-strength Atg1 transgene in this tissue (Fig. 3 D and G). In addition, TEM analyses of these glands revealed that fibrillar inclusions were absent in glands with enhanced degradation (Fig. S8) or reduced in moderately degraded glands (Fig. 3 B and H). These data indicate that the protein inclusion defects in ddlc1 mutant salivary glands are a result of impairment of autophagy.
ddlc1 Enhances Motility Defects in an SBMA Model.
We have previously shown that protein clearance by autophagy plays an important role in a Drosophila eye model of the neuromuscular disease spinal bulbar muscular atrophy (SBMA) (9). SBMA is characterized by progressive loss of motor neurons and muscular atrophy in males, and is caused by polyglutamine expansion in the androgen receptor (AR) in the presence of the AR ligand testosterone (36). Similar to our previous studies in a Drosophila eye model, we observed a polyglutamine length- and ligand-dependent decrease in motor activity when we expressed AR in motor neurons of third instar larvae (Fig. 4A). Either loss of ddlc1 function or knock-down of Atg12 in motor neurons of larvae resulted in motility defects, and loss of ddlc1 enhanced the SBMA defect in motor activity when expanded polyglutamine-containing AR was expressed in motor neurons (Fig. 4B).
Fig. 4.
ddlc1 enhances motility and protein clearance defects in a model of SBMA. (A) Average number (±SEM) of grid lines passed by the posterior end of WT larvae (+/+) or larvae expressing human AR in motor neurons containing 20 glutamines (AR Q20 = w; uas-hAR Q20/d42GAL4), or 52 glutamines (AR Q52 = w; uas-hAR Q52/d42GAL4) and raised on food containing dihydroxytestosterone (DHT; n = 3 × 15 larvae). *P < 0.01 and **P < 0.001, significant difference from control. (B) Average number (±SEM) of grid lines passed by the posterior end of control female sibling (ddlc1/+), ddlc1 mutant (ddlc1/Y), Atg12 RNAi (y w, uas-Atg12-IR/+; d42GAL4/+), AR 52Q-expressing (w; uas-hAR Q52/d42GAL4), or ddlc1 mutant expressing AR 52Q (ddlc1/Y; uas-hAR Q52/d42GAL4) larvae in the absence or presence of DHT (n = 3 × 15 larvae). *P < 0.01 and **P < 0.001, significant difference from ddcl1/+ control; and ***P < 0.001, significant difference from AR Q52 expression + DHT alone. (C–E) Ref(2)P staining (green) in third instar larval motor neurons of (C) WT (n = 8 animals), (D) ddlc1 mutant (n = 8 animals), and (E) ddlc1 mutant expressing hAR Q52 in the presence of DHT (n = 8 animals). (F) Reduced ddlc1 function decreases the turnover of polyQ-expanded AR. Western blots showing the temporal profile of AR Q52 protein level after 1-h pulse of expression. A logarithmic plot of AR Q52/Tubulin ratios was used to determine the line of best fit, and the half-life of AR Q52 was increased 1.73-fold in ddlc1 mutant larvae.
The reduced motility of ddlc1 mutant larvae prompted us to investigate the accumulation of protein inclusions in the nervous and muscular systems of these animals. ddlc1 mutants and ddlc1 mutants expressing AR Q52 with DHT have punctate Ref(2)P staining in a subset of their motor neurons (Fig. 4 D and E), whereas WT motor neurons do not (Fig. 4C). Significantly, reduced ddlc1 function results in decreased clearance of AR Q52 in neurons (Fig. 4F). Interestingly, ddlc1 mutant larval muscles also have numerous Ref(2)P-positive inclusions (Fig. 5B) and attenuated autophagy (Fig. S9), whereas WT larval muscles have a diffuse staining of Ref(2)P (Fig. 5A). These Ref(2)P inclusions in muscles become larger when AR Q52 is expressed in motor neurons in the presence of DHT (Fig. 5C). These data suggest that the motor activity defect in ddlc1 larvae is related to catabolic defects in the neuromuscular tissue, and that expression of AR Q52 in motor neurons enhances this ddlc1 phenotype. In addition, these data also suggest that decreased protein clearance in motor neurons influences the physiology of muscles. Although decreased function of ddlc1 in muscles results in accumulation of Ref(2)P in muscles (Fig. 5D and Fig. S10), knock-down of either ddlc1 or Atg12 specifically in motor neurons also results accumulation of Ref(2)P in muscles (Fig. 5E and Figs. S10 and S11). We investigated this nonautonomous function of ddlc1 and discovered that ddlc1 mutants possess decreased neuromuscular junction bouton numbers (Fig. 5 F, G, and H; P < 0.00001). Combined, these data indicate that decreased ddlc1 function results in decreased autophagy, protein clearance, and synapse number, and that this influences the physiology of muscles and motility.
