Abstract
The presence of an additional 5′ guanosine residue (G-1) is a unique feature of tRNAHis. G-1 is incorporated posttranscriptionally in eukarya via an unusual 3′–5′ nucleotide addition reaction catalyzed by the tRNAHis guanylyltransferase (Thg1). Yeast Thg1 catalyzes an unexpected second activity: Watson–Crick-dependent 3′–5′ nucleotide addition that occurs in the opposite direction to nucleotide addition by all known DNA and RNA polymerases. This discovery led to the hypothesis that there are alternative roles for Thg1 family members that take advantage of this unusual enzymatic activity. Here we show that archaeal homologs of Thg1 catalyze G-1 addition, in vitro and in vivo in yeast, but only in a templated reaction, i.e. with tRNAHis substrates that contain a C73 discriminator nucleotide. Because tRNAHis from archaea contains C73, these findings are consistent with a physiological function for templated nucleotide addition in archaeal tRNAHis maturation. Moreover, unlike yeast Thg1, archaeal Thg1 enzymes also exhibit a preference for template-dependent U-1 addition to A73-containing tRNAHis. Taken together, these results demonstrate that Watson–Crick template-dependent 3′–5′ nucleotide addition is a shared catalytic activity exhibited by Thg1 family members from multiple domains of life, and therefore, that this unusual reaction may constitute an ancestral activity present in the earliest members of the Thg1 enzyme family.
Keywords: archaea, G-1 addition, reverse polymerase, S. cerevisiae
An additional G-residue (G-1) at the 5′ end of tRNAHis is a nearly universal feature, whose loss results in decreased aminoacylation of tRNAHis both in vitro and in vivo, as observed in organisms from E. coli to yeast (1–6). The only organisms known to lack G-1 on tRNAHis are 20 α-proteobacteria (7) in which there is a simultaneous alteration in the histidyl-tRNA synthetase (HisRS) that is suggested to compensate for loss of this universal tRNA feature (8, 9).
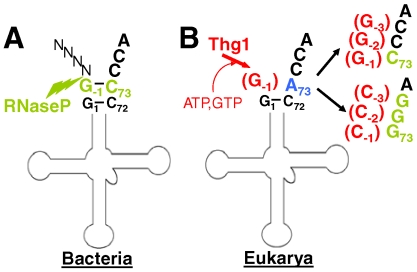
Although the presence of G-1 on tRNAHis is highly conserved, the mechanism by which G-1 is incorporated into tRNA varies in different domains of life. In bacteria, G-1 is genomically encoded and present in the precursor-tRNA transcript (10). The presence of G-1 and a universally conserved C73 discriminator nucleotide results in formation of an unusual 8-base pair acceptor stem in bacterial tRNAHis that directs the tRNA 5′-end maturation enzyme, RNase P, to cleave at an alternate site, leaving G-1 in the mature tRNA (Fig. 1A) (11). In eukarya, the presence of G-1 is the result of a posttranscriptional nucleotide addition reaction, catalyzed in yeast by the essential enzyme tRNAHis guanylyltransferase (Thg1) (Fig. 1B) (12, 13). The widespread conservation of Thg1 throughout eukarya, including humans, suggests a similar mode of G-1 addition.
Fig. 1.
Domain-dependent incorporation of G-1 into tRNAHis. (A) In bacteria, G-1 is genomically encoded and retained by RNase P during 5′-processing. (B) Thg1 adds G-1 to eukaryal tRNAHis, and catalyzes template-dependent 3′–5′ addition to tRNAHis variant substrates. Nucleotides added posttranscriptionally by Thg1 are indicated by parentheses. For clarity, only the altered 3′ end sequences of tRNAHis variant substrates are shown.
Thg1 adds G-1 by an unusual 3′–5′ nucleotide addition reaction, via an unknown molecular mechanism that can not be predicted, due to the lack of sequence similarity between Thg1 and any known enzyme family. In yeast, G-1 is added opposite an A73 discriminator nucleotide that is universally conserved throughout eukarya (Fig. 1B) (14). However, in vitro, wild-type yeast Thg1 exhibits a second biochemical activity with variant tRNAHis substrates that contain alterations at A73. Use of these substrates revealed that wild-type yeast Thg1 adds multiple G or C residues to the 5′ end of the tRNA in a reaction that uses the 3′ acceptor stem nucleotides as a template for 3′–5′ addition (Fig 1B) (15). Although nontemplated (G opposite A) nucleotide addition plays a well-documented physiological role in the maturation of tRNAHis, the biological significance of templated 3′–5′ nucleotide polymerization, in yeast or any other organism, has not been demonstrated. Yet the ability of yeast Thg1 to recognize and add Watson–Crick base-paired nucleotides was completely unexpected for an enzyme that seemingly only functions to add G-1 opposite A73 in tRNAHis. Moreover, the ability of yeast Thg1 to add deoxynucleotides as well as ribonucleotides to tRNA substrates, as well as the observation of templated 3′–5′ addition with other tRNA substrates in vitro suggest that alternative roles for templated addition are a possibility (15).
