Abstract
Ubiquitously expressed seven-transmembrane receptors (7TMRs) classically signal through heterotrimeric G proteins and are commonly referred to as G protein-coupled receptors. It is now recognized that 7TMRs also signal through β-arrestins, which act as versatile adapters controlling receptor signaling, desensitization, and trafficking. Most endogenous receptors appear to signal in a balanced fashion using both β-arrestin and G protein-mediated pathways. Some 7TMRs are thought to be nonsignaling “decoys” because of their inability to activate typical G protein signaling pathways; it has been proposed that these receptors act to scavenge ligands or function as coreceptors. Here we demonstrate that ligand binding to the decoy receptor CXCR7 does not result in activation of signaling pathways typical of G proteins but does activate MAP kinases through β-arrestins in transiently transfected cells. Furthermore, we observe that vascular smooth muscle cells that endogenously express CXCR7 migrate to its ligand interferon-inducible T-cell alpha chemoattractant (ITAC), an effect that is significantly attenuated by treatment with either a CXCR7 antagonist or β-arrestin depletion by siRNA. This example of an endogenous “β-arrestin-biased” 7TMR that signals through β-arrestin in the absence of G protein activation demonstrates that some 7TMRs encoded in the genome have evolved to signal through β-arrestin exclusively and suggests that other receptors that are currently thought to be orphans or decoys may also signal through such nonclassical pathways.
Keywords: biased receptor, chemokine, G protein-coupled receptors, seven-transmembrane receptor
Seven-transmembrane receptors (7TMRs) are the most common class of receptors in the genome and the most common target for medical therapeutics (1). Classically, these receptors were thought to signal exclusively through heterotrimeric G proteins and be desensitized by β-arrestins; therefore, they are frequently referred to as G protein-coupled receptors. It is now appreciated that β-arrestins act as multifunctional adapter proteins that mediate signaling in their own right and also control receptor desensitization and trafficking (2). Most endogenous 7TMRs are thought to signal through G protein- and β-arrestin-mediated pathways in a balanced fashion (3). However, some receptors can be made to signal in a biased fashion toward β-arrestin or G protein signaling by ligand modifications (4), so-called biased ligands (5), or by mutation of key receptor residues involved in coupling to G proteins or β-arrestins, resulting in G protein- or β-arrestin-biased receptors (6). To date, no endogenously expressed biased 7TMRs have been described. We speculated that candidates for such receptors are the “decoy receptors,” which do not appear to signal through G proteins and are thought to scavenge or internalize ligands or act as coreceptors for other chemokine receptors (7).
Seven-transmembrane decoy receptors that have been demonstrated to be unable to activate typical G protein-mediated signaling pathways include C5L2, Duffy antigen receptor for chemokines, D6, and CXCR7 (7). Some of these receptors contain variants of highly conserved residues that are thought to play an important role in signaling to G proteins (8). The decoy receptor CXCR7 (9, 10), which binds the ligands (Stromal-derived factor) SDF-1α/CXCL12 and ITAC/CXCL11, does not activate typical Gαi pathways of a chemokine receptor that would result in GTP hydrolysis or calcium mobilization (9, 11–13). It is currently thought that CXCR7 acts primarily to modulate signaling of CXCR4, an alternate receptor for SDF-1α. A number of mechanisms underlying this function have been proposed. Some propose that CXCR7 may scavenge or sequester SDF-1α, thereby generating gradients of SDF-1α that lead to differential signaling by CXCR4 (12, 14). It has also been proposed that CXCR7 acts as a coreceptor for CXCR4 (11, 13), as the two receptors form heterodimers in the context of overexpression in transiently transfected cells. It has also been proposed that ligand binding to CXCR7 may result in crosstalk with CXCR4 mediated by intracellular signaling molecules (15). More recently, it has been demonstrated that CXCR7 interacts with β-arrestin in a ligand-dependent manner (15–17), resulting in ligand internalization, but without any assessment of changes in signaling. Given these findings we set out to test whether CXCR7 can signal through β-arrestins to determine whether this receptor acts as an endogenous β-arrestin-biased receptor.
Results
CXCR7 Recruits β-Arrestin in a Ligand-Dependent Fashion.