Fig. 5.
ddlc1 mutants accumulate Ref(2)p in muscles and have decreased numbers of synaptic boutons. (A–E) Ref(2)P staining (green) of third instar larval muscles of (A) WT (n = 8 animals), (B) ddlc1 mutant (n = 8 animals), (C) ddlc1 mutant larvae expressing AR Q52 in the presence of dihydroxytestosterone (DHT; n = 8 animals), (D) ddlc1 RNAi (Ddlc1-IR) expression by the muscle c57 GAL4 driver (n = 5 animals), and (E) ddlc1 RNAi (Ddlc1-IR) expression by the motor neuron d42 GAL4 driver (n = 5 animals). (F–H) Quantification of bouton numbers in neuromuscular junctions. (F) WT (n = 9 animals) and (G) ddlc1 mutant (n = 11 animals) stained with HRP antibody. (H) Average number of boutons (±SEM) in the genotypes shown in F and G. *P < 0.00001, significant difference from control.
Discussion
Drosophila ddlc1 is required for cell death of salivary glands. Ddlc1 has been implicated in the regulation of cell death through physical interactions with Bim (21) and p53 binding protein (37). In these cases, Ddlc1 restricts the activity of caspase regulators and caspases. These data are consistent with previous studies in Drosophila indicating that complete loss of ddlc1 function results in ectopic cell death and lethality during embryogenesis (23). By contrast, we found that caspase activity is reduced in ddlc1 mutants. In addition, factors other than caspases are also influenced by ddlc1, and required for salivary gland destruction. Significantly, ddlc1 mutant salivary glands are defective in autophagosome formation. Although autophagy is an important regulator of salivary gland cell death (14, 18), we cannot exclude the possibility that ddlc1 regulates other processes that are required for destruction of this tissue.
Ddlc1 is best known as a dynein motor component, but it also functions in motor-independent processes (21, 38). It is not clear how Ddlc1 regulates autophagy. An intact microtubule network is necessary for proper autophagosome formation (39) and disruption of dynactin impairs autophagy (40), so it is possible that Ddlc1 is part of a dynein motor that is required for autophagy in salivary glands. However, we did not observe any defect in salivary gland degradation in Dynein heavy chain mutants (Fig. S12), and this suggests that Ddlc1 may be regulating autophagy in a motor-independent manner. It remains unclear how Ddlc1 regulates autophagy. Although expression of the autophagy regulator Atg1 was sufficient to rescue salivary gland degradation (Fig. S13), it is possible that Ddlc1 may either influence autophagy signaling or facilitate the localization of autophagosome cargo or autophagy regulatory factors that are needed to form autophagosomes.
Defects in protein clearance have been associated with several neurodegenerative disorders (8, 9, 32). To our surprise, fibrillar protein inclusions that stain with amyloid-specific thioflavin-S and are reminiscent of neuronal amyloids accumulate in ddlc1 mutant salivary glands. We speculate that these protein inclusions form because of attenuated autophagy in ddlc1 mutants, and this conclusion is supported by the fact that activation of autophagy by expression of Atg1 reduces these inclusions. In addition, the levels of Ref(2)P increased and were localized with ubiquitin-positive inclusions in ddcl1 mutant salivary glands.