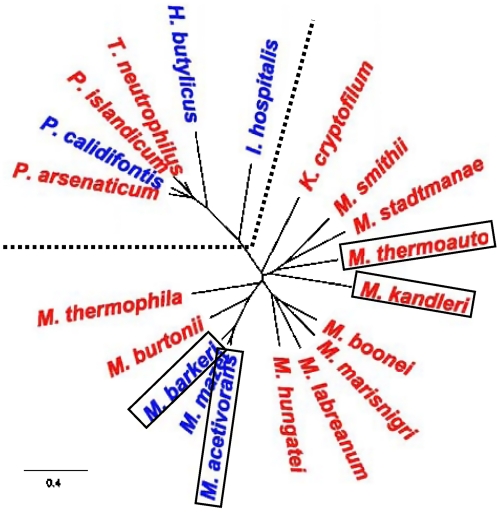
Both bacteria-like and eukarya-like systems for incorporation of G-1 into tRNAHis appear to be present in archaea. Of 55 annotated archaeal tRNAHis genes (16), 40 contain a genomically encoded G-1 residue and a C73 discriminator nucleotide, suggesting that G-1 could be incorporated into tRNAHis using the RNase P-dependent pathway. However, the remaining 15 annotated archaeal tRNAHis genes lack a G residue at the -1 position in their coding sequence. Notably, homologs to yeast Thg1 are present in all of these species with the exception of N. equitans (17), consistent with a requirement for enzymatic G-1 addition in these organisms (Fig. 2). Intriguingly, among the 40 archaea that already contain a genomically encoded G-1 residue, 7 of these also contain a Thg1 homolog (Fig. 2), thus these organisms may use either, or both, pathways for addition of G-1 to tRNAHis.
Fig. 2.
Phylogeny of archaeal Thg1 homologs. An unrooted phylogenetic tree of the archaeal Thg1 homologs was constructed with Geneious Pro software (v.4.7.5) using the neighbor-joining method with a Jukes–Cantor genetic distance model and no outgroup. The tree was built upon a ClustalW alignment (36) of Thg1 sequences from 20 of 21 Thg1-containing archaea (P. aerophilum Thg1 is similar to the other three Pyrobaculum homologs and was omitted for clarity). Organism names are colored according to the presence of G-1 on the annotated tRNAHis; blue, G-1 is genomically encoded; red, G-1 is not present in the genomic sequence. The four archaea investigated in this work are indicated by boxes. The dashed line indicates the separation between crenarchaeal (Upper Left) and euryarchaeal (Lower Right) clades.
The discriminator nucleotide in archaeal tRNAHis is universally a C73, as it is in bacteria (14), raising the possibility that enzymatic G-1 addition in archaea could require a templated 3′–5′ nucleotide addition reaction similar to that previously associated with yeast Thg1 reverse (3′–5′) polymerization activity. Although templated 3′–5′ nucleotide addition activity appears to be required for an unusual form of 5′-tRNA editing in certain protozoan mitochondria (18, 19), the enzyme(s) that catalyze this reaction remain to be identified. Thus, archaeal Thg1 has the potential to be a unique example of a purified enzyme that catalyzes templated 3′–5′ nucleotide addition with a demonstrated biological relevance.
In this study, we investigated the biochemical function of Thg1 homologs from four archaea. The results show that both yeast and archaeal Thg1 enzymes catalyze templated 3′–5′ nucleotide addition, therefore this property appears to be a shared feature of Thg1 enzymes from multiple domains of life. In contrast, the nontemplated reaction catalyzed by eukaryal Thg1 during maturation of tRNAHis may be a derived evolutionary trait unique to eukarya.
Results
Archaeal Thg1 Homologs Do Not Efficiently Add G-1 to A73-Containing tRNAHis.
To test the activities of archaeal Thg1 enzymes, we cloned, expressed, and purified homologs from M. thermoautotrophicus (MtThg1), M. kandleri (MkThg1), M. acetivorans (MaThg1), and M. barkeri (MbThg1) (see SI Text). These four archaea include two that are predicted to require enzymatic addition of G-1 to tRNAHis (Mt and Mk) and two in which G-1 is already encoded in the genome (Ma and Mb) (Fig. 2). The MaThg1 coding sequence is interrupted by an in-frame amber stop codon, and appears to be a previously undocumented case of nonsense suppression by insertion of pyrrolysine at this position, as is seen with other proteins from methanobacteria, although the presence of pyrrolysine is not required for activity (see SI Text) (20).
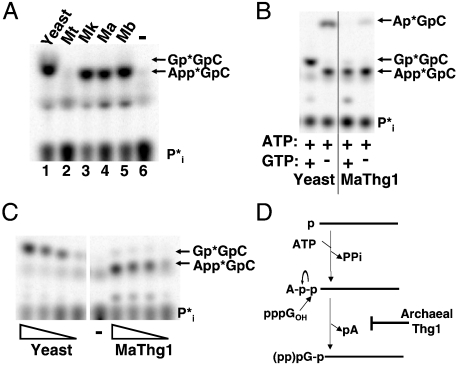
We first tested the ability of each purified archaeal Thg1 to add G-1 to yeast tRNAHis that contains the universally conserved eukaryal A73 discriminator nucleotide. G-1 addition activity was tested using a previously described assay with 5′-32P labeled monophosphorylated tRNAHis (p-tRNAHis) (21). Briefly, following incubation of purified Thg1 with the labeled tRNA, the reactions are treated with RNase A, followed by phosphatase, finally yielding inorganic phosphate ( ) from unreacted substrate, and a labeled G-1p∗GpC trimeric oligonucleotide if the G-1 addition product is formed during the reaction (Fig. 3A). No products were observed in reactions with MtThg1 and this A73-containing tRNA, whereas reaction products were observed with the other three archaeal enzymes (MkThg1, MaThg1, and MbThg1) (Fig. 3A; compare lane 2 with lanes 3–5). However, the primary reaction product of these enzymes’ activity migrates slightly lower in the TLC solvent system used for this assay than the G-1-containing product produced by yeast Thg1. This slower migrating product corresponds to the adenylylated-tRNAHis reaction intermediate (App∗G+1pC) produced by yeast Thg1 (13). The major reaction product observed with all of the archaeal enzymes (a representative analysis with MaThg1 is shown) is only dependent on addition of ATP (Fig. 3B), and migrates identically to the adenylylated tRNAHis species produced by yeast Thg1, as demonstrated by nuclease analysis of isolated reaction products (13).