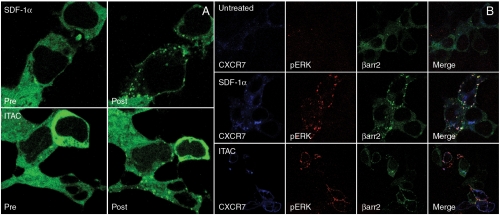
Previous studies have demonstrated the inability of CXCR7 to generate typical G protein responses such as an increase in intracellular calcium after treatment with either SDF-1α or ITAC (9, 11–13). However, we did observe recruitment of β-arrestin to the plasma membrane upon treatment with both ligands in HEK293 cells transiently transfected with CXCR7 and GFP-labeled β-arrestin2 (Fig. 1A), consistent with recent reports of ligand-dependent interactions between these two molecules (15, 16). Notably, we did not observe such recruitment in mock transfected HEK293 cells, which express endogenous CXCR4, an alternate receptor for SDF-1α (Fig. 1B). Recruitment of β-arrestin was rapid with translocation to the plasma membrane by 2 min, but at late time points (30 min) there was a distinct difference in the pattern of recruitment between those cells treated with SDF-1α or ITAC. In those cells treated with SDF-1α for more than 10 min, β-arrestin2 was localized to cytoplasmic vesicles, referred to as a “class B” pattern, whereas in those cells treated with ITAC, β-arrestin2 continued to localize to the plasma membrane, referred to as a “class A” pattern (Fig. 1A) (18). These patterns of β-arrestin recruitment for SDF-1α and ITAC suggest a functional difference in response to these two ligands, because class A interactions are transient in nature and result in rapid recycling of receptors, whereas class B interactions are stronger and result in slow and relatively low levels of recycling (19).
Fig. 1.
CXCR7 recruits β-arrestin resulting in MAP kinase activation in transiently transfected HEK293 cells. (A) β-arrestin recruitment in live HEK 293 cells transiently transfected with CXCR7 and GFP-labeled β-arrestin2 before and 30 min after treatment with SDF-1α and ITAC. (B) Thirty minutes after treatment with either ligand, there is a pattern of pERK formed in CXCR7-transfected cells. Fixed cells were labeled for CXCR7 (blue) and pERK (red) after transfection with CXCR7 and β-arrestin 2-GFP (green). In cells treated with SDF-1α, there is clear localization of all of these components in cytoplasmic vesicles on merged images (white), whereas this change is not as pronounced in those cells treated with ITAC.
CXCR7 Recruitment of β-Arrestin is Associated with phosphoERK Formation.
We observed these same patterns of recruitment in fixed cells that were analyzed for MAP kinase activation, in which we stained for phosphoERK1/2 (pERK) after 30 min of ligand stimulation (Fig. 1B). β-arrestin-mediated pERK formation by class B receptors is a well-characterized response that differs from G protein-mediated pERK phosphorylation in several respects: G protein-mediated activation occurs maximally at short times (2–5 min) and β-arrestin-mediated activity at longer times (≥10 min) (20,21); G protein-mediated pERK distributes evenly in the cytoplasm and nucleus whereas β-arrestin-mediated pERK localizes to cytoplasmic vesicles (21). After 30 min of treatment with SDF-1α, the majority of β-arrestin is present in cytoplasmic vesicles with strong colocalization of CXCR7, β-arrestin-2, and pERK on merged images (Fig. 1B). In cells treated with ITAC, most β-arrestin still localized to the plasma membrane with the class A pattern. These patterns of pERK in transfected cells are consistent with previously characterized β-arrestin-mediated processes, in terms of both spatial and temporal localization (21).
Rat Vascular Smooth Muscle Cells Express CXCR7.
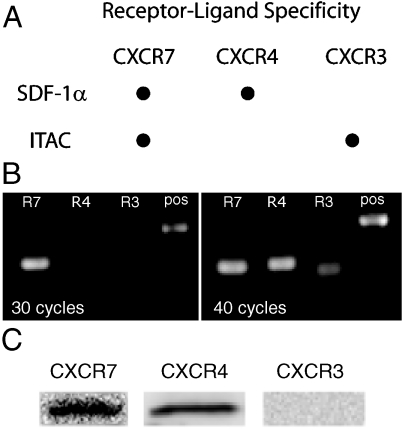
We next sought to address a functional β-arrestin-mediated response by CXCR7 in an endogenous system. Based on analysis of 7TMR mRNA expression in the mouse (22), we chose to study vascular smooth muscle cells (VSMCs), which are predicted to express high levels of CXCR7 and relatively low levels of CXCRs 4 and 3, typical chemokine receptors for SDF-1α and ITAC, respectively, that could confound our analysis (Fig. 2A). Similar to the mouse, we found that rat VSMCs (rVSMCs) expressed orders of magnitude more CXCR7 than CXCR4 and CXCR3 at the mRNA level (Fig. 2B). As expression of CXCR7 at the protein level does not always correlate with its mRNA level (15), we next sought to demonstrate its expression by anti-CXCR7 antibody binding and its unique signature of chemokine binding. At the protein level by immunoblot analysis, we did detect both CXCR7 and CXCR4, but not CXCR3 (Fig. 2C).
