Abstract
Interactions between transmembrane (TM) helices play an important role in the regulation of diverse biological functions. For example, the TM helices of integrins are believed to interact heteromerically in the resting state; disruption of this interaction results in integrin activation and cellular adhesion. However, it has been difficult to demonstrate the specificity and affinity of the interaction between integrin TM helices and to relate them to the activation process. To examine integrin TM helix associations, we developed a bacterial reporter system and used it to define the sequence motif required for helix–helix interactions in the β1 and β3 integrin subfamilies. The helices interact in a novel three-dimensional motif, the “reciprocating large-small motif” that is also observed in the crystal structures of unrelated proteins. Modest but specific stabilization of helix associations is realized via packing of complementary small and large groups on neighboring helices. Mutations destabilizing this motif activate native, full-length integrins. Thus, this highly conserved dissociable motif plays a vital and widespread role as an on-off switch that can integrate with other control elements during integrin activation.
Keywords: interaction motif, transmembrane domain
Integrins are a family of transmembrane (TM) α/β heterodimers that exist in an equilibrium between resting (low-affinity) and active (high-affinity) conformations (1). TM helix–helix associations play an important role in this process for the integrin αIIbβ3, which can be activated physiologically by interactions with cytoplasmic proteins (2–5). The α and β subunit TM domains of αIIbβ3 interact in the resting state, but separate when the integrin assumes a fully activated conformation (3, 4, 6–8). Thus, it appears that the αIIb and β3 TM helices associate in a sufficiently stable interaction to form a clasp maintaining αIIbβ3 in an inactive conformation. If this is the case, one might expect that the TM helices would form an autonomous interaction unit, engaging in a stable interaction even in the absence of extracellular and cytoplasmic domains. Indeed, early modeling studies suggested a specific geometric interaction (4, 9, 10), which has recently been supported by NMR and Cys cross-linking studies of the αIIb and β3 TM helices (11, 12).
Here, we extend genetic methods to examine the association of the isolated TM helices in bacterial membranes. The approach represents a modification on earlier methods to monitor homomeric and heteromeric TM helix–helix association (13–18) that is sufficiently specific to allow measurement of fine changes in heterodimeric TM helix–helix association. We then employed these methods, in conjunction with structural bioinformatics, to identify a unique interaction motif that is conserved between the TM helices of the integrins αIIbβ3, ανβ3, α2β1, and α5β1. Disruption of this motif in full-length integrins resulted in integrin activation and ligand binding in transfected cell lines, demonstrating the functional relevance of the TM domain interactions.
Results
Identification of the β3 TM Helix Motif that Interacts with Its α-TM Partners.
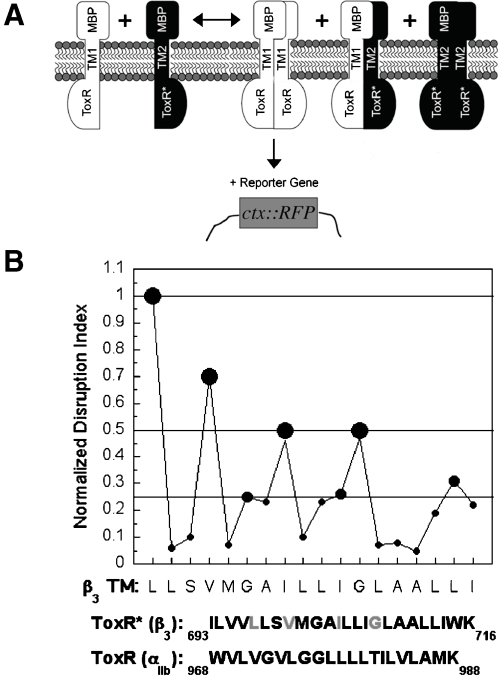
To identify the residues in the β3 TM helix that associate with the TM helices of its complementary α subunits, we reconfigured the homooligomeric TOXCAT assay (Fig. 1A) (19). In the reconfigured assay, there is competition between the homomeric association of a ToxR fusion protein containing a TM helix of interest and the heteromeric association of this fusion protein with a second fusion protein whose ToxR DNA binding domain has been disabled (Fig. 1A). Thus, the disabled ToxR fusion protein acts as a dominant-negative (DN), and the resulting decrease in reporter gene synthesis indicates the extent to which heterooligomerization is favored over homooligomerization (Figs. S2 and S3). We also replaced chloramphenicol acetyl transferase (CAT) as the reporter gene with the red fluorescent protein (RFP) variant mCherry because it can be detected in whole cells without need for an exogenous substrate (DN-ToxRed) (Fig. S1). Lastly, to reduce cell-to-cell variability in reporter synthesis, we prepared a vector in which the wild-type (WT) and disabled fusion proteins were expressed from the same multicopy plasmid under control of the inducible T7 promoter.