) from unreacted substrate, and a labeled G-1p∗GpC trimeric oligonucleotide if the G-1 addition product is formed during the reaction (Fig. 3A). No products were observed in reactions with MtThg1 and this A73-containing tRNA, whereas reaction products were observed with the other three archaeal enzymes (MkThg1, MaThg1, and MbThg1) (Fig. 3A; compare lane 2 with lanes 3–5). However, the primary reaction product of these enzymes’ activity migrates slightly lower in the TLC solvent system used for this assay than the G-1-containing product produced by yeast Thg1. This slower migrating product corresponds to the adenylylated-tRNAHis reaction intermediate (App∗G+1pC) produced by yeast Thg1 (13). The major reaction product observed with all of the archaeal enzymes (a representative analysis with MaThg1 is shown) is only dependent on addition of ATP (Fig. 3B), and migrates identically to the adenylylated tRNAHis species produced by yeast Thg1, as demonstrated by nuclease analysis of isolated reaction products (13).
Fig. 3.
Archaeal Thg1 homologs do not efficiently add G-1 to wild-type yeast tRNAHis. (A) Assays using 5′-32P-labeled tRNAHis and 1 µL each Thg1, as indicated, were performed with 0.1 mM ATP and 1.0 mM GTP as described in Methods. Lane-, buffer only. Reaction products are labeled to the right of each panel, and indicated by arrows. (B) 5′-32P-tRNAHisassay performed with 1 µL yeast or MaThg1. The presence or absence of ATP or GTP (1 mM) is denoted by (+) and (-). (C) Assays of 5′-32P-tRNAHis with titrations (5-fold serial dilutions, starting with approximately ~50 M enzyme) of purified yeast or MaThg1; Lane-, buffer only control reaction. (D) Scheme of steps for G-1 addition to tRNAHis showing first, adenylylation of p-tRNAHis, followed by nucleotidyltransfer to yield G-1-containing tRNA. The apparent block in the 2nd step of the reaction observed with archaeal Thg1 is as indicated.
Adenylylated-tRNAHis is the terminal product of archaeal Thg1 activity with A73-tRNAHis. Measurements of total product formation as a function of decreasing Thg1 concentration reveal similar amounts of App-tRNAHis and G-1-tRNAHis formed at the same low concentrations (0.4 µM) of MaThg1 or yeast Thg1, respectively (Fig. 3C), indicating that the overall activity of MaThg1 is not significantly lower than the yeast enzyme. Instead, the persistent accumulation of the App-tRNAHis intermediate even at the highest concentrations tested (46 µM) suggests that archaeal Thg1 enzymes can not catalyze the second, guanylyltransfer step of the three-step Thg1 reaction with A73-tRNAHis (Fig. 3D) (13).
Archaeal Thg1homologs Efficiently Add G-1 to C73-Containing tRNAHis In Vitro.
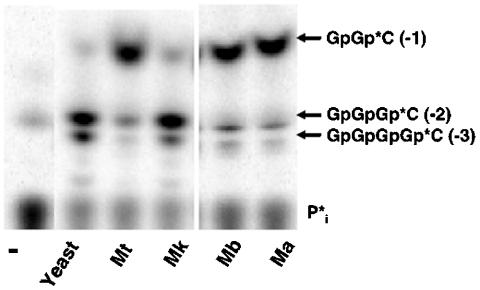
We next tested the ability of archaeal Thg1 to add G-1 to a yeast tRNAHis variant containing C73 instead of A73; this tRNA is a substrate for template-dependent 3′–5′ polymerization by yeast Thg1 in vitro (Fig. 1B). In contrast to the inefficient addition of G-1 to A73-containing tRNA described above, all four archaeal Thg1 enzymes catalyzed efficient addition of G-1 to this C73-containing tRNA substrate (Fig. 4). Whereas yeast and MkThg1 catalyze the addition of multiple G-residues (G-1, G-2, and G-3), the single G-1 addition product is the predominant product observed with Ma, Mb, and MtThg1, and G-2/G-3 are added inefficiently, if at all (Fig. 4). No accumulation of adenylylated tRNA intermediate was observed with any of the archaeal Thg1 enzymes and C73-tRNAHis.
Fig. 4.
Archaeal Thg1 homologs efficiently catalyze G-addition opposite C73 with a tRNAHis variant substrate. Assay of purified yeast and archaeal Thg1 homologs (1 µL each) with 5′-32P labeled C73-tRNAHis variant. Lane-, buffer only. Positions of G-1, G-2, and G-3 reaction products are indicated to the right of the figure.
To quantify the relative catalytic abilities of archaeal Thg1 enzymes, we measured the steady-state kinetic parameters for addition of G-1 to C73-containing tRNAHis catalyzed by MaThg1 and MtThg1, as representative enzymes from archaea that contain or lack, respectively, a genomically encoded G-1 residue. For comparison, kinetic parameters for the same reaction with yeast Thg1 were also determined. Steady-state kinetic assays were performed with triphosphorylated (ppp - tRNAHis) substrates as previously described (21).
With yeast Thg1, kcat/KM for C73-tRNAHis is essentially the same as for wild-type (A73) tRNAHis (Table 1, SI Text). Despite the relatively slow kcat exhibited by MaThg1 for C73-tRNAHis, the significantly lower KM results in only a 10-fold decreased kcat/KM for MaThg1 compared to the yeast enzyme (Table 1, SI Text). Initial determination of MtThg1 kinetic parameters similarly performed at 25 °C indicated very low activity, with a significantly decreased kcat (SI Text). However, because M. thermoautotrophicum is a thermophile that grows optimally at 65 °C (22), MtThg1 activity was measured at 55 °C, where the enzyme exhibits maximal activity in the in vitro assay (SI Text). At this temperature, initial rates for MtThg1 activity at 2 µM and 4 µM tRNA were essentially unchanged, consistent with the KM measured at 25 °C (SI Text). Thus, the kcat/KM calculated for MtThg1 under optimal temperature conditions is within 20-fold of that exhibited for yeast Thg1 (Table 1).