Fig. 2.
Expression of chemokine receptors for SDF-1α and ITAC in rVSMCs. (A) CXCR7 shares ligands with the typical chemokine receptors CXCR4 and CXCR3. Other ligands for CXCR3 that are not shown are IP10 and Mig. (B) mRNA expression levels of CXCR7 (R7), CXCR4 (R4) and CXCR3 (R3) in passage 0 rVSMCs by semiquantitative RT-PCR at 30 and 40 cycles. (C) Immunoblot analysis of receptor expression in rVSMCs using antibodies against CXCR7, 4, and 3.
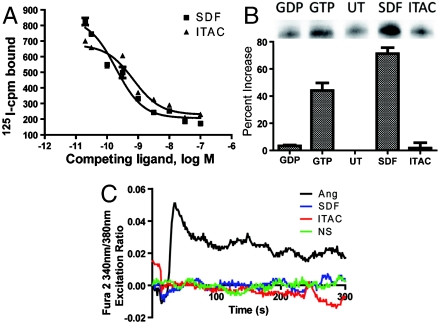
Radioligand binding studies of rVSMCs demonstrate that unlabeled ITAC displaced 125I-SDF-1α, a profile of chemokine binding unique to CXCR7 and not CXCR4 or CXCR3 (Fig. 3A). To assess whether expression of CXCR4 or CXCR3 was at a sufficient level to be functional for G protein activation in rVSMCs, we determined activation of Gαi by pulldown with an antibody against the active conformation of Gαi. Treatment with SDF-1α significantly increased amounts of active Gαi, implying activation of CXCR4, whereas ITAC had no effect on these levels compared to untreated controls, consistent with the absence of CXCR3 in these cells (Fig. 3B). Treatment of rVSMCs with either SDF-1α or ITAC did not result in intracellular calcium flux, a response that would be expected for activation of either CXCR4 or CXCR3 (Fig. 3C) (compared to a positive control of angiotensin, a well-characterized Gαq-coupled receptor) (23). Thus, we conclude that ITAC binds CXCR7 alone without any activation of G proteins, and SDF-1α binds to both CXCR4 and CXCR7, resulting in Gαi activation, albeit with minimal signaling through a typical Gαi-coupled pathway.
Fig. 3.
Chemokine receptor expression and lack of G protein activation by ITAC in rVSMCs. (A) Treatment with unlabeled ITAC or SDF competes off radiolabeled SDF-1α from rVSMCs. (B) Activation of Gαi in SDF-1α-treated rVSMCs (SDF) but not in untreated (UT) or ITAC-treated (ITAC) samples. The negative control was incubated with GDP (GDP) and the positive control incubated with GTP (GTP) for 90 min prior to immunoprecipitation. Data shown are mean ± SEM from three independent experiments. (C) Absence of calcium influx as assessed by change in Fura-2 fluorescence emission ratio upon stimulation of rVSMCs with SDF-1α (blue) or ITAC (red) compared to Angiotensin II (black) from a representative experiment from three independent experiments.
Rat VSMCs Migrate to ITAC in a CXCR7- and β-Arrestin-Mediated Process.
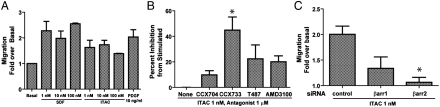
We next addressed whether activation of endogenous CXCR7 in the rVSMCs could result in a functional β-arrestin-mediated response. We first assessed for MAP kinase activation that we had observed in the transiently transfected HEK293 cells, but we were unable to detect significant increases in pERK upon treatment with either SDF-1α or ITAC over the background high basal activation in the rVSMCs even after more than 48 h of serum starvation (Fig. S1). We also did not observe a mitogenic response to these ligands (Fig. S1), suggesting that CXCR7 couples to different signaling pathways in rVSMCs than it does in the context of overexpression in HEK293 cells. We next assessed rVSMC migration to these ligands as β-arrestin-mediated chemotaxis is a well-described phenomenon in a number of 7TMR systems (24–27). We found that SDF-1α and ITAC resulted in significant rVSMC migration across a range of concentrations tested (Fig. 4A), with a response similar to that of other ligands known to induce rVSMC chemotaxis, such as PDGF (28).
