Abstract
The instability of cholesterol clusters and the Umbrella effect of cholesterol in dipalmitoylphosphatidylcholine (DPPC) and dioleoylphosphatidylcholine (DOPC) lipid bilayers were investigated via atomistic Molecular Dynamics (MD) simulation. Cholesterol clusters in phosphatidylcholine (PC) bilayers are found to be very unstable and to readily disperse into cholesterol monomers. This instability results from the difficulty of the bilayer system in preventing water exposure to cholesterol’s bulky hydrophobic bodies in a cluster. The system responds to artificially arranged cholesterol clusters in several interesting manners: (i) Cholesterol clusters quickly form a “frustum” shape to reduce water penetration between cholesterol headgroups; (ii) Many clusters bury themselves deeper into the bilayer interior, causing bilayer deformation; (iii) Cholesterol fluctuates rapidly, both laterally and vertically, in order to escape clusters. These fluctuations result in the disintegration of clusters, and in one incidence, a highly unusual flip-flop event of cholesterol across the DOPC bilayer. Our results show that cholesterols have a strong tendency to avoid forming clusters in lipid bilayers and that the fundamental cholesterol-cholesterol interaction is unfavorable. Furthermore, the radial distribution functions of cholesterol hydroxyl oxygen to various headgroup atoms of PC reveal that the PC headgroups surrounding cholesterol have a clear tendency to reorient and to extend toward cholesterol. This reorientation has a layered structure that extends 2 to 3 nm from the cholesterol molecule. This study demonstrates that the Umbrella hypothesis is valid in both saturated and unsaturated lipid bilayers.
Keywords: the Umbrella Model, cholesterol-lipid interaction, headgroup orientation, frustum shaped cluster, cholesterol flip-flop, molecular dynamics simulation
INTRODUCTION
Among the many lipids that make up cellular membranes, cholesterol is the most important. Knowledge of the molecular-level interactions between cholesterol and neighboring lipids is critical to understanding membrane domains and phase formation,1 and consequently, illuminating their vital roles in cellular functions.2 At present, several models seek to explain the key cholesterol-lipid interaction: the Condensed Complex Model,3 the Superlattice Mode,4,5 and the Umbrella Model.6,7 The three models assume that the cholesterol-cholesterol interaction is unfavorable or repulsive; however, it has been suggested that the interaction may be attractive as well.8 Therefore, it is imperative to verify these fundamental assumptions.
The Condensed Complex Model hypothesizes the existence of low free-energy stoichiometric cholesterol–lipid chemical complexes, which occupy smaller molecular lateral areas than the sum of the component areas.3,9 The Superlattice Model hypothesizes that differences in the cross-sectional area between cholesterol and other lipid molecules can result in a long-range repulsive force among cholesterols and thereby produce superlattice distributions.4,5 The Umbrella Model proposes that the small, polar cholesterol headgroup provides insufficient coverage of the bulky, hydrophobic, sterol body; and therefore, cholesterol will seek association with large-headgroup lipids, such as PC or sphingomyelin.6,10 In water, pure cholesterol forms monohydrate crystals instead of bilayers, because the small hydroxyl group is unable to protect its large non-polar body from water. When cholesterols are incorporated into a phospholipid bilayer, cholesterol’s hydroxyl group interacts with water at the bilayer-aqueous interface to provide partial coverage. This interaction also anchors the cholesterol headgroup to the interface; however, the small hydroxyl headgroup cannot provide complete coverage of the large cholesterol body without the assistance of neighboring phospholipid headgroups. This is illustrated schematically in Fig. 1A. Phospholipid headgroups act like umbrellas by shielding part of the cholesterol non-polar body from the aqueous phase, thus avoiding an unfavorable free energy state. Beneath the phospholipid headgroup “umbrella”, the acyl chains of the phospholipid and the hydrophobic cholesterol bodies share space, thereby causing the long-known “cholesterol condensing effect”. As the cholesterol content of a bilayer rises, small cholesterol clusters begin to form, phospholipids laterally redistribute and their polar headgroups reorient in order to provide more coverage per headgroup for the added cholesterol molecules, as drawn in Fig. 1B. The headgroup umbrellas are “stretched” to provide more coverage area. Beneath the umbrella, acyl chains and cholesterol sterol rings become tightly packed. Understandably, saturated acyl chains would pack well with cholesterol, whereas polyunsaturated chains would pack very poorly, resulting in poor coverage of cholesterol. Thus, cholesterol preferentially seeks phospholipids with saturated chains, rather than those with mono- or polyunsaturated chains. Furthermore, neighboring phospholipids may not be able to provide sufficient coverage to cholesterol in a cluster, particularly to the cholesterol in the center of the cluster, which may not receive any coverage. The free energy cost of the Umbrella coverage must rise rapidly with the increasing size of the cholesterol cluster. Thus, cholesterol molecules have a strong preference against clustering in lipid membranes. Any of the three conceptual models can explain certain experimental results; however, only the Umbrella model is consistent with data from a recent experimental measurement of cholesterol chemical activity in DPPC, POPC, and DOPC bilayers.11
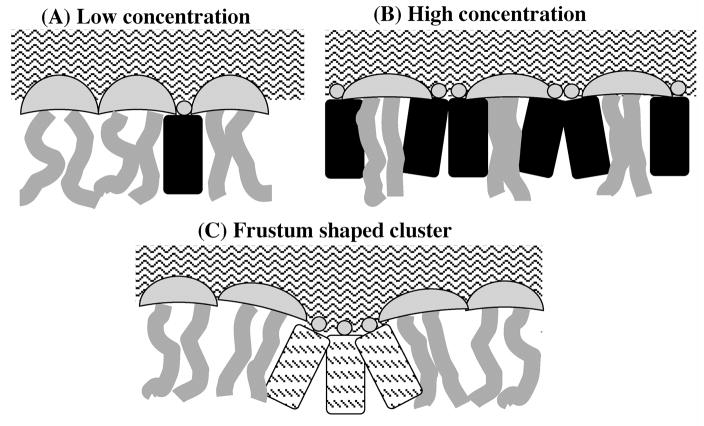
Figure 1.
The Umbrella model: (A) At low cholesterol concentration, headgroups of neighboring PCs easily cover the hydrophobic bodies of cholesterol. Motion of acyl chains next to cholesterol is restricted by the rigid body of cholesterol; (B) At high cholesterol concentration, headgroups of PCs work together to cover more cholesterol. The acyl chains of PCs become highly ordered. (C) The frustum shaped cluster: A cholesterol cluster adopts the “frustum shape” to reduce water penetration between cholesterol headgroups. The cluster also moves deeper into bilayer interior allowing a better coverage of its outskirts by surrounding PC headgroups.