Table 1.
Steady-state kinetic parameters for G-1 addition to pppC73-tRNAHis.
| Substrate |
Enzyme |
kcat (hr-1) |
KM (µM) |
kcat/KM (M-1 s-1) |
%kcat/KM (relative to yeast Thg1) |
| A73-tRNAHis | yeast Thg1 | 8.4*± 0.9 | 0.42*± 0.13 | 5500*± 1200 | — |
| C73-tRNAHis | yeast Thg1 | 20.4 ± 2.4 | 0.99 ± 0.29 | 5670 ± 1200 | 100 |
| C73-tRNAHis | MaThg1 | 0.30 ± 0.03 | 0.12 ± 0.05 | 700 ± 230 | 12 |
| C73-tRNAHis | MtThg1 | 1.1† | 1.1 ±0.85‡ | 300 | 5 |
Archaeal Thg1 Homologs Complement the Essential Function of Yeast Thg1 in Yeast Cells When Provided with an Appropriate tRNA Substrate for Templated 3′–5′ Addition.
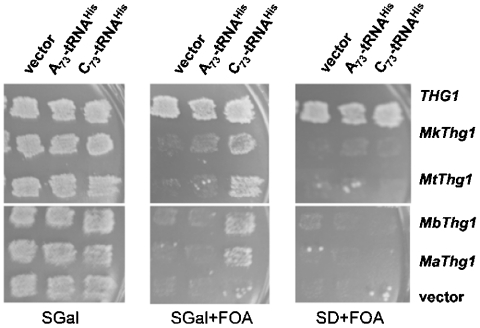
To test in vivo function of the archaeal enzymes, we performed a variation of a previously described plasmid shuffle assay (13, 23). Archaeal Thg1 homologs (CEN LEU2) were introduced into the yeast thg1Δ strain JJY20 [relevant genotype: thg1Δ leu2Δ ura3Δ (CEN URA3 PTHG1-THG1)], and subsequently grown on 5-fluoroorotic acid (FOA)-containing media that selects against the yeast THG1 URA3 covering plasmid, resulting in a strain in which the archaeal Thg1 enzymes are the only potential source of Thg1 activity in the cells. Consistent with the lack of G-1 addition activity exhibited by archaeal Thg1 in vitro with wild-type yeast tRNAHis (Fig. 5), each archaeal Thg1-expressing strain was unable to grow on FOA (Fig. 5, compare growth of strains spotted in the first column for each of the three media shown), indicating that these archaeal Thg1 enzymes are not able to replace the essential function of yeast Thg1 in vivo in yeast.
Fig. 5.
Archaeal Thg1 homologs support growth of the yeast thg1Δ strain in the presence of C73-tRNAHis. Replica plating results with yeast thg1Δ strains transformed with LEU2 plasmids containing various Thg1 homologs (indicated to the right of the figure) and HIS3 plasmids containing various tRNAHis genes (indicated on top of each panel). Growth media are indicated below each panel; images were taken after 3–4 days of growth at 30 °C.
We then tested whether the ability of the archaeal homologs to add G-1 opposite C73 could be exploited to support growth of the yeast deletion mutant. To the thg1Δ strains created above, we simultaneously introduced a third plasmid, containing either an additional copy of wild-type (A73-containing) or variant (C73-containing) tRNAHis, and again tested the ability of the resulting strains to grow on media containing FOA (Fig. 5, second panel). In each strain containing archaeal Thg1, significant growth was observed only in strains that also expressed C73-tRNAHis, and not in those expressing wild-type tRNAHis or a vector control. The ability to complement the thg1Δ growth phenotype was dependent on expression of archaeal Thg1, because the complementation was only observed on galactose-containing media (inducing conditions for expression from the PGAL thg1 promoter) and not in dextrose-containing media (repressing conditions) (Fig. 5, compare second and third panels). Because the presence of G-1 is required for histidylation and thus growth in yeast (4), these results indicate that archaeal Thg1 homologs add G-1 to C73-containing tRNAHis in vivo. The complementation observed in the presence of C73-tRNAHis also demonstrates that the lack of growth observed in strains containing only wild-type (A73) tRNAHis is not due to limiting expression of the archaeal enzymes in yeast.
Archaeal Thg1 Homologs Catalyze Template-Dependent 3′–5′ Addition with Wild-Type tRNAHis.
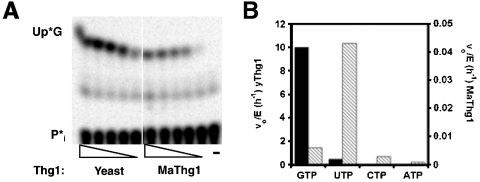
In light of the strong preference for addition of G-1 opposite C73 exhibited by the archaeal Thg1 enzymes, we revisited the lack of G-1 addition previously observed with the wild-type (A73-containing) tRNAHis substrate. We tested the ability of archaeal Thg1 homologs to add U-1 to wild-type tRNAHis, to test the hypothesis that these enzymes might require the ability to form a Watson–Crick base pair for nucleotide addition to occur. Unlike G-1 addition to A73-yeast tRNAHis that is catalyzed by yeast Thg1 but not the archaeal homologs, the addition of U-1 opposite A73 is observed with both yeast and archaeal enzymes (Fig. 6A, representative assay for MaThg1 activity is shown). Therefore, the inability of the archaeal enzymes to add G-1 to wild-type tRNAHis does not reflect an inherent inability to recognize and use this tRNA substrate, but instead reflects an apparent preference of archaeal Thg1 for template-dependent 3′–5′ addition.