Fig. 1.
Use of DN-ToxRed to measure the heteromeric interaction of the β3 TM domain with the TM domain of αIIb. (A) Overview of the DN-ToxRed assay. Two TM domains are coexpressed in the E. coli inner membrane: one (TM1) fused to WT ToxR and able to activate transcription at the ctx promoter upon dimerization; the other (TM2) fused to an inactive ToxR mutant. By interacting heteromerically with TM1, TM2 exerts a DN effect on reporter gene synthesis stimulated by TM1 homooligomerization (see SI Text for details). (B) Effect of Leu- and Ala-scanning mutagenesis of the β3 TM domain on its heteromeric association with αIIb in DN-ToxRed. The results were expressed as a “Normalized Disruption Index,” a previously described measure of the mean fold-change in reporter gene synthesis for each β3 TM domain mutation (21), and were categorized as maximal disruption (0.5–1.0, large filled circles), intermediate disruption (0.25–0.5, intermediate-sized filled circles) and minimal disruption (0–0.25, small filled circles).
The αIIb TM domain has a relatively strong tendency to form homodimers (20, 21), giving a signal approximately half that of the glycophorin A TM helix in DN-ToxRed (Figs. S1 and S2). This signal is attenuated by coexpressing a DN partner containing the WT β3 TM domain (Figs. 1A and S3) that competes for binding to the αIIb TM domain. To identify the interaction interface of the αIIbβ3 TM heterodimer, we measured the effect of a series of Leu and Ala substitutions across the β3 TM helix on its interaction with the αIIb TM domain (Fig. 1B) (19). Mutations most disruptive of the DN effect of the β3 helix occurred at residues L697, V700, I704, and G708, each of which has been identified as important for stabilizing the resting conformation of full-length αIIbβ3 (2–4, 6, 7), and in addition, are in proximity to αIIb based on disulfide-cross-linking and NMR measurements (11, 12).
A Conserved Sequence Motif for Integrin α/β TM Domain Interactions.
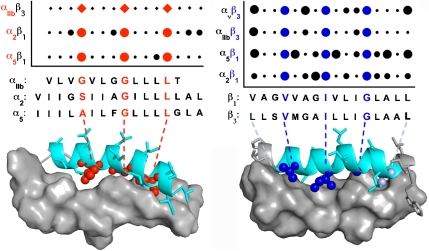
The DN-ToxRed assay was then used to probe the association of the β3 subunit with its other partner, αv, as well as the association of the β1 TM domain with its partners α2 and α5 (Figs. S3 and S4). Scanning mutagenesis indicated that the β3 TM domain uses the same set of residues to engage both αIIb and αν. Furthermore, equivalent positions in the β1 TM domain are critical for its interaction with α2 and α5, revealing a consensus β-subunit motif, V/L-x2-V-x3-I-x3-G-x3-L (Fig. 2). Each residue of this consensus motif is located at the helix–helix interface in a model for the αIIbβ3 TM heterodimer (Fig. 2). More variability in the ability to disrupt the DN effect was observed for mutations near the ends of the TM sequences or at residues neighboring the common conserved motif. Thus, V-x3-I-x3-G provides a conserved framework comprised of two large, hydrophobic residues and a small glycine residue for interaction, while the more variable regions likely serve to modulate the affinity and specificity of TM domain pairs.
Fig. 2.