Fig. 4.
rVSMC migration in response to ITAC is a CXCR7- and β-arrestin-dependent process. (A) Migration of rVSMCs to 1–100 nM of SDF-1α and ITAC compared to PDGF positive control. Shown is representative data from at least three independent experiments. (B) Effects of CCX704 (inactive control), CCX733 (CXCR7 antagonist), T497 (CXCR3 antagonist), and AMD3100 (CXCR4 antagonist) on ITAC-stimulated migration. Only treatment with CCX733 resulted in a significant inhibitory effect (∗ p < 0.05). (C) Significant inhibition of ITAC-stimulated rVSMC migration by β-arrestin2 depletion (∗ p < 0.05) but not β-arrestin1 depletion. Data shown are mean ± SEM from at least four independent experiments.
To avoid possible confounding with CXCR4-mediated processes, we then further characterized the response of rVSMCs to ITAC alone. To assess whether this migration response was mediated by CXCR7, cells were treated with selective chemokine receptor antagonists. Treatment with the CXCR3 antagonist T487 (29) or the CXCR4 antagonist AMD3100 (30) had no significant effect on migration, however, the CXCR7 antagonist CCX733 had a significant inhibitory effect on migration, whereas a structurally related compound which does not bind CXCR7, CCX704, had none (Fig. 4B). Therefore, we conclude that the majority of the migration response to ITAC is mediated by CXCR7 and not the typical chemokine receptors CXCR3 and CXCR4. To determine whether this CXCR7-mediated migration was β-arrestin-dependent, we depleted β-arrestins by siRNA knockdown. Although depletion of β-arrestin1 had no statistically significant effect on migration, depletion of β-arrestin2 resulted in a marked attenuation of rVSMC migration in response to ITAC compared to control siRNA-transfected cells (Fig. 4C). Although it is impossible to exclude signaling by the dozens of different Gα and Gβγ subunits (31), we did find that this response was not sensitive to treatment with pertussis toxin and thus independent of Gαi (Fig. S2). The specificity of CXCR7 antagonism and effect of β-arrestin depletion demonstrate that CXCR7-mediated rVSMC migration is a β-arrestin-mediated process and that CXCR7 acts as a β-arrestin-biased receptor.
Discussion
Although virtually all reports in the literature have agreed that CXCR7 does not couple to G proteins, functional roles for the receptor that belie a signaling event suggest a nonclassical positive signaling role for this receptor. In the initial characterization of CXCR7, its expression increased the adhesion of cancer cells to endothelia and enhanced their survival in vitro and in vivo (9). In the zebrafish, polarized expression of CXCR4 and CXCR7 is essential for primordial germ cell migration (12, 14). These reports suggested that CXCR7 may scavenge the CXCL12 in the stationary, nonmigrating cells. Prostate cancer cells expressing CXCR7 suggest a role for this receptor in adhesion, invasiveness, and survival, where activation of Akt is linked to CXCR7 and results in the regulation of a number of genes associated with the NF-κB and MAP kinase pathways (32). Our current data, in which CXCR7 is an active receptor capable of signaling through β-arrestin, provide a mechanistic basis for understanding these earlier studies.
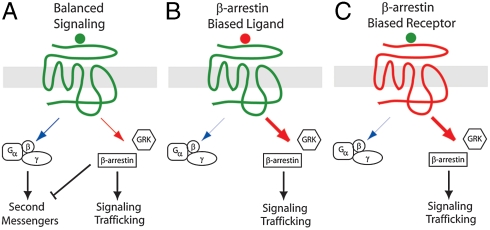
For the vast majority of 7TMR signaling, it is thought that there is a balance between G protein- and β-arrestin-mediated pathways (Fig. 5A). Agonist binding typically results in signaling mediated by G proteins and β-arrestins accompanied by receptor desensitization and internalization mediated by β-arrestins. This is in contrast to a system with biased signaling, where signaling is mediated selectively through only one of these two pathways. Two limit cases of biased signaling can be considered. In the case of a biased ligand, treatment of a balanced receptor with a biased agonist results in a biased response (Fig. 5B), whereas, in the case of a biased receptor, binding of a balanced agonist results in a biased response (Fig. 5C). Before the present study, receptors that have been demonstrated to be biased have been genetically engineered from balanced receptors by mutation of key residues involved in G protein coupling, such as mutations of the highly conserved DRY motif of the AT1AR to generate the variant AT1AR(DRY/AAY) (33) or mutations of three highly conserved residues in the β2 adrenergic receptor to generate the β-arrestin-biased β2AR(TYY) (6). Surprisingly, similar mutations are observed in decoy receptors such as CXCR7, which lacks a typical sequence surrounding the DRY motif (7), suggesting that biased receptors have evolved in our genome.