Fig. 6.
Archaeal Thg1 catalyzes template-dependent U-1 addition activity with wild-type tRNAHis. (A) U-1 addition assay with 5′-32P labeled tRNAHis and titrations (5-fold serial dilutions) of yeast or MaThg1. Lane-, buffer only. Identity of observed reaction products indicated to the left of the panel. (B) Initial rates of N-1 addition to wild-type tRNAHis (2 µM) with yeast Thg1 (0.2 µM) (solid bars) or MaThg1 (0.15 µM) (hatched bars), and 1 mM of each NTP, as indicated. Note separate scale on Y-axis for each enzyme's activity.
To further probe the template-dependent nature of the reaction catalyzed by MaThg1 with yeast tRNAHis, steady-state initial rates of 3′–5′ nucleotide addition were measured using each of the four possible NTPs and ppp-tRNAHis with yeast and MaThg1. For yeast Thg1, it was previously demonstrated that any nucleotide can be added to the 5′ end of wild-type tRNAHis after prolonged incubation at high concentrations of Thg1 (15), although, consistent with the physiological role of Thg1 in yeast, the rate of G-1 addition under steady-state conditions is significantly faster than the rate of addition of any other N-1 residue (Fig. 6B). In contrast, for MaThg1 the NTP preference was essentially reversed, with UTP added most efficiently to the A73-containing tRNAHis substrate, consistent with a requirement for formation of a Watson–Crick base pair for catalysis of 3′–5′ nucleotide addition (Fig. 6B). Thus, template-dependent 3′–5′ addition activity of archaeal Thg1 is not limited to formation of G-C base pairs. The rate of addition of U-1(vo/[E] = 0.04 hr-1) is nearly 10-fold slower than the kcat for G-1 addition to C73-tRNAHis by MaThg1 (Table 1), suggesting an additional effect of the identity of the base pair being formed on templated activity with tRNAHis substrates. The slower rate of MaThg1-catalyzed U-A vs. G-C addition likely contributes to the lack of complementation of the yeast thg1Δ phenotype observed by archaeal Thg1 homologs in the presence of A73-tRNAHis (Fig. 5).
Discussion
In this study, we demonstrated that archaeal homologs of yeast Thg1 catalyze solely template-dependent 3′–5′ nucleotide addition to tRNAHis substrates, and do not catalyze the nontemplated G opposite A addition reaction that is the hallmark reaction of eukaryal Thg1 enzymes. Purified Thg1 homologs from four different archaea do not efficiently add G-1 to wild-type yeast tRNAHis (Fig. 3), but instead add G-1 to a tRNAHis variant that contains C73 and is thus a substrate for template-dependent G-1 addition (Fig. 4). Moreover, archaeal Thg1 homologs substitute for the essential G-1 addition function of yeast Thg1 in vivo, as long as they are provided with the C73-containing tRNAHis substrate that reflects the preference of these enzymes to catalyze templated 3′–5′ nucleotide addition (Fig. 5). 3′–5′ addition catalyzed by archaeal Thg1 is not restricted to forming G-C base pairs, but also forms U-A base pairs, as evidenced by the preference for MaThg1 to efficiently add U-1 over other non-Watson–Crick pairing N-1 residues to wild-type (A73-containing) tRNAHis, thus distinguishing these enzymes from yeast Thg1 that prefers to add the nontemplated G-1 to A73-tRNAHis (Fig. 6). However, because yeast Thg1 is also capable of a previously described template-dependent 3′–5′ addition activity (15), these results demonstrate that the ability to catalyze Watson–Crick base pair-dependent nucleotide addition is a universal property of Thg1 family members from multiple domains of life. In contrast, the ability to add G-1 opposite A73 with eukaryal tRNAHis is so far a unique feature of eukaryal Thg1 enzymes.
Biological Function for Template-Dependent Thg1 Addition Activity.
Although template-dependent 3′–5′ nucleotide addition was originally discovered as a property of yeast Thg1, the physiological relevance of this activity in yeast remains unknown (15). Nonetheless, unexplained interactions of yeast Thg1 with the Orc2 component of the origin recognition complex (24) and cell-cycle progression defects associated with decreased Thg1 expression in yeast and human cells (24, 25) suggest that the role of the enzyme in eukarya may be more complicated than is currently understood. However, this work provides an opportunity to explore possible physiological roles for template-directed nucleotide addition activity in other organisms. Certain archaea (represented here by M. thermoautotrophicus and M. kandleri) lack a genomically encoded G-residue at the -1 position of the tRNAHis gene (Fig. 2), and thus can not incorporate G-1 via an RNase P-dependent pathway like the one used in bacteria. Because the discriminator nucleotide in archaeal tRNAHis is universally a C73, our results are consistent with a role for archaeal Thg1 in this G-1 addition reaction, thus this work provides unique evidence for a likely physiological role for Thg1-catalyzed template-dependent 3′–5′ nucleotide addition in any organism.
