Abstract
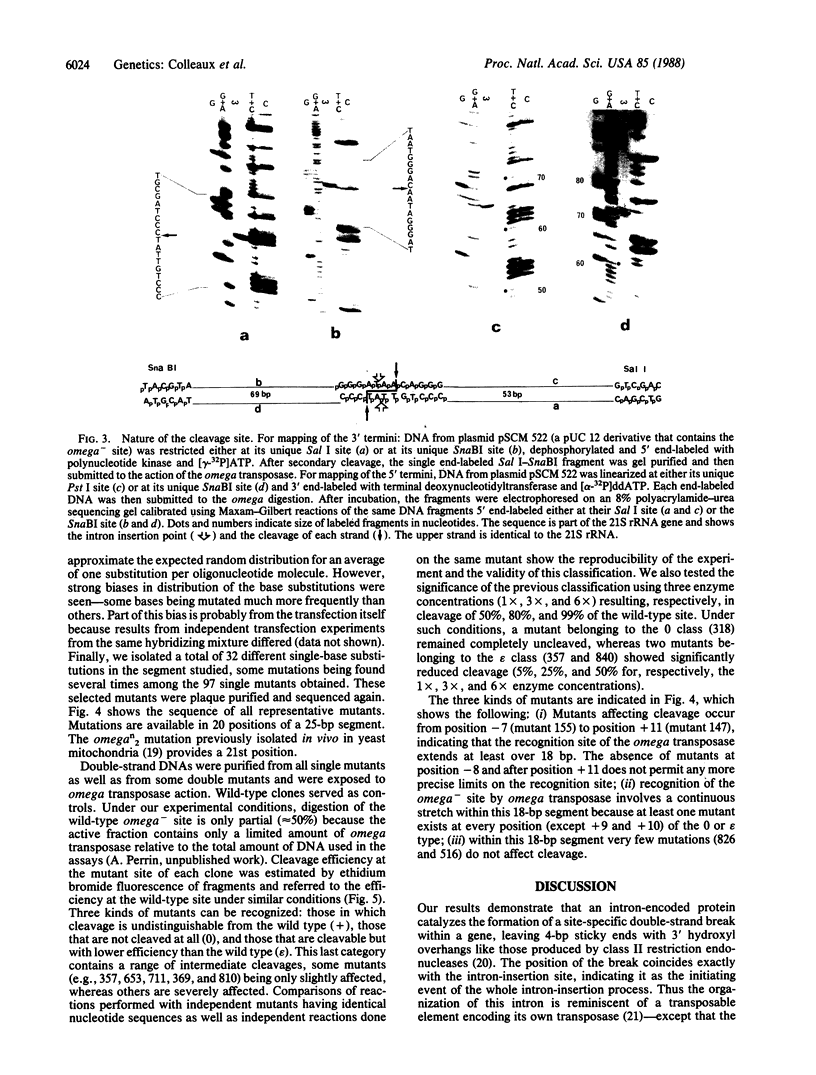
The optional group I intron of the mitochondrial 21S rRNA gene of Saccharomyces cerevisiae contains a 235-codon-long open reading frame the translation product of which (the omega transposase) catalyzes the formation of a double-strand break within the intron-minus (omega-) copies of the same gene. Purified omega transposase generates in vitro a 4-base-pair staggered cut with 3' hydroxyl overhangs at the exact position where the intron eventually inserts in the gene. Using randomly mutagenized synthetic oligonucleotides, single-base mutants were produced at 21 positions around the cleavage site. Experiments with these oligonucleotides show that the recognition site extends over an 18-base pair-long sequence within which minimal sequence degeneracy is tolerated. The intron-encoded omega transposase is, therefore, one of the most specific restriction endonucleases known to date.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Colleaux L., d'Auriol L., Betermier M., Cottarel G., Jacquier A., Galibert F., Dujon B. Universal code equivalent of a yeast mitochondrial intron reading frame is expressed into E. coli as a specific double strand endonuclease. Cell. 1986 Feb 28;44(4):521–533. doi: 10.1016/0092-8674(86)90262-x. [DOI] [PubMed] [Google Scholar]
- Dujon B., Bolotin-Fukuhara M., Coen D., Deutsch J., Netter P., Slonimski P. P., Weill L. Mitochondrial genetics. XI. Mutations at the mitochondrial locus omega affecting the recombination of mitochondrial genes in Saccharomyces cerevisiae. Mol Gen Genet. 1976 Jan 16;143(2):131–165. doi: 10.1007/BF00266918. [DOI] [PubMed] [Google Scholar]
- Dujon B. Sequence of the intron and flanking exons of the mitochondrial 21S rRNA gene of yeast strains having different alleles at the omega and rib-1 loci. Cell. 1980 May;20(1):185–197. doi: 10.1016/0092-8674(80)90246-9. [DOI] [PubMed] [Google Scholar]
- Flory S. S., Tsang J., Muniyappa K., Bianchi M., Gonda D., Kahn R., Azhderian E., Egner C., Shaner S., Radding C. M. Intermediates in homologous pairing promoted by RecA protein and correlations of recombination in vitro and in vivo. Cold Spring Harb Symp Quant Biol. 1984;49:513–523. doi: 10.1101/sqb.1984.049.01.058. [DOI] [PubMed] [Google Scholar]
- Jacquier A., Dujon B. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell. 1985 Jun;41(2):383–394. doi: 10.1016/s0092-8674(85)80011-8. [DOI] [PubMed] [Google Scholar]