Fig. 5.
Balanced and biased signaling by 7TMRs. (A) In a balanced system, signaling is mediated by G proteins and β-arrestins, while β-arrestins also control desensitization and receptor trafficking. (B) Treatment of an unbiased receptor with a biased agonist results in a biased response, in this case, β-arrestin-mediated signaling only. (C) In the case of a β-arrestin-biased receptor, treatment of the receptor with an unbiased ligand results in β-arrestin-mediated signaling only.
β-arrestins were initially discovered as negative regulators of G protein-mediated signaling by 7TMRs (34), but it was subsequently discovered that they themselves were capable of positively regulating cell signaling (35, 36). The discovery of an endogenous β-arrestin-biased receptor has important implications for our basic understanding of 7TMR biology. It inverts the classical paradigm of signaling from the view that all signaling by 7TMRs is mediated by heterotrimeric G proteins, to one where receptors may only signal through non-G protein-mediated mechanisms. This suggests that other receptors that have been considered to be “nonsignaling” may also signal through such nonclassical pathways. Many drug discovery and deorphanization strategies are based solely on assays for G protein-mediated actions and may not recognize β-arrestin-mediated processes. In the future, a more complete understanding of 7TMR pharmacology can only be gained by looking at G protein and β-arrestin activity in concert.
Materials and Methods
Materials.
A plasmid encoding CXCR7 in a modified version of pcDNA3 (pFIRES) was a gift from Chemocentryx (9). Recombinant chemokines were obtained from R&D Systems. Anti-CXCR7 monoclonal antibodies were obtained from R&D systems, anti-rat CXCR4 and anti-rat CXCR3 antibodies from Abcam, antiphosphoERK antibody from Cell Signaling Technologies, and antitotalERK antibody from Upstate Biotechnology. Chemokine inhibitors CCX704 and CCX733 were obtained from Chemocentryx. T487 was obtained from Tularik, and AMD3100 was purchased from Sigma.
Transient Transfection.
HEK-293 cells were maintained in MEM media supplemented with 10% FBS and 1% penicillin/streptomycin. For DNA transfection, FuGene transfection reagent (Roche Applied Science) was added at a ratio of 5-μl to 1-μg plasmid DNA to a solution of plasmid DNA in 500-μl serum-free MEM. This transfection mixture was incubated for 30–45 min prior to addition to HEK293 cells of 50% confluence. For a 10-cm cell culture dish, 5 μg of plasmid DNA encoding CXCR7 was used. Cells were split and used for assays 48 h after transfection. For rVSMCs, 80–90% confluent early passage (< 3) VSMCs were transfected with siRNA using Lipofectamine 2000 transfection reagent (Invitrogen) according to the modified manufacturer’s instructions (37). The siRNA sequence targeting β arrestin1 was 5-AGCCUUCUGUGCUGAGAAC-3, corresponding to position 431–459 relative to the start codon. The siRNA sequences targeting β arrestin2 were 5-GGACCGCAAAGUGUUUGUG-3 and 5-CCAACCTCATTGAATTCGA-3, corresponding to positions 150–168 and 1115–1133 relative to the start codon.
Confocal Microscopy.
HEK-293 cells were split into 35-mm glass-bottom dishes (MatTek) and transfected with 1 μg of CXCR7 and 0.2 μg of β-arrestin-2-GFP. Forty-eight hours after transfection, cells were starved for at least 5 h in serum-free media prior to treatment with ligand. For live cell imaging, cells were maintained at 37 °C with a heating plate while confocal microscopy was performed. SDF-1α and ITAC were added to a final concentration of 100 nM and images were taken from 0 to 30 min. For immunostaining, cells were treated for 30 min with 100 nM ligand, followed by aspiration of serum and fixation. Samples were then washed followed by permeabilization and blocking. Cells were then incubated overnight with primary antibody and then washed prior to incubation with secondary antibody at 1∶500 dilution. Samples were then washed and visualized.
PCR.
RT-PCR primers for rat CXCR7, CXCR4, and CXCR3 were obtained from Qiagen. Total RNA was extracted from passage 0 rVSMC by phenol-chloroform extraction using Ultraspec reagent (Bio-X). Qiagen RT-PCR kit was used for 30 and 40 cycles for semiquantitative RT-PCR. Samples were run on 1% agarose gel and stained with ethidium bromide prior to digital imaging.