We note that, whereas the measured kcat values for archaeal Thg1 activity are quite low (0.3–1.1 hr-1) (Table 1), the ability of archaeal enzymes to support growth of the yeast thg1Δ strain in the presence of C73-tRNAHis (Fig. 5) demonstrates that these enzymes catalyze G-1 addition on a biologically relevant timescale. Because the tRNA substrates used here are derived from yeast tRNAHis sequences, it is possible that the in vitro activities of the archaeal enzymes could be improved using tRNAHis substrates derived from archaea. However, because the kcat/KM values measured for yeast Thg1 activity are similarly low (Table 1), it is possible that the measured in vitro activities of both yeast and archaeal enzymes do not accurately reflect the in vivo rates. Such a discrepancy could be caused by the use of nonoptimal in vitro reaction conditions, lack of important tRNA modifications on the in vitro transcribed substrates, or lack of additional components that stimulate Thg1 activity in vivo. While this remains a possibility, investigations of alternative assay conditions have not yet yielded increased rates with the yeast enzyme, and Thg1 purified from E. coli in the absence of any other yeast proteins is catalytically active and not stimulated by inclusion of yeast crude extracts in the assay.
A role for archaeal Thg1 homologs in G-1 addition to C73-containing tRNAHis in archaea accounts for the presence of the enzyme in the majority of archaea in which it has been identified, including M. thermoautotrophicus and M. kandleri. However, a second group of archaea, including M. barkeri and M. acetivorans, is particularly intriguing in that, whereas we have shown here that enzymes from these species catalyze G-1 addition in vitro and in vivo, these organisms also contain a genomically encoded G-1 residue in tRNAHis (Fig. 2). This raises the possibility that, in some archaea, G-1 addition could occur by either or both of the two known G-1 incorporation pathways. It is also possible that the presence of Thg1 in these organisms is required for another activity that is independent of a role for the enzyme in maturation of tRNAHis.
Alternative Biological Roles for Template-Dependent 3′–5′ Nucleotide Addition.
Possible biological roles for templated nucleotide addition activity in the 3′–5′ direction, opposite to that of all known DNA and RNA polymerases, are not limited to tRNAHis maturation. For example, templated 3′–5′ addition is required for a tRNA editing activity that functions in the mitochondria of certain lower eukarya, where it is used to repair up to three mismatches found in many of the mitochondrial tRNA genes (18, 19). The editing reaction requires stepwise 3′–5′ addition, using the nucleotides of the 3′ half of the aminoacyl-acceptor stem as a template (19, 26–28). Similarities between the enzymatic activities required for G-1 addition, now known to be catalyzed by Thg1, and 5′-tRNA editing, for which the identity of the enzyme(s) involved remains a mystery, were noted at the time of the discovery of the editing activity (18, 19). It is tempting to speculate that Thg1 enzymes from archaea that do not require enzymatic addition of G-1 to tRNAHis may participate in a similar tRNA editing or repair activity in their respective organisms, which would necessarily take advantage of their ability to catalyze template-dependent 3′–5′ addition. Although 5′-editing or repair of tRNA has not been identified in archaea, the existence of such activities may mirror tRNA 3′-end surveillance mechanisms that are found in multiple domains of life (29–31). The example of the ubiquitous 3′-CCA addition enzyme is particularly intriguing in this respect, because it catalyzes the essential de novo addition of the 3′-CCA end to the majority of eukaryal tRNAs, but in bacteria and archaea, predominantly functions in 3′-end repair pathways, because the 3′-CCA sequence is normally encoded in the genome (32, 33).
Implications for Thg1 3′–5′ Nucleotide Addition Mechanism.
As the only known enzyme family that catalyzes nucleotide addition in the opposite direction to all known DNA and RNA polymerases, the Thg1 molecular mechanism is of great interest, but is currently unknown. The results presented here add an additional dimension to this issue, raising the question of how enzymes from archaea and yeast exhibit distinct biochemical properties despite sharing a significant degree of overall sequence similarity (even the most distantly related archaeal enzymes share approximately 30% identity with yeast Thg1), including many residues that play critical roles in yeast Thg1 activity (23). A second issue to be explained is the apparently inefficient addition of multiple G nucleotides to C73-tRNAHis observed with most archaeal Thg1 enzymes. Although consistent with the presumed biological need for only a single G-1 residue at the 5′ end of tRNAHis, this suggests the existence of additional mechanism(s) used by some archaeal Thg1 enzymes to limit the addition of multiple nucleotides to the 5′ end of tRNAHis. It remains to be seen whether multiple nucleotide additions analogous to yeast reverse polymerase activity are catalyzed by these archaeal Thg1 homologs with other substrates.
Implications for Evolution of 3′–5′ Addition Activities.
Despite the fact that eukaryal G-1 addition required for tRNAHis maturation was the first Thg1 activity to be described, these results indicate that this is not likely the ancestral activity of Thg1 family members. Instead, template-dependent nucleotide addition is catalyzed by diverse Thg1 family members from multiple domains of life, and therefore these results suggest a scenario in which template-dependent 3′–5′ addition was a property of the earliest Thg1 family members. Further investigation of Thg1 homologs from other organisms, including > 20 bacteria in which Thg1 homologs can now be found, will be important for understanding the evolution of 3′–5′ nucleotide addition activities. Interestingly, Thg1-containing bacteria all contain a genomically encoded G-1 residue, suggesting that they are similar to the 7 archaea that do not require enzymatic G-1 addition to tRNAHis (Fig. 2), and therefore the function of bacterial Thg1 may be the same as in these archaea. Particularly, it will be interesting to determine the evolutionary basis for the apparent acquisition of the ability to catalyze nontemplated G-1 addition opposite A73 by eukaryal Thg1 family members.
Methods
Expression and Purification of Thg1 Homologs.
Cloned archaeal Thg1 enzymes were expressed in and affinity-purified from E. coli, essentially as previously described for yeast Thg1. For detailed descriptions of cloning, expression and purification protocols, see SI Text.
3′–5′ Nucleotide Addition Assays with p-tRNA Substrates.