Molecular Dynamics (MD) simulation has recently become a powerful investigative tool for testing and verifying the conceptual models of cholesterol-lipid interaction.12–15 Zhang et al. studied the possibility of cholesterol-sphingolipid 1:1 and 1:2 complexes through a comparison of 16:0–18:1PC (POPC) – cholesterol interaction energies with sphingolipid – cholesterol interaction energies.16 They concluded that the difference between the two energies was too small to indicate the formation of a complex. On the other hand, calculation of the free energy associated with complex formation turned out to be challenging and inconclusive.12 At present, no image of a cholesterol-phospholipid complex at the molecular level exists. Zhu et al. used MD simulation to investigate the cholesterol superlattice in POPC bilayers containing 40 mol % of cholesterol.17 By artificially arranging cholesterol into a superlattice distribution, they discovered that the lateral distribution of cholesterol in the superlattice bilayer is more stable than that in a random bilayer; however, at the completion of the 200 ns simulation, the final lateral distribution of cholesterol lacks long-range order, which is the proposed signature characteristic of a superlattice. Thus, a much longer simulation time might be needed to simulate a cholesterol superlattice. Nonetheless, a number of MD investigations into the arrangement and orientation of phospholipid headgroups near cholesterol make it possible to examine and test the Umbrella Model. Despite the direct interaction of the cholesterol headgroup with water, Pitman et al. showed that the cholesterol body is quite dry.18 They constructed a probability isosurface for phosphate and choline atoms of PC near cholesterol, and demonstrated that the hydrophilic choline groups of PC cover the cholesterol surface. These observations are in a good agreement with the Umbrella hypothesis. Kucerka et al. analyzed the locations of choline nitrogen and phosphate atoms relative to cholesterol’s hydroxyl group, and showed that the Umbrella Effect is largest for the first nearest neighbor PCs and smaller for the second nearest neighbors, and no Umbrella Effect can be detected for the third nearest-neighbor PCs.13 Interestingly, a recent coarse grain simulation study by de Meyer and Smit demonstrated that no cholesterol condensing effect could be observed if they slightly modified the cholesterol structure with the addition of an extra hydrophilic head group or a reduction in size of the hydrophobic body.19 This study confirmed that the cholesterol condensing effect originates directly from the mismatch between cholesterol’s small polar headgroup and its large nonpolar body.
The studies described here examined two critical claims of the Umbrella Model hypothesis at the molecular level: (i) Large cholesterol clusters are unfavorable in a lipid bilayer and the instability arises directly from the inability of the system to sufficiently cover the hydrophobic bodies of cholesterol in a cluster; and (ii) Neighboring phospholipids should exhibit a tendency to maneuver their headgroups towards cholesterol in order to provide coverage to the non-polar bodies of cholesterol. MD simulations were performed for di16:0PC (DPPC) and di18:1PC (DOPC) lipid bilayers, both containing 20 mol % cholesterol. For each PC system, we performed two 200 ns simulations: the first involved artificially arranged cholesterol clusters; the second featured randomly located cholesterols. In order to compare the simulation results to pure PC bilayers, a third 200 ns simulation of a system without cholesterol was performed. Our results show that cholesterol clusters in PC bilayers are very unstable and readily disperse into cholesterol monomers. The systems responded to the artificially arranged, large, cholesterol clusters in several interesting manners: cholesterol clusters quickly adopted a “frustum” shape in order to reduce water penetration between cholesterol headgroups; large clusters submerged deeper into the bilayer interior, causing some bilayer deformation; cholesterols fluctuated rapidly, both laterally and vertically, in order to escape the clusters. These fluctuations resulted in the eventual disintegration of the cholesterol clusters. A highly unusually flip-flop event of cholesterol across the DOPC bilayer was also observed, further demonstrating that cholesterol clusters are exceptionally unfavorable structures. Furthermore, the radial distribution functions of cholesterol hydroxyl oxygen to various headgroup atoms of PC reveal that the PC headgroups surrounding cholesterol have a clear tendency to reorient and extend toward cholesterol. This Umbrella Effect can influence the headgroup orientation of PCs that are 2 to 3 nm away from a cholesterol headgroup. Our results show that the Umbrella hypothesis is valid in both saturated and unsaturated lipid bilayers.
SIMULATION METHODS
Six independent 200 ns simulations were performed at 323 °K, with a time step of 2.0 fs. In the simulation of pure PC bilayers, there were 256 PC molecules in each leaflet. In the simulation of PC bilayers containing 20 mol % of cholesterol, the total number of lipids in each leaflet was also kept at 256, in which 204 were phospholipids and 52 were cholesterols. All systems contained 14656 water molecules.
The phospholipid force field was from Berger et al.20 and the force field for cholesterol was based on the GROMOS force field from Holtje et al.21 The simple point charge (SPC) model was used for water molecules. The force field parameter files for phospholipids were obtained from Dr. Peter Tieleman’s website and the file for cholesterol was obtained from the GROMACS website. The MD simulations were performed using GROMACS 4.0.322 and the analysis was done using the GROMACS tools as well as our own programs. The LINCS algorithm was used to keep the lengths of all bonds constant. The cut-off distance for Lennard-Jones interactions and electrostatic interactions were both set at 1.0 nm. Long-range electrostatic interactions were handled with the particle mesh Ewald (PME) method.23 All systems were run for 200 ns in the NPT ensemble using Berendsen’s thermostat and barostat with a coupling time constant of 0.1 ps and 1.0 ps, respectively. The pressure normal and parallel to the bilayer were coupled separately at 1 bar.
The final structure (i.e., run E) obtained from a previous simulation study24 was used as the starting point for constructing the DPPC systems. The original structure was a pure DPPC bilayer with 128 lipids (i.e., 64 lipids in each leaflet) and 3655 water molecules. Thirteen randomly chosen DPPC molecules in each leaflet were replaced with cholesterols so that the center of mass (COM) of cholesterol coincided with the COM of the replaced DPPC. Nearby molecules were translated to avoid bad contacts between atoms using VMD and energy minimization. This structure was first run for 200ns and was then enlarged four times, using the GROMACS tool “genconf”, to create the initial random DPPC system of 512 lipids. Additionally, another copy of the equilibrated 128-lipid random system was used to construct the cluster system. Cholesterol clusters were constructed by exchanging DPPCs near cluster sites with some distant cholesterols. Two clusters were built in each leaflet: one contained 6 cholesterols while the other contained 7 cholesterols. VMD and energy minimization were used again to eliminate bad contacts. Finally, this 128-lipid cluster system containing 2 cholesterol clusters in each leaflet was duplicated four times to obtain a 512-lipid system with 8 cholesterol clusters in each leaflet.