Active Gαi Pulldown Assay.
rVSMCs were starved for 48 h prior to 5 min of stimulation with SDF-1α or ITAC (positive control). After 5 min, the cells were placed on ice, washed with ice cold PBS and lysed, solubilized for 1 h, and then incubated for 1 h with an antiactive Gαi antibody and Protein A-agarose beads (New East Biosciences). Beads were washed with lysis buffer three times and incubated with SDS-PAGE buffer at 95 °C for 10 min and run on a 4–20% gradient polyacrylamide gel, transferred to nitrocellulose membrane, and incubated with anti-Gαi antibody.
Intracellular Calcium Flux Assay.
rVSMCs were split into glass-bottom dishes at least 12 h before experiments. Smooth muscle cells (SMCs) were loaded with 5 μmol/L Fura-2/acetoxymethyl ester (Invitrogen) for 30 min. Intracellular calcium levels were quantitated by the Fura-2 excitation ratio at 340 and 380 nm on an epifluorescence microscope. Agonist-stimulated calcium release was calculated as the change in the Fura-2 excitation ratio from baseline.
Radioactive Ligand Binding.
Cells were grown in 24-well plates until confluent. Binding was assessed by incubation with 0.02 nM 125I-SDF-1α or ITAC in 25 mM Hepes, pH 7.5, containing 140 mM NaCl, 1 mM CaCl2, 5 mM MgCl2, and 0.2% BSA for 2 h at 22 °C in the presence of 0–300 nM unlabeled ligand. In some experiments, AMD3100 (50 mM) or CCX733 (1 mM) were also included. Cells were washed three times with binding buffer containing 0.5 M NaCl, solubilized with 0.3 mL of 0.5 M NaOH, and counts per minute bound determined by gamma counting.
VSMC Migration.
Rat aortas were stripped of adventitia and endothelial cells, and then digested with 0.1% collagenase II and 15 U/mL elastase in the presence of 0.1% soybean trypsin inhibitor. Released SMCs were cultured in DMEM with 10% FBS and 1% penicillin/streptomycin (Life Technologies). SMCs were used during passages 1–2. The migration of serum-starved SMCs was assessed using Transwell™ membranes (Costar; 8-μm pore). Filters were coated with 10 μg/mL fibronectin (in PBS overnight at 4 °C), rinsed once with PBS, and placed in 24-well dish wells that contained serum-free DMEM supplemented with agonists. SMCs suspended in serum-free DMEM containing 0.1% BSA were added to the upper chamber (1 × 105 cells/well). Cells were allowed to migrate for 5 h at 37 °C. Nonmigrated SMCs were removed from the top filter surface with a cotton swab. Migrated SMCs, attached to the bottom surface, were fixed in 4% paraformaldehyde and dyed with crystal violet. Dye intensity was quantified by densitometry. For rVSMCs transfected with fluorescent siRNAs, images were obtained on an epifluorescence microscope and the number of cells quantified.
Supplementary Material
Acknowledgments.
We thank Thomas Schall and Juan Jaen for valuable discussions and advice. We thank Donna Addison and Elizabeth Hall for secretarial assistance. We thank Chemocentryx for a construct encoding CXCR7 and selective CXCR7 antagonists. This work was supported in part by National Institutes of Health (NIH) Grants HL16037 and HL70631 (to R.J.L.). R.J.L. is an Investigator with The Howard Hughes Medical Institute. S.R. is supported by NIH T32 training Grant HL07101-34. Drs. Gerard are supported by NIH Grants HL69511 and 36162.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912852107/DCSupplemental.