p62 [Ref(2)P in flies] and ubiquitin accumulate in aging brains of autophagy-deficient mice and flies (33, 41). In addition, p62-associated protein inclusions were degraded by autophagy in a model of Huntington disease (32), and disruption of the LC3/Atg8 binding domain in p62 resulted in the formation of ubiquitin-positive inclusions (42). These studies suggest similarities between the role of autophagy in neuronal protein aggregation disorders and in ddlc1 mutant salivary glands, and this prompted us to study the influence of ddlc1 mutations on the nervous system. Both ddlc1 mutant larvae and the SBMA polyglutamine disorder model larvae exhibited decreased motor activity. In addition, expression of AR Q52 in motor neurons of ddlc1 mutant larvae enhanced motility defects, and reduced ddlc1 function slowed the clearance of AR Q52 protein. Significantly, Ref(2)P-associated inclusions are present in the motor neurons and muscles of ddlc1 mutant larvae in the presence and absence of expression of AR Q52 in motor neurons. However, the size of Ref(2)P-positive inclusions increases in the muscles of ddlc1 mutants expressing AR Q52 in motor neurons. These observations suggest that the increased size of protein inclusions in ddlc1 muscle is triggered in a cell-nonautonomous manner by altered motor neuron function. Significantly, we observed a nonautonomous influence of ddlc1 on autophagy in muscle, and this was associated with decreased neuromuscular junction synapse numbers. These data are consistent with the nonautonomous influence of neuronal function on muscle physiology in a Caenorhabditis elegans model of polyglutamine disease (43). In addition, these data are in accordance with the recent demonstration that autophagy in motor neurons regulates synapse development via clearance of Highwire in flies (44).
We have identified ddlc1 as a factor that is required for autophagy in dying cells, and show that loss of ddlc1 leads to accumulation of p62-associated aggregates in salivary glands and muscles. These tissues are composed of endo-replicating cells with extensive cytoplasm, and it is possible that they rely more heavily on autophagy for clearance of unwanted or damaged proteins than diploid cells, and that decreased autophagy leads to protein aggregates. Alternatively, and like neurons, these are relatively long-lived cells, and it is possible that, at stages approaching death, autophagy begins to promote clearance of cytoplasm. Future studies should resolve how Ddlc1 functions in the regulation of protein clearance, autophagy, and cell death.
Materials and Methods
Drosophila Strains.
A collection of 1,475 single P-element–lacZ insertion mutations were screened to identify lines that possess defects in salivary gland degradation, and l(1)G0371 was identified as an insertion in ddlc1. The ddlc1 precise excision line was generated by crossing the l(1)G0371 line to Δ 2–3 transposase and precise excision was confirmed by DNA sequencing. UAS-p35 (45) was used to inhibit caspases. UAS-Atg1GS10797 (35) was used for ectopic induction of autophagy, and UAS-Atg12-IR (9) was used to knock down autophagy. fkhGAL4, daGAL4, d42GAL4, c57GAL4, and elavGS were used to drive gene expression. The UAS-AR Q20 and UAS-AR Q52 lines were used for the SBMA model (9). UAS-GFP-LC3 (27) was used as a marker of autophagosomes, and tubulin-GFP-LAMP1 was used as a marker of lysosomes. WT Canton-S or the precise excision of the l(1)G0371 P-element were used as controls.
UAS-Ddlc1 Transgenic Flies.
The ddlc1 ORF was inserted in the pUAST Drosophila transformation vector, sequenced, and used to generate transgenic Drosophila (Best Gene).
Salivary Gland Histology.
Animals were staged, fixed, embedded in paraffin, sectioned, and stained as previously described (14). DNA fragmentation was detected by TUNEL using the Apoptotag kit (Chemicon). To detect amyloidosis with Thioflavin-S, paraffin sections were rehydrated, stained with Mayer hematoxylin (Fluka Chemie) for 1 min, and stained with 1% thioflavin-S (Sigma) for 5 min, followed by dehydration and mounting in Vectashield with DAPI (Vector Laboratories). Sections were examined with a Zeiss Axio Imager.Z1 microscope.
Caspase Substrate Assays.
The EnzChek Caspase-3 Assay kit (Molecular Probes) was used according to manufacturer protocols. Animals were staged as white prepupae, aged 4 h, and whole animal lysates were analyzed as previously described (14).
Fluorescence Microscopy.
For autophagosome and lysosome detection, salivary glands were dissected in PBS solution, stained with Hoechst 33342 to detect DNA, mounted in PBS solution, imaged with a Zeiss Axio Imager.Z1 microscope with apotome, and quantified using Zeiss automatic measurement software. The average number of puncta per field was determined from 3 independent fields within a salivary gland.
Immunohistochemistry.