The initial structures for the DOPC systems were constructed differently. The pure DOPC bilayer was constructed by first obtaining the PDB coordinates file of a single DOPC molecule from the Dundee PRODRG2 Server.25 A single DOPC was placed inside a solvent box and a simulation was performed to relax the lipid. Afterwards, a bilayer of 128 DOPCs was built from this relaxed DOPC. After running the128 DOPC bilayer for a short period of time, “genconf” was used to create a bilayer of 512 DOPC. To construct the random system, 52 DOPC in each leaflet were randomly selected and replaced with cholesterol. During the replacement, the COM of cholesterol coincided with the COM of the replaced DOPC and the cholesterol orientation was chosen at random. The VMD program was then used to remove any bad links or overlaps that could happen after cholesterol insertion. For the cluster system, 8 randomly chosen DOPCs in each leaflet were first replaced with cholesterol and the system was run for a short period of time. 8 cholesterol clusters were then built by replacing DOPCs surrounding the original 8 cholesterols with other cholesterol using the VMD program. Four of the clusters had 7 cholesterols and the others had 6 cholesterols. All initial structures went through energy minimization using both the steepest descent and conjugate gradient algorithms; each ran for 10,000 steps to eliminate any bad contacts between atoms.
RESULTS AND DISCUSSION
Area Per Lipid
The area per lipid for a pure DPPC or DOPC bilayer arrived at a stable value almost immediately after the start of the simulation. This was largely for the reason that our initial structures were established using the final structures of previous simulations (see the Simulation Methods section). Initially, the area per lipid decreased sharply in the four systems containing 20 mol % cholesterol before becoming stable after 100 ns. The average values for the pure, random and cluster DPPC systems are 65.4±0.5, 48.1±0.4, and 48.3±0.5 Å2, respectively; the average values for the pure, random and cluster DOPC systems are 67.1±0.5, 51.9±0.4, and 51.9±0.4 Å2, respectively. These values were determined using the last 50 ns of simulation.
Dispersing of Large Cholesterol Clusters
Fig. 2 depicts the initial and final snapshots of a single leaflet of a bilayer in both the cluster and the random systems. For clarity, cholesterol is represented by space-filling models and water molecules are not shown. Upon considering the snapshots for the cluster systems, it becomes clear that by the end of the 200 ns simulation, the original, large cholesterol clusters, which were artificially arranged, have dispersed, primarily into cholesterol monomers. For ease of comparison, snapshots of the initial and final lateral distributions of both random systems are also shown in Fig. 2. By the end of the 200 ns simulation, cholesterol molecules are more uniformly distributed over the bilayer in the random systems than in the corresponding arranged cluster systems.
Figure 2.
The initial and final snapshots of the cholesterol distribution in the cluster and the random systems. Cholesterol (in yellow) is represented by the space-filling model. Only the lipids on the top leaflet are shown.
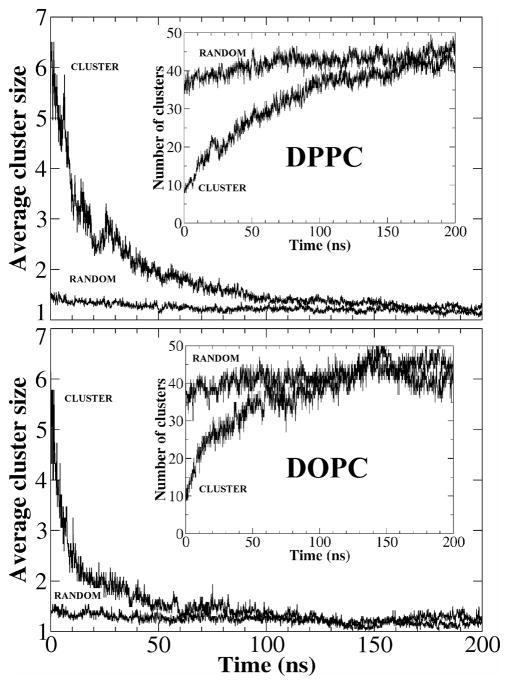
For the purposes of studying the time course of cluster size evolution, we performed a cluster size analysis. We arbitrarily defined a cluster counting criterion: if the distance between the centers of mass of two neighboring cholesterol molecules was less than 0.9 nm, the two molecules would be considered members of the same cluster. Cholesterol monomers were classified as a cluster of size 1. Fig. 3 shows the average cholesterol cluster size versus time in both the cluster and the random systems. Initially, the average cluster sizes of the two systems were very different: about 1.4 in the random system and about 6 in the cluster system. As the simulation progressed, the average cluster size in both random DPPC and DOPC systems only changed slightly, to a final value of 1.2. In contrast, the average cluster size in the artificially arranged cluster systems initially decreased sharply and then arrived at values similar to those of the random systems at 100–150 ns. The decrease in average cluster size appears to be slower in the DPPC system than in the DOPC system; however, this could have resulted from the different initial cluster arrangements: two 7-cholesterol clusters were closely paired in the DPPC system, almost as a single 14-cholesterol cluster, whereas the two clusters were more divided in the DOPC system (Fig. 2).
Figure 3.
The average cluster size vs. time for the cluster and the random systems. The inserts: The total number of cholesterol clusters vs. time in the two systems.
The total number of cholesterol clusters must increase as larger clusters disperse into smaller clusters. The inserts in Fig. 3 graph the total count of cholesterol clusters verse time in both the DPPC and the DOPC systems. Analogous to the decrease in cholesterol cluster size of the cluster systems, the total cluster count increases rapidly before approaching values that correspond to the cluster count of random systems, at 100 – 150ns.
Previous Monte Carlo simulations have shown that some clustering of cholesterol is expected in a PC bilayer containing 20 mol % of cholesterol, even when the mixing is ideal. Considering that in both the DPPC and the DOPC systems, all large cholesterol clusters dispersed into cholesterol monomers so quickly (<150 ns), and the randomly distributed cholesterol did not spontaneously reform into large clusters, it can be concluded that the artificially arranged cholesterol clusters are unfavorable structures, and that the general cholesterol-cholesterol interaction is a strongly repulsive one. If the concentration permits, cholesterol has a strong tendency remain as monomers in both saturated and unsaturated PC bilayers.