References
- 1.Ma P, Zemmel R. Value of novelty? Nat Rev Drug Discovery. 2002;1(8):571–572. doi: 10.1038/nrd884. [DOI] [PubMed] [Google Scholar]
- 2.Lefkowitz RJ, Shenoy SK. Transduction of receptor signals by beta-arrestins. Science. 2005;308(5721):512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- 3.Benovic JL, Staniszewski C, Mayor F, Jr., Caron MG, Lefkowitz RJ. beta-Adrenergic receptor kinase. Activity of partial agonists for stimulation of adenylate cyclase correlates with ability to promote receptor phosphorylation. J Biol Chem. 1988;263(8):3893–3897. [PubMed] [Google Scholar]
- 4.Kenakin T. Collateral efficacy in drug discovery: taking advantage of the good (allosteric) nature of 7TM receptors. Trends Pharmacol Sci. 2007;28(8):407–415. doi: 10.1016/j.tips.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 5.Gesty-Palmer D, et al. Distinct beta-arrestin- and G-protein-dependent pathways for parathyroid hormone receptor-stimulated ERK1/2 activation. J Biol Chem. 2006;281(16):10856–10864. doi: 10.1074/jbc.M513380200. [DOI] [PubMed] [Google Scholar]
- 6.Shenoy SK, et al. beta-arrestin-dependent, G protein-independent ERK1/2 activation by the beta2 adrenergic receptor. J Biol Chem. 2006;281(2):1261–1273. doi: 10.1074/jbc.M506576200. [DOI] [PubMed] [Google Scholar]
- 7.Graham GJ. D6 and the atypical chemokine receptor family: Novel regulators of immune and inflammatory processes. Eur J Immunol. 2009;39(2):342–351. doi: 10.1002/eji.200838858. [DOI] [PubMed] [Google Scholar]
- 8.Okinaga S, et al. C5L2, a nonsignaling C5A binding protein. Biochemistry. 2003;42(31):9406–9415. doi: 10.1021/bi034489v. [DOI] [PubMed] [Google Scholar]
- 9.Burns JM, et al. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J Exp Med. 2006;203(9):2201–2213. doi: 10.1084/jem.20052144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balabanian K, et al. The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J Biol Chem. 2005;280(42):35760–35766. doi: 10.1074/jbc.M508234200. [DOI] [PubMed] [Google Scholar]
- 11.Levoye A, Balabanian K, Baleux F, Bachelerie F, Lagane B. CXCR7 heterodimerizes with CXCR4 and regulates CXCL12-mediated G protein signalling. Blood. 2009;113(24):6085–6093. doi: 10.1182/blood-2008-12-196618. [DOI] [PubMed] [Google Scholar]
- 12.Boldajipour B, et al. Control of chemokine-guided cell migration by ligand sequestration. Cell. 2008;132(3):463–473. doi: 10.1016/j.cell.2007.12.034. [DOI] [PubMed] [Google Scholar]
- 13.Sierro F, et al. Disrupted cardiac development but normal hematopoiesis in mice deficient in the second CXCL12/SDF-1 receptor, CXCR7. Proc Natl Acad Sci USA. 2007;104(37):14759–14764. doi: 10.1073/pnas.0702229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dambly-Chaudiere C, Cubedo N, Ghysen A. Control of cell migration in the development of the posterior lateral line: Antagonistic interactions between the chemokine receptors CXCR4 and CXCR7/RDC1. BMC Dev Biol. 2007;7:23. doi: 10.1186/1471-213X-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zabel BA, et al. Elucidation of CXCR7-Mediated Signaling Events and Inhibition of CXCR4-Mediated Tumor Cell Transendothelial Migration by CXCR7 Ligands. J Immunol. 2009;183(5):3204–3211. doi: 10.4049/jimmunol.0900269. [DOI] [PubMed] [Google Scholar]
- 16.Kalatskaya I, et al. AMD3100 is a CXCR7 ligand with allosteric agonist properties. Mol Pharmacol. 2009;75(5):1240–1247. doi: 10.1124/mol.108.053389. [DOI] [PubMed] [Google Scholar]
- 17.Luker KE, Gupta M, Steele JM, Foerster BR, Luker GD. Imaging ligand-dependent activation of CXCR7. Neoplasia. 2009;11(10):1022–1035. doi: 10.1593/neo.09724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oakley RH, Laporte SA, Holt JA, Caron MG, Barak LS. Differential affinities of visual arrestin, beta arrestin1, and beta arrestin2 for G protein-coupled receptors delineate two major classes of receptors. J Biol Chem. 2000;275(22):17201–17210. doi: 10.1074/jbc.M910348199. [DOI] [PubMed] [Google Scholar]
- 19.DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. Beta-arrestins and cell signaling. Annu Rev Physiol. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- 20.Ren XR, et al. Different G protein-coupled receptor kinases govern G protein and beta-arrestin-mediated signaling of V2 vasopressin receptor. Proc Natl Acad Sci USA. 2005;102(5):1448–1453. doi: 10.1073/pnas.0409534102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahn S, Shenoy SK, Wei H, Lefkowitz RJ. Differential kinetic and spatial patterns of beta-arrestin and G protein-mediated ERK activation by the angiotensin II receptor. J Biol Chem. 2004;279(34):35518–35525. doi: 10.1074/jbc.M405878200. [DOI] [PubMed] [Google Scholar]