- Jacquier A., Dujon B. The intron of the mitochondrial 21S rRNA gene: distribution in different yeast species and sequence comparison between Kluyveromyces thermotolerans and Saccharomyces cerevisiae. Mol Gen Genet. 1983;192(3):487–499. doi: 10.1007/BF00392195. [DOI] [PubMed] [Google Scholar]
- Kostriken R., Strathern J. N., Klar A. J., Hicks J. B., Heffron F. A site-specific endonuclease essential for mating-type switching in Saccharomyces cerevisiae. Cell. 1983 Nov;35(1):167–174. doi: 10.1016/0092-8674(83)90219-2. [DOI] [PubMed] [Google Scholar]
- Krasnow M. A., Cozzarelli N. R. Site-specific relaxation and recombination by the Tn3 resolvase: recognition of the DNA path between oriented res sites. Cell. 1983 Apr;32(4):1313–1324. doi: 10.1016/0092-8674(83)90312-4. [DOI] [PubMed] [Google Scholar]
- Macreadie I. G., Scott R. M., Zinn A. R., Butow R. A. Transposition of an intron in yeast mitochondria requires a protein encoded by that intron. Cell. 1985 Jun;41(2):395–402. doi: 10.1016/s0092-8674(85)80012-x. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Michel F., Dujon B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2(1):33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Jacquier A., Dujon B. Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie. 1982 Oct;64(10):867–881. doi: 10.1016/s0300-9084(82)80349-0. [DOI] [PubMed] [Google Scholar]
- Nickoloff J. A., Chen E. Y., Heffron F. A 24-base-pair DNA sequence from the MAT locus stimulates intergenic recombination in yeast. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7831–7835. doi: 10.1073/pnas.83.20.7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J. Restriction enzymes and their isoschizomers. Nucleic Acids Res. 1987;15 (Suppl):r189–r217. doi: 10.1093/nar/15.suppl.r189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Watabe H., Iino T., Kaneko T., Shibata T., Ando T. A new class of site-specific endodeoxyribonucleases. Endo.Sce I isolated from a eukaryote, Saccharomyces cerevisiae. J Biol Chem. 1983 Apr 25;258(8):4663–4665. [PubMed] [Google Scholar]
- Zinn A. R., Butow R. A. Nonreciprocal exchange between alleles of the yeast mitochondrial 21S rRNA gene: kinetics and the involvement of a double-strand break. Cell. 1985 Apr;40(4):887–895. doi: 10.1016/0092-8674(85)90348-4. [DOI] [PubMed] [Google Scholar]