Effect of mutations on the heteromeric association of α and β integrin subunit TM domains. The TM domains of β1, β2, αv, α2, and α5 were scanned with Leu and Ala mutations and the effect on the heteromeric association of the TM domains with their complementary subunit TM domains was measured either using DN-ToxRed (circles) or functional data with the intact integrin for αIIb (diamonds). The extent of disruption was quantified and shown as filled circles as described in Fig. 1. The data for αIIb are a composite of functional data reported by Luo et al. (3) and Li et al.(2). αIIb residues 969–981, α2 residues 1005–1020, α5 residues 960–975, β1 residues 713–728, and β3 residues 697–712 are shown below the graph with critical residues shown in red and blue for the alpha and beta TM sequences, respectively. Residues that affect heterooligomerization are also highlighted on a previously described model of αIIbβ3 (9).
We also scanned the TM domains of α2, α5, and αIIb with leucine and alanine to determine how these mutations affected interaction with their complementary β subunit TM domains (Fig. S4). A common α consensus sequence was observed (Fig. 2) and featured a small-x3-G-x3-L motif, where “small” represents residues with small side chains such as Gly, Ala, or Ser. The critical residues in this motif lie along one face of the α subunit TM helix where they interact extensively with the β subunit TM, as illustrated by a model of the αIIbβ3 heterodimer (Fig. 2). Thus, the small residues present in the α/β heterodimer interface provide complementary packing for the large, hydrophobic residues present on the adjacent helix. Together, they define a consensus motif that drives stable and specific interactions between isolated integrin TM helices.
Mutation of Interfacial Residues in β1 TM Cause Constitutive Integrin Activation.
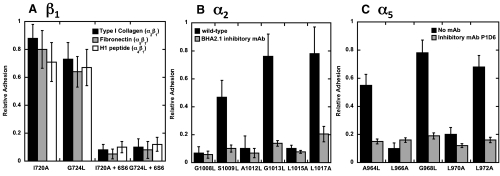
Next, we asked whether mutations that disrupt TM heterodimer formation would activate full-length α2β1 and α5β1. Jurkat A1 cells are β1-null, but endogenously express a number of α subunits that pair with β1, giving rise to integrins with specificities for the extracellular matrix proteins collagen, laminin, fibronectin, and vascular cell adhesion molecule-1 (VCAM-1) (22). We transfected these cells with full-length β1 containing specific TM domain mutations, confirmed that there was comparable expression of the resulting β1-containing integrins on the cell surface by flow cytometry (Fig. S5), and assessed integrin function by measuring cell adhesion to the various immobilized adhesion substrates. The β1 mutations I720A and G724L, each of which disrupts heterodimer formation in DN-ToxRed, caused a substantial increase in cell adhesion to type I collagen, fibronectin, and the α4β1-specific peptide H1 (23) (Fig. 3A). The increased adhesion was comparable to that of WT Jurkat cells treated with MnCl2 and could be blocked by pretreating the cells with EDTA or the β1-specific inhibitory mAb 6S6. These results were confirmed by flow cytometry using soluble type I collagen and fibronectin as ligands for α2β1 and α5β1, respectively (Figs. S5 and S6).Thus, cells expressing the interfacial β1 mutants I720A and G724L constitutively bound soluble type I collagen (Fig. S5), consistent with the presence of activated α2β1 on the cell surface. On the other hand, there was no specific binding of soluble ligands to cells expressing the noninterfacial mutants A718L and G719A (Fig. S5).
Fig. 3.
Effect of β1, α2, and α5 TM domain mutations on the function of β1 integrins. (A) Adhesion of Jurkat cells expressing β1 TM domain mutations to type I collagen fibronectin and H1 peptide (specific for α4β1). Adhesion specific for β1 integrins was assessed by performing the assay in the presence of the β1-specific mAb 6S6. (B) Adhesion of HEK293 cells expressing α2 TM domain mutations to type I collagen. Specific adhesion was assessed using the α2-specific mAb BHA2.1. (C) Adhesion of HEK293 cells expressing α5 TM domain mutations to fibronectin. Specific adhesion was assessed using the α5-specific mAb P1D6.