- 22.Regard JB, Sato IT, Coughlin SR. Anatomical profiling of G protein-coupled receptor expression. Cell. 2008;135(3):561–571. doi: 10.1016/j.cell.2008.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajagopal K, et al. Beta-arrestin2-mediated inotropic effects of the angiotensin II type 1A receptor in isolated cardiac myocytes. Proc Natl Acad Sci USA. 2006;103(44):16284–16289. doi: 10.1073/pnas.0607583103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Y, Cheng Z, Ma L, Pei G. Beta-arrestin2 is critically involved in CXCR4-mediated chemotaxis, and this is mediated by its enhancement of p38 MAPK activation. J Biol Chem. 2002;277(51):49212–49219. doi: 10.1074/jbc.M207294200. [DOI] [PubMed] [Google Scholar]
- 25.Ge L, Ly Y, Hollenberg M, DeFea K. A beta-arrestin-dependent scaffold is associated with prolonged MAPK activation in pseudopodia during protease-activated receptor-2-induced chemotaxis. J Biol Chem. 2003;278(36):34418–34426. doi: 10.1074/jbc.M300573200. [DOI] [PubMed] [Google Scholar]
- 26.Ge L, Shenoy SK, Lefkowitz RJ, DeFea K. Constitutive protease-activated receptor-2-mediated migration of MDA MB-231 breast cancer cells requires both beta-arrestin-1 and -2. J Biol Chem. 2004;279(53):55419–55424. doi: 10.1074/jbc.M410312200. [DOI] [PubMed] [Google Scholar]
- 27.DeFea KA. Stop that cell! Beta-arrestin-dependent chemotaxis: A tale of localized actin assembly and receptor desensitization. Annu Rev Physiol . 69:535–560. doi: 10.1146/annurev.physiol.69.022405.154804. [DOI] [PubMed] [Google Scholar]
- 28.Wu JH, et al. The platelet-derived growth factor receptor-beta phosphorylates and activates G protein-coupled receptor kinase-2. A mechanism for feedback inhibition. J Biol Chem. 2005;280(35):31027–31035. doi: 10.1074/jbc.M501473200. [DOI] [PubMed] [Google Scholar]
- 29.Heise CE, et al. Pharmacological characterization of CXC chemokine receptor 3 ligands and a small molecule antagonist. J Pharmacol Exp Ther. 2005;313(3):1263–1271. doi: 10.1124/jpet.105.083683. [DOI] [PubMed] [Google Scholar]
- 30.Donzella GA, et al. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat Med. 1998;4(1):72–77. doi: 10.1038/nm0198-072. [DOI] [PubMed] [Google Scholar]
- 31.Cabrera-Vera TM, et al. Insights into G protein structure, function, and regulation. Endocr Rev. 2003;24(6):765–781. doi: 10.1210/er.2000-0026. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, et al. The role of CXCR7/RDC1 as a chemokine receptor for CXCL12/SDF-1 in prostate cancer. J Biol Chem. 2008;283(7):4283–4294. doi: 10.1074/jbc.M707465200. [DOI] [PubMed] [Google Scholar]
- 33.Wei H, et al. Independent beta-arrestin 2 and G protein-mediated pathways for angiotensin II activation of extracellular signal-regulated kinases 1 and 2. Proc Natl Acad Sci USA. 2003;100(19):10782–10787. doi: 10.1073/pnas.1834556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lohse MJ, Benovic JL, Codina J, Caron MG, Lefkowitz RJ. beta-Arrestin: A protein that regulates beta-adrenergic receptor function. Science. 1990;248(4962):1547–1550. doi: 10.1126/science.2163110. [DOI] [PubMed] [Google Scholar]
- 35.DeFea KA, et al. The proliferative and antiapoptotic effects of substance P are facilitated by formation of a beta-arrestin-dependent scaffolding complex. Proc Natl Acad Sci USA. 2000;97(20):11086–11091. doi: 10.1073/pnas.190276697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luttrell LM, et al. Beta-arrestin-dependent formation of beta2 adrenergic receptor-Src protein kinase complexes. Science. 1999;283(5402):655–661. doi: 10.1126/science.283.5402.655. [DOI] [PubMed] [Google Scholar]
- 37.Kim J, Ahn S, Rajagopal K, Lefkowitz RJ. Independent beta-arrestin2 and Gq/protein kinase Czeta pathways for ERK stimulated by angiotensin type 1A receptors in vascular smooth muscle cells converge on transactivation of the epidermal growth factor receptor. J Biol Chem. 2009;285(18):11953–11962. doi: 10.1074/jbc.M808176200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.