Salivary glands were fixed and stained as described (19). Neuromuscular junction preparations of wandering third instar larvae were performed as described (46). Rabbit anti-Ref(2)P was used at 1:1,000, mouse anti–mono- and poly-ubiquitinylated proteins (FK2; Biomol) were used at 1:500 concentration, and rabbit anti-HRP conjugated to FITC (Jackson Immunoresearch) was used at 1:800. All preparations were imaged with a Zeiss Axio Imager.Z1 microscope with apotome. Neuromuscular junctions that innervate muscles 6 and 7 of segment A3 were analyzed, and the total bouton numbers on type 1 neurons are reported.
Electron Microscopy.
Salivary glands were fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4, postfixed in buffered 1% osmium tetroxide for 1 h at room temperature, dehydrated, treated with propylene oxide, and infiltrated for embedding in SPI-pon/Araldite. Ultrathin sections were stained with uranyl acetate and lead citrate before examination in a Philips CM12 TEM.
Larval Motility Assay.
Fifteen wandering third instar larvae of the indicated genotypes were washed in PBS solution and placed on a 1% agarose gel in an 80-mm Petri dish with gridlines spaced by 5 mm. The larvae were allowed to acclimate for 1 min, and the number of grid lines that the posterior end of larvae passed in 2 min was determined.
Protein Turn-Over Analysis.
Third instar larvae were washed with water and ethanol and transferred to 0.5 mL of 5 mM RU486 and 1 mM DHT for 1 h (9, 47). Larvae were then transferred to vials containing yeast paste. AR and tubulin protein levels were determined as described (9). Quantitation of Western blots was performed using the Odyssey Infrared Imaging System (LI-COR Biosciences). The mean AR/tubulin ratios and SEM from ≥3 replicates were plotted on a logarithmic scale and used to determine the line of best fit by regression analysis (y = Ae-Kx). The slope of the best fit line was used to estimate half-life with the equation t1/2 = 0.693/K.
Supplementary Material
Acknowledgments
We thank V. Budnik, M. Freeman, T.S. Hays, H. Kramer, R.K. Murphey, J. Nambu, T.P. Neufeld, I. Nezis, T.E. Rusten, H. Stenmark, the Bloomington Stock Center, and the Developmental Studies Hybridoma Bank for flies and antibodies; K. Kerr for advice on neuromuscular junction staining; and R. Simin and T. Fortier for technical support. This work was supported by National Institutes of Health Grants NS053825 and AG031587 (to J.P.T.) and GM079431 (to E.H.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907967107/DCSupplemental.
References
- 1.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 2.Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 4.Thumm M, et al. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- 5.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baehrecke EH. Autophagy: dual roles in life and death? Nat Rev Mol Cell Biol. 2005;6:505–510. doi: 10.1038/nrm1666. [DOI] [PubMed] [Google Scholar]
- 7.Boya P, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boland B, et al. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. J Neurosci. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandey UB, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 10.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 11.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 12.Schweichel JU, Merker HJ. The morphology of various types of cell death in prenatal tissues. Teratology. 1973;7:253–266. doi: 10.1002/tera.1420070306. [DOI] [PubMed] [Google Scholar]
- 13.Clarke PG. Developmental cell death: morphological diversity and multiple mechanisms. Anat Embryol (Berl) 1990;181:195–213. doi: 10.1007/BF00174615. [DOI] [PubMed] [Google Scholar]
- 14.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qu X, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 16.Mohseni N, McMillan SC, Chaudhary R, Mok J, Reed BH. Autophagy promotes caspase-dependent cell death during Drosophila development. Autophagy. 2009;5:329–338. doi: 10.4161/auto.5.3.7444. [DOI] [PubMed] [Google Scholar]
- 17.Hou YC, Chittaranjan S, Barbosa SG, McCall K, Gorski SM. Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J Cell Biol. 2008;182:1127–1139. doi: 10.1083/jcb.200712091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee CY, Baehrecke EH. Steroid regulation of autophagic programmed cell death during development. Development. 2001;128:1443–1455. doi: 10.1242/dev.128.8.1443. [DOI] [PubMed] [Google Scholar]