Assays contained approximately 1 nM 5′-32P-tRNA (specific activity 6,000 Ci/mmol, prepared as described in SI Text) in 25 mM Hepes, 10 mM MgCl2, 3 mM DTT, 125 mM NaCl, 0.2 mg/mL BSA, pH 7.5 buffer, 0.1 mM ATP and 1 mM GTP, unless otherwise indicated, according to the previously published protocol (21). Reactions were initiated by addition of either 1 µl enzyme (final concentration 0.1–1 μL depending on concentration of the purified stock), or 5-fold serial dilutions of enzyme starting with the same purified stock, as indicated, and were performed for 2 h at 25 °C.
U-1 addition activity was tested using a variation of the previously described nuclease protection assay with 5′-32P-labeled tRNAHis. Assays were performed identically to those described above, except that reactions were treated with RNase T1 (Ambion) in place of RNase A, to release a labeled U-1p ∗ G+1 dimer product if U-1 is added, but still yield labeled phosphate if no nucleotide has been added. The identity of the Up*G reaction product was confirmed by further subjecting the reaction products to RNase T2 digestion that released Up* after reaction of the tRNA with Thg1.
Steady-State Kinetic Parameters for Thg1 Activity.
Triphosphorylated 32PppC73-tRNAHis (SI Text) was assayed as described previously (21) in Thg1 assay buffer, in the presence of 1 mM GTP at room temperature, unless otherwise indicated. Steady-state reactions contained 0.05–8 µM C73-tRNAHis and 1–500 nM of each purified Thg1 such that at least 5-fold excess tRNA was maintained in each reaction. Time courses measured linear initial rates (< 10%) of the release of labeled 32PPi due to G-1 addition to the 32Ppp-tRNA for each [tRNA]. Steady-state kinetic parameters kcat, KM, and kcat/KM for each enzyme were determined by a fit of the initial velocities obtained as a function of [C73-tRNAHis] to the Michaelis–Menten equation using KaleidaGraph (Synergy). Initial rates of N-1 addition to A73-tRNAHis (Fig. 6) were measured in the same way, with ppp-tRNAHis substrate.
Analysis of In Vivo Thg1 Function of Archaeal Thg1 Homologs.
Plasmids for galactose-inducible expression of archaeal Thg1 [CEN LEU2 PGAL - thg1∗] (SI Text), along with analogous yeast THG1-containing and empty vector control plasmids, were transformed into yeast strain JJY20 [relevant genotype MATα thg1Δ::kanMX his3–1 leu2Δ met15Δ ura3Δ (CEN URA3 PTHG1 - THGI)] (34). Positive (Leu+) transformants were selected at 30 °C on synthetic dextrose (SD) media (35), and tested by replica plating at 30 °C to synthetic galactose (SGal) media containing FOA, to induce expression of the various Thg1 homologs and select against the wild-type yeast THG1 URA3 plasmid. To test the tRNA requirement for Thg1 function, CEN HIS3 plasmids containing either wild-type (A73)-tRNAHis, C73-tRNAHis, or no tRNA were transformed into each of the Leu+ strains using the same method. Positive transformants were selected on SD media lacking leucine and histidine, and further subjected to replica plating as described above.
Supplementary Material
Acknowledgments.
We thank Juan Alfonzo, Jonatha Gott, Mike Gray, and Anita Hopper for valuable discussions and comments during preparation of the manuscript. We also thank Melanie Baker and Eric Phizicky for providing tRNAHis yeast expression plasmids.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910961107/DCSupplemental.
References
- 1.Nameki N, Asahara H, Shimizu M, Okada N, Himeno H. Identity elements of Saccharomyces cerevisiae tRNA(His) Nucleic Acids Res. 1995;23(3):389–394. doi: 10.1093/nar/23.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosen AE, Musier-Forsyth K. Recognition of G-1:C73 atomic groups by Escherichia coli histidyl-tRNA synthetase. J Am Chem Soc. 2004;126(1):64–65. doi: 10.1021/ja0381609. [DOI] [PubMed] [Google Scholar]
- 3.Rudinger J, Florentz C, Giege R. Histidylation by yeast HisRS of tRNA or tRNA-like structure relies on residues -1 and 73 but is dependent on the RNA context. Nucleic Acids Res. 1994;22(23):5031–5037. doi: 10.1093/nar/22.23.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu W, Hurto RL, Hopper AK, Grayhack EJ, Phizicky EM. Depletion of Saccharomyces cerevisiae tRNA(His) guanylyltransferase Thg1p leads to uncharged tRNAHis with additional m(5)C. Mol Cell Biol. 2005;25(18):8191–8201. doi: 10.1128/MCB.25.18.8191-8201.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan W, Francklyn C. Cytosine 73 is a discriminator nucleotide in vivo for histidyl-tRNA in Escherichia coli. J Biol Chem. 1994;269(13):10022–10027. [PubMed] [Google Scholar]
- 6.Himeno H, et al. Role of the extra G-C pair at the end of the acceptor stem of tRNA(His) in aminoacylation. Nucleic Acids Res. 1989;17(19):7855–7863. doi: 10.1093/nar/17.19.7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang C, Sobral BW, Williams KP. Loss of a universal tRNA feature. J Bacteriol. 2007;189(5):1954–1962. doi: 10.1128/JB.01203-06. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connolly SA, Rosen AE, Musier-Forsyth K, Francklyn CS. G-1:C73 recognition by an arginine cluster in the active site of Escherichia coli histidyl-tRNA synthetase. Biochemistry. 2004;43(4):962–969. doi: 10.1021/bi035708f. [DOI] [PubMed] [Google Scholar]