We used a similar strategy to examine the activating effect of α2 and α5 TM mutations, choosing HEK293 cells for these experiments because they constitutively express WT β1. We found that the interfacial α2 mutations S1009L, G1013L, and L1017A increased cell adhesion to type I collagen, whereas the noninterfacial mutations G1008L, A1012L and L1015A did not (Fig. 3B). Similarly, only the interfacial α5 mutations A964L, G968L, and L972A increased cell adhesion to fibronectin (Fig. 3C). In both cases, adhesion was integrin-specific, as it was inhibited by EDTA and the monoclonal antibodies BHA2.1 and P1D6 specific for α2β1 and α5β1, respectively. Comparable results were seen by flow cytometry, where the interfacial α2 mutations S1009L and G1013L and the interfacial α5 mutation G968L induced constitutive binding of soluble collagen and fibronectin, respectively, and noninterfacial mutations did not (Figs. S5 and S6). Together these results imply that the consensus α and β TM helix interaction motifs are sufficient to restrain β1 integrins in their resting conformations.
Complementary Packing of Small and Large Residues in the Interaction Motif.
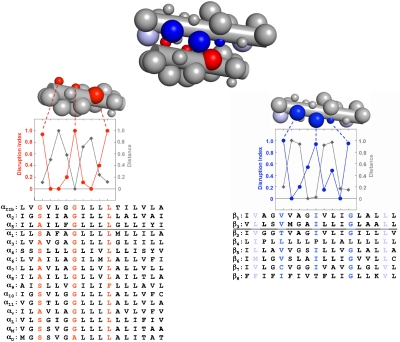
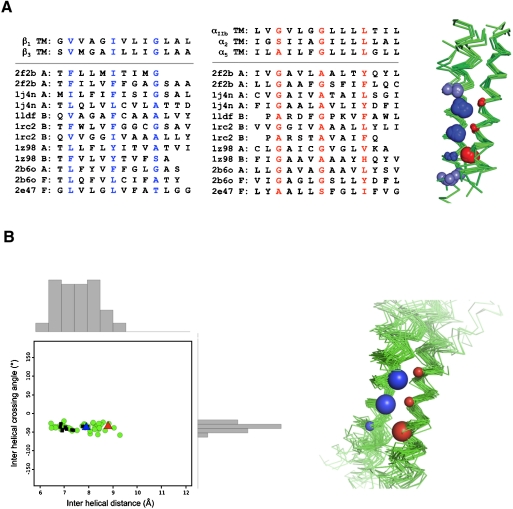
The consensus sequences for the α and β integrin subunit TM interfaces have critical side chains spaced at approximately four-residue intervals, indicative of the right-handed helical crossings often observed in interacting TM helices (24–26). However, the consensus sequence motifs are distinct from high-affinity homodimers such as GpA (27), in which a small-x3-small motif in each monomer forms the homodimer interface. Instead, a small-x3-G-x3-L motif in the α subunit TM domain interacts with V-x3-I-x3-G in the β subunit, forming a previously unrecognized zipper-like TM heterodimerization motif that is conserved across the entire integrin superfamily (Fig. 4). If this motif is energetically favorable, it might be expected to occur in other proteins. To address this question, we searched a library of 188 right-handed interacting parallel heterodimeric TM pairs excised from crystal structures of membrane proteins deposited in the protein data bank (Fig. S7). As described in SI Text, a sequence-directed search for dimers with the integrin consensus sequence pattern identified 24 helical pairs; geometric clustering revealed 12 dimers whose sequences and structures at conserved, interfacial positions were similar (Fig. 5A).
Fig. 4.
A reciprocal large-small motif mediates the heteromeric association of integrin TM domains. The integrin heterodimer is represented as an idealized pair of helices with large spheres denoting hydrophobic (L, I, V) residues and small spheres representing small (G, A, S) residues. The disruption index for each residue and its corresponding interhelical distance (the minimum Cα to Cα distance between helices, normalized from 0 to 1 to allow comparison with the disruption index) are plotted for the α (red) and β (blue) helices and show that these values are negatively correlated. The most disruptive mutants have the smallest interhelical distance and thus form the helix–helix interface. Further, as shown by the aligned TM domain sequences of the entire integrin superfamily, the core interfacial residues (highlighted according to subunit: red for α subunits and blue for β subunits) are conserved. The residues occur at the maximum of the experimentally determined disruption index and with a four-residue periodicity.