The angles at which cholesterol molecules faced each other in a cluster were randomly chosen (see SIMULATION METHODS section). The sterol ring structure of a cholesterol molecule contains a smooth face and a rough face. By visually inspecting the initial cluster systems, the numbers of cholesterol pairs having smooth-smooth, smooth-rough, and rough-rough contacts in the initial DPPC cluster system were determined to be 8, 4, and 8, respectively. Similarly, the number of cholesterol pairs having smooth-smooth, smooth-rough, and rough-rough contacts in the initial DOPC cluster system were 6, 4, and 2, respectively. We monitored the average distances between these cholesterol pairs as a function of time to determine which pair group is more stable than others. As expected, all three average distances increased with time as cholesterol clusters dispersed. Nonetheless, there is no clear evidence that any particular pair group is more stable than another (data not shown). This result should be noted with caution as we did not attempt to construct the pairs in the most stable fashion. It is entirely possible that a certain pair type is more stable at a higher cholesterol concentration, at which cholesterol clustering cannot be avoided.
Frustum Shaped Cholesterol Clusters
The Umbrella model proposes that cholesterol clusters in a lipid bilayer are unfavorable because it is difficult for neighboring PC headgroups to effectively shield cholesterol’s large hydrophobic body from water, most notably for cholesterols at the center of a cluster. Thus, in the systems with artificially arranged clusters, the cholesterol molecules actively dispersed into surrounding PCs. The substantial cholesterol clustering created in the initial stages of the simulation granted us an opportunity to observe the interactions between the bilayer and the clusters at the molecular level, thereby illuminating the nature of the unfavorable cholesterol – cholesterol interaction.
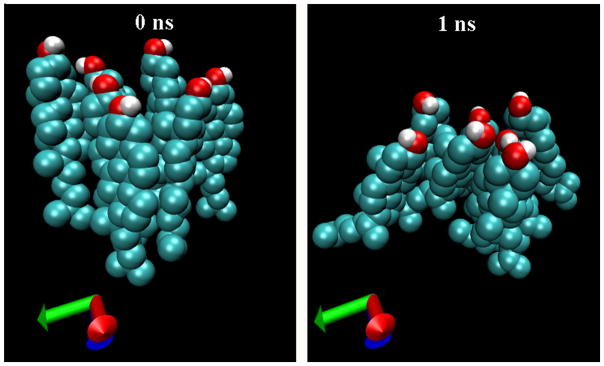
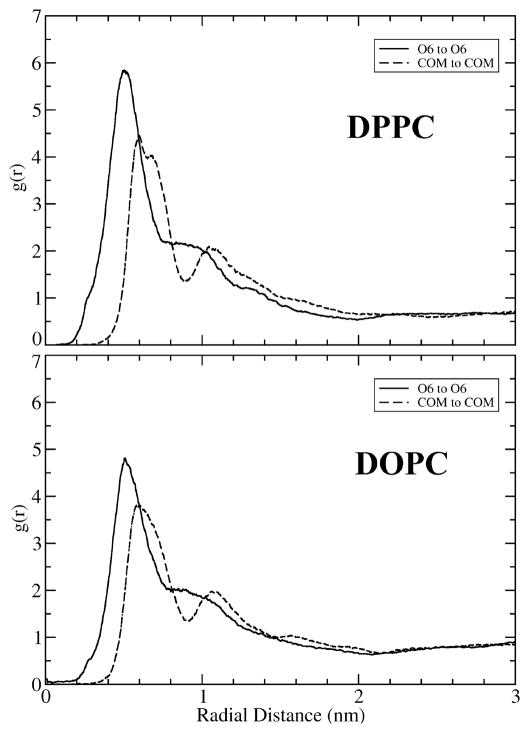
Promptly after the start of the simulation, the hydroxyl headgroups of the clustered cholesterol gathered together in a pinching action, causing their large, bulky sterol rings to fan out within the bilayer, creating a ‘frustum’ (truncated cone) shaped cluster. The bunched hydroxyl headgroups, which face the aqueous phase, form the top of the frustum, while the larger sterol rings, situated in the hydrophobic core of the membrane, spread apart to form the base. Fig. 4 illustrates the shape transition of a particular cholesterol cluster in the DOPC cluster system. Seven cholesterol molecules were artificially grouped to form the cluster. The closely packed hydrophobic bodies were parallel to one another at the start of the simulation. Strikingly, the ‘frustum’ shape depicted in Fig 4 is formed just 1ns after the simulation began. In both the DPPC and DOPC systems, numerous ‘frustum’ shaped clusters were observed in the early stages of simulation. It is intriguing to note that a number of inverse examples can also be found among the snapshots; therefore, statistical data are needed in order to confirm the observation of frustum shaped clusters. For this purpose, two radial distribution functions (RDFs) were computed: one between cholesterol hydroxyl oxygen (O6) atoms, and a second between centers of mass (COM) of cholesterol molecules. The center of mass of cholesterol is located near the center of the sterol rings. If there were no tendency for the headgroups of clustered cholesterol to gather closer together than their sterol bodies, the two RDFs should overlap. However, if the observation of frustum shaped clusters has statistical significance, then RDF between hydroxyl O6 atoms should have a higher density at short distances than the COM-to-COM RDF. Fig. 5 shows the two RDFs over the first 20 ns in the simulation of the DPPC and the DOPC cluster systems; during these 20 ns, many cholesterol clusters remain intact. Clearly, the two RDFs are very different at short distances. In the DPPC cluster system, the headgroup O6-to-O6 RDF has a large sharp peak at about 0.5 nm, while the body COM-to-COM RDF has a smaller peak at about 0.7 nm. Additionally, the magnitude of O6-to-O6 RDF is significantly greater than that of the COM-to-COM RDF for distances < 0.7 nm. Therefore, the statistical data clearly supports the conclusion that cholesterol clusters do generally have a frustum shape. We were unable to study the shape of cholesterol clusters in the late stages of simulation as an insufficient number of cholesterol clusters remained. This was the case even in the cluster systems. Nonetheless, a degree of cholesterol clustering in a lipid membrane of high cholesterol content cannot be avoided. It would be interesting to investigate whether these clusters also adopt a frustum shape.
Figure 4.
Left: An artificially arranged cholesterol cluster in DOPC cluster system at the start of the simulation. Right: At 1ns, the cluster has already adopted the frustum shape.
Figure 5.
The radial distribution functions (RDF) in the cluster systems over the first 20 ns. Solid line: The RDF between cholesterol oxygen O6. Dashed line: The RDF between the center of mass (COM) of cholesterol.