- 19.Martin DN, Baehrecke EH. Caspases function in autophagic programmed cell death in Drosophila. Development. 2004;131:275–284. doi: 10.1242/dev.00933. [DOI] [PubMed] [Google Scholar]
- 20.Ravikumar B, et al. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat Genet. 2005;37:771–776. doi: 10.1038/ng1591. [DOI] [PubMed] [Google Scholar]
- 21.Puthalakath H, Huang DC, O’Reilly LA, King SM, Strasser A. The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol Cell. 1999;3:287–296. doi: 10.1016/s1097-2765(00)80456-6. [DOI] [PubMed] [Google Scholar]
- 22.King SM, et al. Brain cytoplasmic and flagellar outer arm dyneins share a highly conserved Mr 8,000 light chain. J Biol Chem. 1996;271:19358–19366. doi: 10.1074/jbc.271.32.19358. [DOI] [PubMed] [Google Scholar]
- 23.Dick T, Ray K, Salz HK, Chia W. Cytoplasmic dynein (ddlc1) mutations cause morphogenetic defects and apoptotic cell death in Drosophila melanogaster. Mol Cell Biol. 1996;16:1966–1977. doi: 10.1128/mcb.16.5.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phillis R, Statton D, Caruccio P, Murphey RK. Mutations in the 8 kDa dynein light chain gene disrupt sensory axon projections in the Drosophila imaginal CNS. Development. 1996;122:2955–2963. doi: 10.1242/dev.122.10.2955. [DOI] [PubMed] [Google Scholar]
- 25.Neufeld TP, Baehrecke EH. Eating on the fly: function and regulation of autophagy during cell growth, survival and death in Drosophila. Autophagy. 2008;4:557–562. doi: 10.4161/auto.5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee CY, et al. Genome-wide analyses of steroid- and radiation-triggered programmed cell death in Drosophila. Curr Biol. 2003;13:350–357. doi: 10.1016/s0960-9822(03)00085-x. [DOI] [PubMed] [Google Scholar]
- 27.Rusten TE, et al. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell. 2004;7:179–192. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 28.Biyasheva A, Do TV, Lu Y, Vaskova M, Andres AJ. Glue secretion in the Drosophila salivary gland: a model for steroid-regulated exocytosis. Dev Biol. 2001;231:234–251. doi: 10.1006/dbio.2000.0126. [DOI] [PubMed] [Google Scholar]
- 29.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 30.Maezawa I, et al. Congo red and thioflavin-T analogs detect Abeta oligomers. J Neurochem. 2008;104:457–468. doi: 10.1111/j.1471-4159.2007.04972.x. [DOI] [PubMed] [Google Scholar]
- 31.Ross CA, Pickart CM. The ubiquitin-proteasome pathway in Parkinson’s disease and other neurodegenerative diseases. Trends Cell Biol. 2004;14:703–711. doi: 10.1016/j.tcb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 32.Bjørkøy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nezis IP, et al. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol. 2008;180:1065–1071. doi: 10.1083/jcb.200711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 35.Scott RC, Juhász G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 37.Lo KW, et al. The 8-kDa dynein light chain binds to p53-binding protein 1 and mediates DNA damage-induced p53 nuclear accumulation. J Biol Chem. 2005;280:8172–8179. doi: 10.1074/jbc.M411408200. [DOI] [PubMed] [Google Scholar]
- 38.Barbar E. Dynein light chain LC8 is a dimerization hub essential in diverse protein networks. Biochemistry. 2008;47:503–508. doi: 10.1021/bi701995m. [DOI] [PubMed] [Google Scholar]
- 39.Köchl R, Hu XW, Chan EY, Tooze SA. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic. 2006;7:129–145. doi: 10.1111/j.1600-0854.2005.00368.x. [DOI] [PubMed] [Google Scholar]
- 40.Kimura S, Noda T, Yoshimori T. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct Funct. 2008;33:109–122. doi: 10.1247/csf.08005. [DOI] [PubMed] [Google Scholar]
- 41.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 42.Ichimura Y, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 43.Garcia SM, Casanueva MO, Silva MC, Amaral MD, Morimoto RI. Neuronal signaling modulates protein homeostasis in Caenorhabditis elegans post-synaptic muscle cells. Genes Dev. 2007;21:3006–3016. doi: 10.1101/gad.1575307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen W, Ganetzky B. Autophagy promotes synapse development in Drosophila. J Cell Biol. 2009;187:71–79. doi: 10.1083/jcb.200907109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- 46.Budnik V, Gorczyca M, Prokop A. Selected methods for the anatomical study of Drosophila embryonic and larval neuromuscular junctions. Int Rev Neurobiol. 2006;75:323–365. doi: 10.1016/S0074-7742(06)75015-2. [DOI] [PubMed] [Google Scholar]
- 47.Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci USA. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.