Fig. 5.
A family of helical dimers containing the integrin TM heterodimer structural motif. (A) Alignment of integrin sequences and representative sequences from a sequence-directed search of a library of 188 right-handed interacting parallel heterodimeric TM pairs deposited in the protein data bank The structures for the dimers are shown aligned by the sequence motif for each helix. In the accompanying superimposed structures, blue spheres indicate the integrin consensus motif on the β subunits (V-x3-I-x3-G), and the red spheres indicate the integrin consensus motif on the α subunits (small-x3-small-x3-L). Sphere size is used to indicate the size of the side chain at that position along the helix, with large spheres denoting large, hydrophobic residues (V, I, L) and small spheres denoting small, polar residues (G, A, S). (B) Results of a search of the Orientations of Proteins in Membranes crystallographic database for helical dimers using a functional perturbation index. All 69 helix pairs show a tight interhelical crossing angle and distance distribution (shown in histograms), consistent with the recently determined αIIbβ3 TM NMR structure (11) (red triangle) and a high-resolution structure based on Cys cross-linking results (12) (blue triangle). The black squares correspond to 10 structures that were also identified by the sequence-directed search shown in A.
In a related method, we used correlation analysis to identify TM pairs having structures consistent with our β1 and β3 integrin mutagenesis results. Disruptive mutations generally occur at sites of interaction between neighboring helices. Thus, it should be possible to identify structural candidates for the integrin TM heterodimer by correlating the proximity of a given side-chain with the perturbation that occurs when the side-chain is mutated. To quantify the proximity of a given residue to neighboring helices, we computed dmin, the distance between the Cα atom at a given position and the closest Cα atom of a neighboring helix (see SI Materials and Methods for a detailed description of the method used). Using correlation analysis and a subsequent structure-directed search, 69 helical pairs were identified, all of which had right-handed helical crossing angles but varying interhelical distances (Fig. 5B). We investigated the sequence propensities at the interface of this family of structures and found biases that match well to the integrin consensus motif (Fig. 4). Ten members of this cluster also displayed the exact integrin consensus sequence and were among the 12 best structures identified by the first method (Fig. 5A). Thus, two distinct methods converged to define the integrin heterodimer motif as a subset of interacting helical pairs found in the crystal structures of unrelated proteins. Furthermore, when we compared the recent NMR structure for the αIIbβ3 TM heterodimer (pdbid 2K9J) (11, 12) to our family of structures, we found that 2 of the NMR models had an RMSD of approximately 1.0 Å and interhelical geometry consistent with our structures (-32.7° and 8.8 Å, Fig. 5B). Similarly, the recent high-resolution Cys cross-linking–based model of the resting state of αIIbβ3 (11, 12) also gave interhelical geometries similar to the proposed model (-35.6° and 7.9 Å, Fig. 5B). Thus, our proposed model of the integrin TM heterodimer family is in excellent agreement with previous structural characterizations of αIIbβ3 as well.
Discussion
This work addresses the structural motifs, as well as the interaction strength, underlying the association of the TM helices in the integrin superfamily. Although there is considerable evidence indicating that the TM helices of integrins separate when these heterodimeric proteins are fully activated, it has been unclear whether the helices interact tightly in a unique geometry that helps stabilize the resting state or whether they are merely held in place by other regions of the protein that provide the primary restraints in the resting state. Our data clearly demonstrate that the heterodimeric TM domain interaction is an autonomously favorable and geometrically specific unit. The strength of the interaction of the TM helices in the DN-ToxRed assay is intermediate between that observed for weakly associating helices (such as the destabilizing GpA mutant G83I) and constitutively associated structural dimers (such as GpA). This intermediate affinity would be most appropriate for a switchable system, since too-weak or too-tight interactions would impede facile conformational transitions. The relatively weak TM interaction energies might also reflect an energetic balance with contributions from additional extracellular and cytoplasmic domain interactions. For example, a salt bridge in the membrane-proximal cytoplasmic domain has previously been shown to be important for stabilizing the integrin resting state (28) and is one of many interactions disrupted upon full activation. Likewise, mutations at the TM-cytoplasmic interface are sufficient to impair signaling events that occur during activation (29). Thus, the TM heterodimeric pair should be considered as a full partner, acting in concert with cytoplasmic and extracellular domains of the integrin to establish the thermodynamic stability of the resting state and facilitate the orchestration and integration of the diverse inputs required during activation (4).