The hydrophobic interaction is the driving force behind the tendency for cholesterol to adopt a frustum shape. As cholesterol forms a cluster, it is very difficult, and sometimes impossible, for neighboring PC headgroups to fill the spaces between cholesterol headgroups. Water is then able penetrate through the gaps between cholesterol headgroups, and make unfavorable contact with the hydrophobic body of cholesterol. The pinching action of cholesterol headgroups mitigates the situation by reducing the gaps, thereby decreasing the unfavorable water penetration. Ultimately, unfavorable cholesterol clusters disperse into cholesterol monomers through lateral diffusion, as shown in Fig. 2 and 3; however, this is a relatively time-consuming process (> 100 ns). The formation of ‘frustum’-shaped cholesterol clusters is the system’s immediate response (< 1ns) to the unfavorable situation.
Location of Cholesterol Clusters
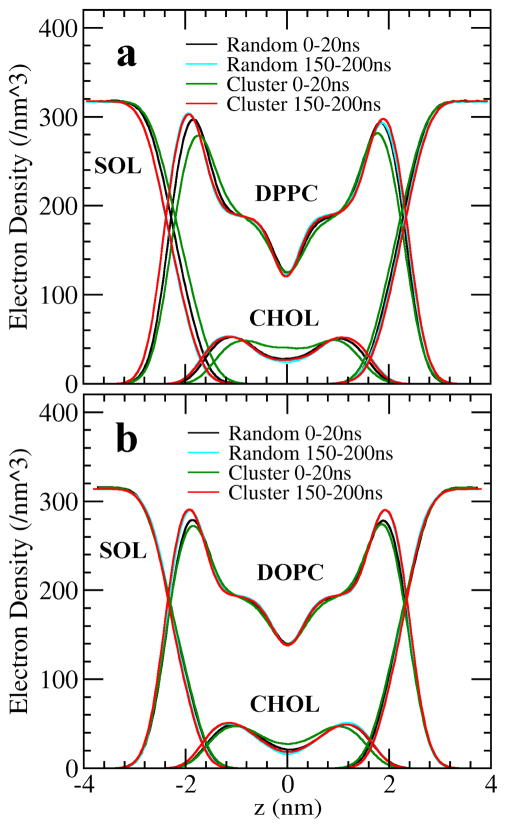
Along with adopting a frustum conformation, cholesterol clusters also showed a clear tendency to bury themselves deeper into the lipid bilayer than a cholesterol monomer does. In numerous cases, the cholesterol tails even burrowed into the opposite leaflet of DOPC or DPPC bilayers. Fig. 6 depicts the electron density profiles of cholesterol, PC and water across the bilayer in both cluster and random systems. During the first 20 ns of simulation, the cholesterol electron density in the cluster systems (green curves) is distributed closer to the bilayer center (i.e., z = 0) than in other distributions. This phenomenon was more pronounced in the DPPC bilayer. We calculated the average locations of PC’s phosphate residue and of cholesterol’s hydroxyl in the direction of the bilayer normal, and discovered that relative to PC’s phosphate, the average position of cholesterol hydroxyls in a cholesterol cluster is approximately 2 Å deeper into the bilayer interior than the average position of cholesterol monomers in the random system. The average position of clustered cholesterol gradually rose higher in the bilayer, as the clusters dispersed into monomers. During the final 50 ns of simulation, the difference in the electron density profiles between the cluster system (red curves) and the random system (cyan curves) became very small. It appears that cholesterol clusters try to move away from the aqueous phase and that by burying cholesterol clusters deeper into the bilayer interior, neighboring PCs can cover the outskirts of the frustum-shaped clusters, in an “Umbrella action”, more effectively (Fig. 1C).
Figure 6.
The electron density profiles of PC, cholesterol and solvent across the bilayer in the cluster and the random systems.
An Unusual Cholesterol Flip-Flop Event
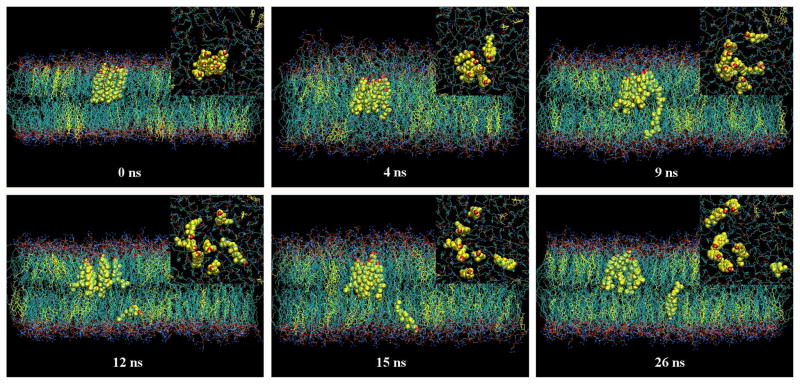
A peculiar flip-flop of a cholesterol molecule in a cluster was observed in the simulation of 20 mol % of cholesterol in a DOPC bilayer. Fig. 7 shows the time sequence snapshots of the event. The inserts in Fig. 7 are corresponding snapshots of the membrane surface when viewed from the top aqueous phase. For purposes of clarity, water molecules are not shown and only cholesterol molecules of the cluster in question are represented by the space-filling model. The initial arrangement of the cluster is shown in the snapshot at 0 ns. At 4ns, the cluster has clearly adopted a frustum conformation and has submerged approximately 4 Å into the lipid bilayer from its initial position. Cholesterol in the cluster fluctuated rapidly in the direction of the bilayer normal; one cholesterol molecule was buried nearly halfway into the opposite leaflet of the bilayer. At 9 ns, that particular cholesterol molecule had submerged completely into the opposite leaflet with an ‘upside down’ orientation: the headgroup was at the center of the bilayer, while the hydrocarbon tail was exposed to the aqueous surface. From 12 ns to 26 ns, the orientation of the cholesterol molecule fluctuated back and forth, eventually completing the flip-flop.
Figure 7.
The snapshots of an unusual flip-flop of cholesterol across the DOPC bilayer in the cluster system. Inserts: the snapshots of the membrane surface viewed from the top aqueous phase. 0 ns: An artificially arranged cholesterol cluster at the start of the simulation. 4 ns: The cluster adopted the frustum shape and also moved deeper into the bilayer. One cholesterol molecule was half way into the bottom leaflet. 9 ns: The cholesterol was completely inside the bottom leaflet, but was also up side down. 12 to 26 ns: The cholesterol completed the flip entirely within the bottom leaflet. At the same time, the cluster dispersed into smaller pieces.