- 9.Ardell DH, Andersson SG. TFAM detects co-evolution of tRNA identity rules with lateral transfer of histidyl-tRNA synthetase. Nucleic Acids Res. 2006;34(3):893–904. doi: 10.1093/nar/gkj449. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orellana O, Cooley L, Soll D. The additional guanylate at the 5′ terminus of Escherichia coli tRNAHis is the result of unusual processing by RNase P. Mol Cell Biol. 1986;6(2):525–529. doi: 10.1128/mcb.6.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirsebom LA, Svard SG. The kinetics and specificity of cleavage by RNase P is mainly dependent on the structure of the amino acid acceptor stem. Nucleic Acids Res. 1992;20(3):425–432. doi: 10.1093/nar/20.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooley L, Appel B, Soll D. Post-transcriptional nucleotide addition is responsible for the formation of the 5′ terminus of histidine tRNA. Proc Natl Acad Sci USA. 1982;79(21):6475–6479. doi: 10.1073/pnas.79.21.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu W, Jackman JE, Lohan AJ, Gray MW, Phizicky EM. tRNAHis maturation: An essential yeast protein catalyzes addition of a guanine nucleotide to the 5′ end of tRNAHis. Genes Dev. 2003;17(23):2889–2901. doi: 10.1101/gad.1148603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26(1):148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackman JE, Phizicky EM. tRNAHis guanylyltransferase catalyzes a 3′–5′ polymerization reaction that is distinct from G-1 addition. Proc Natl Acad Sci USA. 2006;103(23):8640–8645. doi: 10.1073/pnas.0603068103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider KL, Pollard KS, Baertsch R, Pohl A, Lowe TM. The UCSC Archaeal Genome Browser. Nucleic Acids Res. 2006;34((Database issue)):D407–410. doi: 10.1093/nar/gkj134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lonergan KM, Gray MW. Editing of transfer RNAs in Acanthamoeba castellanii mitochondria. Science. 1993;259(5096):812–816. doi: 10.1126/science.8430334. [DOI] [PubMed] [Google Scholar]
- 19.Lonergan KM, Gray MW. Predicted editing of additional transfer RNAs in Acanthamoeba castellanii mitochondria. Nucleic Acids Res. 1993;21(18):4402. doi: 10.1093/nar/21.18.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Longstaff DG, Blight SK, Zhang L, Green-Church KB, Krzycki JA. In vivo contextual requirements for UAG translation as pyrrolysine. Mol Microbiol. 2007;63(1):229–241. doi: 10.1111/j.1365-2958.2006.05500.x. [DOI] [PubMed] [Google Scholar]
- 21.Jackman JE, Phizicky EM. tRNAHis guanylyltransferase adds G-1 to the 5′ end of tRNAHis by recognition of the anticodon, one of several features unexpectedly shared with tRNA synthetases. RNA. 2006;12(6):1007–1014. doi: 10.1261/rna.54706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith DR, et al. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: Functional analysis and comparative genomics. J Bacteriol. 1997;179(22):7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackman JE, Phizicky EM. Identification of critical residues for G-1 addition and substrate recognition by tRNA(His) guanylyltransferase. Biochemistry. 2008;47(16):4817–4825. doi: 10.1021/bi702517q. [DOI] [PubMed] [Google Scholar]
- 24.Rice TS, Ding M, Pederson DS, Heintz NH. The highly conserved tRNAHis guanylyltransferase Thg1p interacts with the origin recognition complex and is required for the G2/M phase transition in the yeast Saccharomyces cerevisiae. Eukaryotic Cell. 2005;4(4):832–835. doi: 10.1128/EC.4.4.832-835.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo D, et al. Identification and characterization of a novel cytoplasm protein ICF45 that is involved in cell cycle regulation. J Biol Chem. 2004;279(51):53498–53505. doi: 10.1074/jbc.M406737200. [DOI] [PubMed] [Google Scholar]
- 26.Bullerwell C, Gray M. In vitro characterization of a tRNA editing activity in the mitochondria of Spizellomyces punctatus, a Chytridiomycete fungus. J Biol Chem. 2004;280(4):2463–2470. doi: 10.1074/jbc.M411273200. [DOI] [PubMed] [Google Scholar]
- 27.Price DH, Gray MW. A novel nucleotide incorporation activity implicated in the editing of mitochondrial transfer RNAs in Acanthamoeba castellanii. RNA. 1999;5(2):302–317. doi: 10.1017/s1355838299981840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laforest MJ, Bullerwell CE, Forget L, Lang BF. Origin, evolution, and mechanism of 5′ tRNA editing in chytridiomycete fungi. RNA. 2004;10(8):1191–1199. doi: 10.1261/rna.7330504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levinger L, Morl M, Florentz C. Mitochondrial tRNA 3′ end metabolism and human disease. Nucleic Acids Res. 2004;32(18):5430–5441. doi: 10.1093/nar/gkh884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reichert AS, Morl M. Repair of tRNAs in metazoan mitochondria. Nucleic Acids Res. 2000;28(10):2043–2048. doi: 10.1093/nar/28.10.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schurer H, Schiffer S, Marchfelder A, Morl M. This is the end: Processing, editing, and repair at the tRNA 3′-terminus. Biol Chem. 2001;382(8):1147–1156. doi: 10.1515/BC.2001.144. [DOI] [PubMed] [Google Scholar]
- 32.Li F, et al. Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and its complexes with ATP or CTP. Cell. 2002;111(6):815–824. doi: 10.1016/s0092-8674(02)01115-7. [DOI] [PubMed] [Google Scholar]
- 33.Zhu L, Deutscher MP. tRna nucleotidyltransferase is not essential for Escherichia coli viability. EMBO J. 1987;6(8):2473–2477. doi: 10.1002/j.1460-2075.1987.tb02528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 35.Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- 36.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.