The heteromeric motif uncovered represents a dissection of the broader class of parallel right-handed crossing motifs discovered in previous studies of helical pairs in membrane proteins (24). Within the family of right-handed parallel crossings were examples of previously well-characterized motifs such as the homodimeric GpA-like packing (30) and glycine-zippers (31), as well as ones in which small-x3-small residues occurred on only one of the two helices. The subfamily we identified here also shares key interfacial residues, as well as similar interhelical geometries, with the experimentally determined αIIbβ3 TM NMR structure (11, 12) and the high-resolution model based on Cys cross-linking results (11, 12). Thus, the zipper-like reciprocating small/large motifs we identified not only defines another subfamily within this grouping, but its role in signaling and membrane protein assembly. Indeed, the synergistic combination of experimental and computational methods employed here, including bacterial assays to assess association, cellular studies to assess function, and structural bioinformatics to assess structure, has considerable potential to uncover the grammatical and syntactical rules relating amino acid sequence to three-dimensional structure, as well as the higher order context imparted by a cell or organism that define function.
Materials and Methods
Design, Cloning and Characterization of ToxRed and DN-ToxRed Constructs.
A detailed discussion of cloning and characterization for DN-ToxRed is provided in SI Text. ToxR and ToxR* constructs containing integrin TM domains were amplified from their respective pccKAN plasmids and subcloned into pCDF-Duet. A bacterial codon-optimized form of mCherry was amplified, ligated in-frame with the ctx promoter region, and subcloned into pCDF-Duet. Constructs were transformed into chemically competent MM39 (DE3) cells and 1 mM IPTG was added to induce expression of the ToxR and ToxR* chimeras. Levels of mCherry expression were used to calculate a disruption index, a measure of the change in mCherry synthesis at each position due to mutation, as previously described (21). Data for cell lysates was collected using a Molecular Dynamics plate reader with an excitation wavelength of 587 nm and an emission wavelength of 615 nm.
Integrin Expression and Functional Analysis in Jurkat and HEK293 Cell Lines.
A detailed discussion of the methods used is provided in SI Text. Briefly, full-length integrin α2, α5 and β1 constructs were subcloned into pCDNA3.1 Hyg/Zeo and transfected transiently for 72 h. Integrin function was measured using static cell adhesion and flow cytometry assays. For cell adhesion, 96 well plates were coated with either fibronectin, laminin, type I collagen, type IV collagen, or H1-peptide as previously described (32). Specific cell adhesion was determined by performing the assays in the presence of the monoclonal antibodies 6S6, JBS5, or BHA2.1. Adhesion of cells expressing WT integrins incubated with 1 mM MnCl2 was a positive control for the assays. For flow cytometry, α2β1 and α5β1 function was assayed using FITC-labeled type I collagen, soluble fibronectin, and the mAb HUTS-4 specific for activated conformations of β1.
Computational Search for TM Helix Pairs Containing the Conserved Integrin Association Motif.
A database of proteins was used to search for interacting helices that fit the conserved sequence pattern and the functional perturbation data. For details on creation and searching of the database, see SI Text.
Supplementary Material
Acknowledgments.
We thank Steven Sandler (University of Massachusetts) for pMK8 expressing E. coli codon-optimized mCherry; Andrew Tsorkas for firefly luciferase vector pGL3-Basic; David Boettinger and Prof. Yoji Shimizu (University of Minnesota) for Jurkat A1 cell line; Lawrence Brass for HEK293 freestyle cell line; Mark Goulian for assistance with fluorescence imaging; and Dieter Langosch, William Russ, and Shalom Goldberg for advice. This work was supported by the National Institutes of Health (GM60610, HL40387, HL62250, and HL81012) and a National Research Service Award postdoctoral fellowship (to B.W.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910873107/DCSupplemental.