This cholesterol flip-flop event is noteworthy for several reasons. First, it has been reported that no cholesterol flip-flop has ever been observed in atomistic molecular dynamics simulations of a DPPC or a DOPC bilayer, making this event as particularly unique.26 The estimated half-time for such a cholesterol molecule flip-flop ranged from seconds to hours.27 It follows that the chances of observing a flip-flop in a small system during a 200 ns simulation is exceptionally low. Second, unlike other observed flip-flops, the motions of the flip and the flop were completely decoupled from each other. The cholesterol molecule did not gradually rotate and lay flat at the center of the bilayer, as expected in a normal membrane flip-flop.13,26,28 Instead, the cholesterol molecule submerged directly into the destination leaflet without changing orientation. When fully integrated into the destination leaflet, the cholesterol molecule completed the flip-flop and adopted the proper orientation. In a normal cholesterol flip-flop, a cholesterol molecule is surrounded by PC molecules. As the cholesterol gradually rotates toward the center of the bilayer, the cavity created by the rotation at the bilayer-aqueous interface is quickly filled by surrounding PCs. This can be clearly seen in the snapshots of normal cholesterol flipflop in other studies.13,26,28 However, there are few surrounding PCs in a cholesterol cluster, and the cavity created by a rotating cholesterol would not be easily covered by other cholesterols, as cholesterol is a largely hydrophobic molecule. Additionally, the surrounding PC molecules would not be physically close enough to fill the cavity. Thus, a higher free energy penalty would result if a cholesterol molecule within a cluster flip-flopped through a bilayer in the normal fashion. Submerging directly into the other leaflet without changing headgroup orientation could be a quick manner to complete the flip-flop with a low free energy cost.
There have been a number of reports on sterol flip-flops in other bilayer systems. Kucerka et al. have observed several cholesterol flip-flops in short chain diC14:1PC bilayers, which are more disordered, have higher molecular tilts and greater lateral area.13 Rog et al. showed that replacing the cholesterol hydroxyl group with a ketone group can result in some flip-flops of ketosterone across DPPC bilayers.26 Ketone groups are less hydrophilic than hydroxyl groups are, and produce a greater tilt angle for ketosterone with respect to the membrane normal. These factors facilitate ketosterone flip-flops. In both cases, flip-flops were encouraged by destabilizing the sterol from the normal “upright” position, causing the sterol to lie flat at the center of bilayer for a certain period of time during the flip-flop. The flip-flop event we observed in the DOPC bilayer was an unusual event and was promoted by artificially arranging cholesterol into an unfavorable situation: cholesterol clusters. In such a cluster, it is very difficult for a bilayer system to shield the large, bulky hydrophobic body of cholesterol from water. In response, clustered cholesterol molecules fluctuate rapidly in all directions in order to escape the cluster. The flip-flop event we observed is a quick way to escape. This situation bears similarity to cholesterol in a polyunsaturated PC bilayer. Compared to monounsaturated PCs, polyunsaturated PCs are less suitable as “umbrella providers” for cholesterol. The multiple double bonds in the acyl chains prevent cholesterol from approaching a polyunsaturated PC. Ergo, such a bilayer cannot effectively cover cholesterol. It has been reported that in a di20:4PC bilayer, cholesterol is not in an “upright” orientation, but instead lies flat in the middle, between the leaflets.29 A recent coarse grained simulation also confirmed that a large population of cholesterol lies flat between the leaflets in di20:4PC bilayer.28 However, it has been shown experimentally that adding either 40 mol% POPC or just 5 mol% di14:0PC (DMPC) reverted cholesterol to the upright orientation.30 POPC has one saturated chain and one monounsaturated chain; therefore, POPC should be considered a good “umbrella provider”. Similarly, DMPC has two saturated chains, and is an even better umbrella provider. These finding demonstrate that the orientation of cholesterol in the normal ‘upright’ position is directly related to whether the cholesterol can be effectively covered in that position. The flip-flop event is a clear indicator that cholesterol in clusters cannot be effectively covered and that cholesterol clusters are therefore an unfavorable arrangement.
The Umbrella Effect
In order to provide partial coverage for its large hydrophobic body, cholesterol’s small hydroxyl group can form hydrogen bonds with water molecules at the bilayer-aqueous interface. A unique aspect of the Umbrella Model is that it specifically predicts that headgroups of neighboring PCs should rearrange in order to assist cholesterol in covering the rest of its hydrophobic body.6,7 Therefore, PC headgroups are expected to relocate closely around cholesterol hydroxyl groups, and thus cover the hydrophobic body of cholesterol. Additionally, some form of preferred PC headgroup orientation is expected for PCs close to cholesterol.
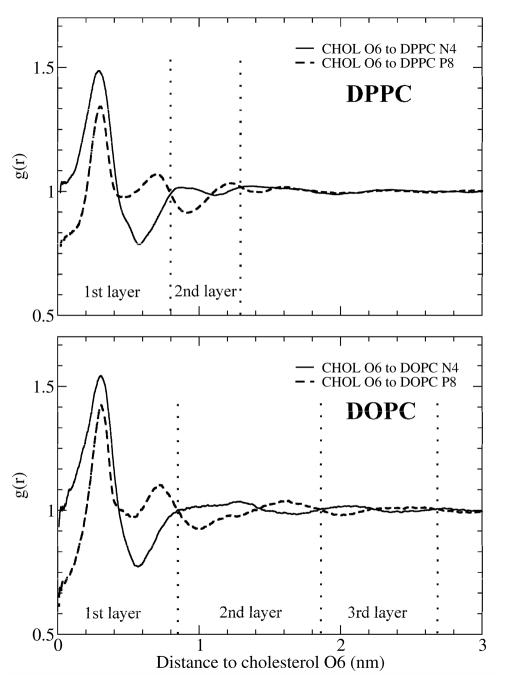
As discussed earlier, discoveries that are solely based on snapshot examples may not be reliable. In order to confirm or else invalidate the Umbrella Model, we must seek statistical data. For this purpose, two 2D RDFs were calculated in the plane parallel to the bilayer surface: one between the cholesterol hydroxyl oxygen (O6) and the nitrogen of PC’s choline group (N4); and a second between cholesterol hydroxyl oxygen (O6) and PC’s phosphate residue (P8). If the orientation of PC headgroups were not affected by the presence of cholesterol, the two RDFs would overlap. Fig. 8 shows the 2D, O6-N4 and O6-P8 RDFs for the random systems over the last 50 ns of the simulation. As depicted in Fig. 4, the systems were relaxed and stable during this period. In Fig. 8, we see clear evidence of the reorientation of PC headgroups surrounding cholesterol. In DOPC/cholesterol bilayers, the density of N4 has a large, single peak at 0.3 nm from a cholesterol hydroxyl O6; the density of P8 has a smaller peak at about 0.3 nm and another smaller, but broader, peak at about 0.7 nm. The two RDFs first cross at 0.4 nm from a cholesterol hydroxyl O6, and cross once again at 0.85 nm. The data indicate that either N4 or P8 can get quite close to the cholesterol hydroxyl O6. Correspondingly, within 0.4 nm of a cholesterol hydroxyl O6, the density of PC nitrogen (N4) is statistically significantly greater than the density of PC phosphate (P8). In contrast, at distances between 0.4 nm and 0.85 nm, the density of PC phosphate is greater. Because each PC headgroup has one nitrogen and one phosphate, and the area per lipid in this DOPC/cholesterol bilayer is 51.9 Å2, the RDFs indicate that the first layer of PC surrounding cholesterol have a statistical preference to orient their headgroups (i.e., the P-N vectors) radially inward toward cholesterol. Furthermore, judging by the distances, the PC headgroups are closely packed around cholesterol’s hydroxyl group and above the large, hydrophobic body of cholesterol. These findings are in favorable agreement with the predictions of the Umbrella Model.
Figure 8.
RDFs over the last 50 ns in the random system. Solid line: The RDF between cholesterol oxygen O6 and the nitrogen atom of PC choline group. Dashed line: The RDF between cholesterol oxygen O6 and PC phosphate residue P8.
A rigorous examination of Fig. 8 also reveals that the alternating change in magnitude of O6-N4 and O6-P8 RDFs occurred at least five times in the DPPC/cholesterol system, and six times in the DOPC/cholesterol system before the two RDFs overlapped with each other. The implication of this is that cholesterol can affect PC headgroup orientation up to a distance of 2 or 3 nm, and that PC headgroups form a layered structure around cholesterol hydroxyls, like the rings of a tree. This is an important new finding. Understandably, the difference in RDF magnitude decreases with distance from a cholesterol hydroxyl. Thus, the ordering effect is largest for the first layer of nearby PC headgroups, and becomes progressively smaller for the second and the third layers of PC headgroups. This begs the question of how cholesterol can affect the orientation of PC headgroups in the second or the third layer. Clearly, only the PCs in the first layer can physically participate in the covering action; however, this action requires a preferential orientation of PC headgroups in the first layer. This ordering of PC headgroups in the first layer also induces a weaker ordering of orientation in PC headgroups farther away, possibly through the electric dipole interactions between PC headgroups. The induced ordering is gradually relaxed over distance, which resulted in the alternating magnitude change of O6-N4 and O6-P8 RDFs in Fig. 8.
We can define the range of the Umbrella Effect as the distance from cholesterol O6 at which the two above RDFs overlap. In Fig. 8, it is interesting that the range of the Umbrella Effect in the DPPC bilayer is shorter (~1.6 nm) than the range in the DOPC bilayer (~2.7 nm). Several factors could contribute to the extended range of umbrella effect in unsaturated PC bilayers. First, as headgroups of DOPC in the first layer cover a cholesterol molecule, the cis double bonds in DOPC chains make close contact with the cholesterol sterol body more difficult; consequently, this could induce a packing stress in the acyl chain region. This packing stress also needs to be relaxed over distance. Additionally, the area per lipid for the DOPC bilayer is larger than that of the DPPC bilayer. In other words, each DOPC headgroup must cover more lateral area than a DPPC headgroup does. These constraints on DOPC could make DOPC less efficient in relaxing the ordering of headgroup orientation, resulting in a longer range of the Umbrella Effect in DOPC bilayers. On the other hand, saturated PCs, such as DPPC, are better umbrella providers and can cover cholesterol more effectively. Less packing constraints make saturated PCs more effective in relaxing the headgroup orientation order over distance. Thus, the range of the Umbrella effect is shorter in saturated PC bilayers. We also computed the 3D RDFs of O6-N4 and O6-P8 (data not shown). The Umbrella Effect appears more obvious in the 2D RDFs than in 3D RDFs. This is likely because 2D RDFs reduce the P-N vector fluctuation in the direction of bilayer normal.
Another research group has previously investigated the Umbrella Effect. Kucerka et al. first separated PCs surrounding cholesterol in diC22:1PC or diC14:1PC bilayers into first, second, and third nearest neighbor groups.13 The 2D RDFs of O6-N4 and O6-P8 were calculated for each group. They showed that the Umbrella Effect is largest for the first nearest neighbor PCs and smaller for the second nearest neighbors. However they also concluded that no Umbrella Effect can be detected for the third nearest-neighbor PCs. A snapshot that clearly captures two PC headgroups in “Umbrella action” was shown in their paper. It should be noted that they classified PCs into groups according to the actual distance from each cholesterol headgroup. If there were three PCs within 0.85 nm from a cholesterol headgroup, they would classify the three PCs according to distance, as the first, the second and the third nearest neighbor. However, in our analysis, we would placel all three PCs in the first layer of PCs from a cholesterol headgroup. Consequently, their first, second and third nearest neighbor PCs are subsets of our first layer PCs. Therefore, their conclusion is in good agreement with our result: there is a large Umbrella Effect for the first layer of PCs surrounding a cholesterol headgroup. Additionally, our data indicates that the range of the umbrella effect (i.e., headgroup orientation order) is farther than what has been previously reported: it can extend up to 2–3 nm (or 2 to 3 lipid distance) from a cholesterol headgroup.
CONCLUSIONS
Our results demonstrate that cholesterol clusters are very unstable in DPPC and DOPC lipid bilayers containing 20 mol % of cholesterol, and that cholesterol-cholesterol interaction is generally unfavorable (repulsive). During 200 ns simulations of DPPC and DOPC bilayers, a very brief period when compared to traditional ‘wet’ experiments, artificially arranged cholesterol clusters dispersed into cholesterol monomers. At the outset of the simulations, numerous statistical quantities, such as average cluster sizes and electron density profiles, differed substantially between the random and the cluster systems; however, these differences significantly diminished by the end of the simulation. Additional indicators of cholesterol cluster instability arise from observations that cholesterol clusters are often frustum shaped, are buried deeper in the lipid bilayer than cholesterol monomers are, and can even cause an unusual cholesterol flip-flop across the DOPC bilayer. These results suggest that cholesterol cluster instability stems from the inability of the bilayer system to cover the bulky hydrophobic bodies of cholesterol in a cluster. Among the three major models of cholesterol-lipid interaction, only the Umbrella Model suggests that the unfavorable cholesterol-cholesterol interaction is attributed to the hydrophobic effect. Necessarily, a frustum shaped cholesterol cluster sitting deep in a lipid bilayer must create some degree membrane deformation and lipid packing stresses, which are highly unfavorable. For the system to respond in this manner clearly implies that it has done so only to avoid an even more unfavorable situation i.e., large cholesterol clusters in their “normal” shape and position in the bilayer. In cell plasma membranes, local cholesterol clusters can form during natural membrane turnover, as newly synthesized cholesterol is delivered to cell membranes via vesicle fusion. Our results indicate that these cholesterol clusters could be quite short-lived: dissipating almost immediately upon formation. A more accurate estimation of cluster lifetime should be made for a larger multi-component bilayer system, where the heterogeneous environment of biomembranes can be mimicked.