References
- 1.Bennett JS. Structure and function of the platelet integrin αIIbβ3. J Clin Invest. 2005;115(12):3363–3369. doi: 10.1172/JCI26989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li R, et al. Activation of integrin αIIbβ3 by modulation of transmembrane helix associations. Science. 2003;300(5620):795–798. doi: 10.1126/science.1079441. [DOI] [PubMed] [Google Scholar]
- 3.Luo BH, Springer TA, Takagi J. A specific interface between integrin transmembrane helices and affinity for ligand. PLoS Biol. 2004;2(6):776–786. doi: 10.1371/journal.pbio.0020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li W, et al. A push-pull mechanism for regulating integrin function. Proc Natl Acad Sci USA. 2005;102(5):1424–1429. doi: 10.1073/pnas.0409334102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu J, et al. Requirement of alpha and beta subunit transmembrane helix separation for integrin outside-in signaling. Blood. 2007;110(7):2475–2483. doi: 10.1182/blood-2007-03-080077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo BH, Carman CV, Takagi J, Springer TA. Disrupting integrin transmembrane domain heterodimerization increases ligand binding affinity, not valency or clustering. Proc Natl Acad Sci USA. 2005;102(10):3679–3684. doi: 10.1073/pnas.0409440102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Partridge AW, Liu S, Kim S, Bowie JU, Ginsberg MH. Transmembrane domain helix packing stabilizes integrin αIIbβ3 in the low affinity state. J Biol Chem. 2005;280(8):7294–7300. doi: 10.1074/jbc.M412701200. [DOI] [PubMed] [Google Scholar]
- 8.Yin H, et al. Activation of platelet αIIbβ3 by an exogenous peptide corresponding to the transmembrane domain of αIIb. J Biol Chem. 2006;281(48):36732–36741. doi: 10.1074/jbc.M605877200. [DOI] [PubMed] [Google Scholar]
- 9.Metcalf DG, Law PB, DeGrado WF. Mutagenesis data in the automated prediction of transmembrane helix dimers. Proteins. 2007;67(2):375–384. doi: 10.1002/prot.21265. [DOI] [PubMed] [Google Scholar]
- 10.Gottschalk KE. A coiled-coil structure of the αIIbβ3 integrin transmembrane and cytoplasmic domains in its resting state. Structure. 2005;13(5):703–712. doi: 10.1016/j.str.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 11.Kim C, Lau TL, Ulmer TS, Ginsberg MH. Interactions of platelet integrin {α}IIb and {β}3 transmembrane domains in mammalian cell membranes and their role in integrin activation. Blood. 2009;113(19):4747–4753. doi: 10.1182/blood-2008-10-186551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu J, et al. The structure of a receptor with two associating transmembrane domains on the cell surface: Integrin αIIbβ3. Mol Cell. 2009;34(2):234–249. doi: 10.1016/j.molcel.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider D, Finger C, Prodohl A, Volkmer T. From interactions of single transmembrane helices to folding of alpha-helical membrane proteins: Analyzing transmembrane helix-helix interactions in bacteria. Curr Protein Pept Sci. 2007;8(1):45–61. doi: 10.2174/138920307779941578. [DOI] [PubMed] [Google Scholar]
- 14.Russ WP, Engelman DM. TOXCAT: A measure of transmembrane helix association in a biological membrane. Proc Natl Acad Sci USA. 1999;96(3):863–868. doi: 10.1073/pnas.96.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurezka R, Langosch D. In vitro selection of membrane-spanning leucine zipper protein-protein interaction motifs using POSSYCCAT. J Biol Chem. 2001;276(49):45580–45587. doi: 10.1074/jbc.M105362200. [DOI] [PubMed] [Google Scholar]
- 16.Karimova G, Ullmann A, Ladant D. A bacterial two-hybrid system that exploits a cAMP signaling cascade in Escherichia coli. Methods Enzymol. 2000;328:59–73. doi: 10.1016/s0076-6879(00)28390-0. [DOI] [PubMed] [Google Scholar]