In addition, our results show that the Umbrella Effect of cholesterol exists in both saturated DPPC and unsaturated DOPC bilayers. The radial distribution functions of cholesterol hydroxyl oxygen to various headgroup atoms of PC reveal that the PC headgroups surrounding cholesterol have a clear tendency to reorient and extend toward cholesterol’s hydroxyl group. The orientation of the PC headgroups has a layered structure, which can extend 2 to 3 nm from a cholesterol molecule. The results of this study strongly support the Umbrella Model, and provide new insights into the fascinating mechanics of cholesterol-lipid interactions.
Acknowledgments
This work was supported by the National Institute of Health grant 1 R01 GM077198-01A1 subaward 49238-8402 and National Science Foundation grant MCB-0344463. The authors thank Dr. Gerald W. Feigenson for critical reading of the manuscript, and Mr. Charlie Huang for assistance in editing. The authors also acknowledge the High Performance Computing Center (HPCC) at Texas Tech University for providing HPC resources for this work.
References
- 1.Feigenson GW. Biochim Biophys Acta. 2009;1788:47. doi: 10.1016/j.bbamem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mesmin B, Maxfield FR. Biochim Biophys Acta. 2009;1791:636. doi: 10.1016/j.bbalip.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radhakrishnan A, McConnell HM. Biophys J. 1999;77:1507. doi: 10.1016/S0006-3495(99)76998-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Somerharju P, Virtanen JA, Cheng KH, Hermansson M. Biochim Biophys Acta. 2009;1788:12. doi: 10.1016/j.bbamem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Chong PL. Proc Natl Acad Sci U S A. 1994;91:10069. doi: 10.1073/pnas.91.21.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang J, Feigenson GW. Biophys J. 1999;76:2142. doi: 10.1016/S0006-3495(99)77369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang J. Methods Enzymol. 2009;455:329. doi: 10.1016/S0076-6879(08)04212-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pata V, Dan N. Biophys J. 2005;88:916. doi: 10.1529/biophysj.104.052241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radhakrishnan A, McConnell HM. Biochemistry. 2000;39:8119. doi: 10.1021/bi0005097. [DOI] [PubMed] [Google Scholar]
- 10.Parker A, Miles K, Cheng KH, Huang J. Biophys J. 2004;86:1532. doi: 10.1016/S0006-3495(04)74221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ali MR, Cheng KH, Huang J. Proc Natl Acad Sci U S A. 2007;104:5372. doi: 10.1073/pnas.0611450104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berkowitz ML. Biochim Biophys Acta. 2009;1788:86. doi: 10.1016/j.bbamem.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Kucerka N, Perlmutter JD, Pan J, Tristram-Nagle S, Katsaras J, Sachs JN. Biophys J. 2008;95:2792. doi: 10.1529/biophysj.107.122465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niemela PS, Hyvonen MT, Vattulainen I. Biochim Biophys Acta. 2009;1788:122. doi: 10.1016/j.bbamem.2008.08.018. [DOI] [PubMed] [Google Scholar]
- 15.Pandit SA, Chiu SW, Jakobsson E, Grama A, Scott HL. Langmuir. 2008;24:6858. doi: 10.1021/la8004135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z, Bhide SY, Berkowitz ML. J Phys Chem B. 2007;111:12888. doi: 10.1021/jp074037i. [DOI] [PubMed] [Google Scholar]
- 17.Zhu Q, Cheng KH, Vaughn MW. J Phys Chem B. 2007;111:11021. doi: 10.1021/jp070487z. [DOI] [PubMed] [Google Scholar]
- 18.Pitman MC, Suits F, Mackerell AD, Jr, Feller SE. Biochemistry. 2004;43:15318. doi: 10.1021/bi048231w. [DOI] [PubMed] [Google Scholar]
- 19.de Meyer F, Smit B. Proc Natl Acad Sci U S A. 2009;106:3654. doi: 10.1073/pnas.0809959106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berger O, Edholm O, Jahnig F. Biophys J. 1997;72:2002. doi: 10.1016/S0006-3495(97)78845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holtje M, Forster T, Brandt B, Engels T, von Rybinski W, Holtje HD. Biochim Biophys Acta. 2001;1511:156. doi: 10.1016/s0005-2736(01)00270-x. [DOI] [PubMed] [Google Scholar]
- 22.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. J Comput Chem. 2005;26:1701. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 23.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. Journal of Chemical Physics. 1995;103:8577. [Google Scholar]
- 24.Tieleman DP, Berendsen HJC. Journal of Chemical Physics. 1996;105:4871. [Google Scholar]
- 25.Schuettelkopf AW, van Aalten DMF. Acta Crystallographica. 2004;D60:1355. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 26.Rog T, Stimson LM, Pasenkiewicz-Gierula M, Vattulainen I, Karttunen M. J Phys Chem B. 2008;112:1946. doi: 10.1021/jp075078h. [DOI] [PubMed] [Google Scholar]
- 27.Sapay N, Bennett WFD, Tieleman DP. Soft Matter. 2009;5:3295. [Google Scholar]
- 28.Marrink SJ, de Vries AH, Harroun TA, Katsaras J, Wassall SR. J Am Chem Soc. 2008;130:10. doi: 10.1021/ja076641c. [DOI] [PubMed] [Google Scholar]
- 29.Harroun TA, Katsaras J, Wassall SR. Biochemistry. 2008;47:7090. doi: 10.1021/bi800123b. [DOI] [PubMed] [Google Scholar]
- 30.Kucerka N, Nieh MP, Marquardt D, Harroun TA, Wassall SR, Katsaras J. Journal of Physics: Conference Series. 2009 [Google Scholar]