- 17.Gerber D, Shai Y. In vivo detection of hetero-association of glycophorin-A and its mutants within the membrane. J Biol Chem. 2001;276(33):31229–31232. doi: 10.1074/jbc.M101889200. [DOI] [PubMed] [Google Scholar]
- 18.Schneider D, Engelman DM. GALLEX, a measurement of heterologous association of transmembrane helices in a biological membrane. J Biol Chem. 2003;278(5):3105–3111. doi: 10.1074/jbc.M206287200. [DOI] [PubMed] [Google Scholar]
- 19.Yin H, et al. Computational design of peptides that target transmembrane helices. Science. 2007;315(5820):1817–1822. doi: 10.1126/science.1136782. [DOI] [PubMed] [Google Scholar]
- 20.Li R, et al. Oligomerization of the integrin αIIbβ3: Roles of the transmembrane and cytoplasmic domains. Proc Natl Acad Sci USA. 2001;98(22):12462–12467. doi: 10.1073/pnas.221463098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li R, et al. Dimerization of the transmembrane domain of Integrin αIIb subunit in cell membranes. J Biol Chem . 2004;279(25):26666–26673. doi: 10.1074/jbc.M314168200. [DOI] [PubMed] [Google Scholar]
- 22.Romzek NC, et al. Use of a β1 integrin-deficient human T cell to identify β1 integrin cytoplasmic domain sequences critical for integrin function. Mol Biol Cell. 1998;9(10):2715–2727. [PMC free article] [PubMed] [Google Scholar]
- 23.Mould AP, Humphries MJ. Identification of a novel recognition sequence for the integrin α4β1 in the COOH-terminal heparin-binding domain of fibronectin. Embo J. 1991;10(13):4089–4095. doi: 10.1002/j.1460-2075.1991.tb04985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walters RF, DeGrado WF. Helix-packing motifs in membrane proteins. Proc Natl Acad Sci USA. 2006;103(37):13658–13663. doi: 10.1073/pnas.0605878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore DT, Berger BW, DeGrado WF. Protein-protein interactions in the membrane: Sequence, structural, and biological motifs. Structure. 2008;16(7):991–1001. doi: 10.1016/j.str.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Senes A, Engel DE, DeGrado WF. Folding of helical membrane proteins: The role of polar, GxxxG-like and proline motifs. Curr Opin Struct Biol. 2004;14(4):465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Lemmon MA, Flanagan JM, Treutlein HR, Zhang J, Engelman DM. Sequence specificity in the dimerization of transmembrane alpha-helices. Biochemistry. 1992;31(51):12719–12725. doi: 10.1021/bi00166a002. [DOI] [PubMed] [Google Scholar]
- 28.Hughes PE, et al. Breaking the integrin hinge. A defined structural constraint regulates integrin signaling. J Biol Chem. 1996;271(12):6571–6574. doi: 10.1074/jbc.271.12.6571. [DOI] [PubMed] [Google Scholar]
- 29.Armulik A, Velling T, Johansson S. The integrin β1 subunit transmembrane domain regulates phosphatidylinositol 3-kinase-dependent tyrosine phosphorylation of Crk-associated substrate. Mol Biol Cell. 2004;15(6):2558–2567. doi: 10.1091/mbc.E03-09-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacKenzie KR, Prestegard JH, Engelman DM. A transmembrane helix dimer: Structure and implications. Science. 1997;276(5309):131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 31.Kim S, et al. Transmembrane glycine zippers: Physiological and pathological roles in membrane proteins. Proc Natl Acad Sci USA. 2005;102(40):14278–14283. doi: 10.1073/pnas.0501234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller MW, et al. Small-molecule inhibitors of integrin α2β1 that prevent pathological thrombus formation via an allosteric mechanism. Proc Natl Acad Sci USA. 2009;106(3):719–724. doi: 10.1073/pnas.0811622106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.