Abstract
Antigen-specific T cell cytokine expression is dictated by the context in which T cell receptor (TCR) engagement occurs. Recently it has become clear that epigenetic changes play a role in this process. DNA Methyltransferase-3a (DNMT3a) is a de novo methyltransferase important to the epigenetic control of cell fate. We have determined that DNMT3a expression is increased following TCR engagement and that costimulation mitigates DNMT3a protein expression. T cells lacking DNMT3a simultaneously express IFN-γ and IL-4 after expansion under non-biasing conditions. While global methylation of DNA from WT and KO T cells is similar, DNMT3a null T cells demonstrate selective hypomethylation of both the Il4 and Ifng loci after activation. Such hypomethylated KO Th2 cells retain a greater capacity to express IFN-γ protein when they are subsequently exposed to Th1-biasing conditions. Based on these findings we propose that DNMT3a is a key participant in regulating T cell polarization at the molecular level by promoting stable selection of a context specific cell fate through methylation of selective targets in T cells.
This is an author-produced version of a manuscript accepted for publication in The Journal of Immunology (The JI). The American Association of Immunologists, Inc. (AAI), publisher of The JI, holds the copyright to this manuscript. This version of the manuscript has not yet been copyedited or subjected to editorial proofreading by The JI; hence, it may differ from the final version published in The JI (online and in print). AAI (The JI) is not liable for errors or omissions in this author-produced version of the manuscript or in any version derived from it by the U.S. National Institutes of Health or any other third party. The final, citable version of record can be found at www.jimmunol.org.
Keywords: Th1/Th2 Cells, Cell Differentiation, Gene Regulation
Introduction
The fate of a T cell after encounter with antigen is critically dependent on the context in which activation occurs. The particular cytokines present during activation promote expansion and differentiation of T cells with restricted and stable patterns of selective cytokine expression. Such specification of cell fate is strongly associated with induction of particular transcription factors by local cytokines, e.g. T-bet, GATA-3, ROR-γt, and FOXP3 are critical to Th1, Th2, Th17, and Treg differentiation, respectively (1–4). But epigenetic changes through chromatin remodeling are also important to the stability of the differentiated phenotype (5).
Epigenetic regulation is in part controlled by DNA methylation at CpG dinucleotides near transcription initiation sites, resulting in a chromatin structure termed heterochromatin that represses transcription. DNA methylation is catalyzed by DNA Methyltransferases (DNMTs) including the maintenance methyltransferase DNMT1 that acts on hemi-methylated substrates to maintain methylation patterns after DNA replication and the de novo methyltransferases DNMT3a and DNMT3b that catalyze methylation of unmethylated DNA (6). The structurally related DNMT3L lacks intrinsic catalytic activity; however it can directly bind to DNMT3a and DNMT3b, increasing their catalytic activity (7). Interestingly, DNMT3a also appears to play a role in the active demethylation of DNA at some promoters during cyclical demethylation and remethylation that occur surrounding transcription of these genes (8).
Alterations in DNMT expression that correlate with changes in genomic DNA methylation are well described in many cancers, including leukemias. Mizuno et al. reported that DNMT1, DNMT3a, and DNMT3b transcripts were all overexpressed in AML and acute phase CML compared to normal bone marrow or chronic phase CML. Furthermore, the cell cycle regulator p15(INAK4B) was methylated to a greater extent in cells with higher DNMT1 or DNMT3B expression (9). More recently, several reports have examined genomic methylation and DNMT expression in autoimmune disease. In ITP patients, Tao et al. found decreased DNMT3a and DNMT3b transcripts in peripheral blood mononuclear cells compared to healthy controls (10). Luo et al. specifically examined T cell methylation and DNMT expression in patients with lupus and found that CD4 T cells had hypomethylation that was associated with lower DNMT1 and DNMT3a transcript levels (11). While Balada et al. also observed genomic hypomethylation in CD4 T cells in their cohort of lupus patients, they did not find any consistent changes in DNMT transcript levels (12). Taken together, these studies suggest that altering genomic methylation through modulation of DNMT expression may be a factor contributing to multiple diseases.
Detailed examination of the DNA methylation patterns in T cells have been reported for the prototypical Th1 cytokine gene Ifng and the Th2 cytokine gene cluster consisting of Il4, Il13, and Il5. In naïve CD4 T cells, the Ifng locus is largely demethylated and remains so following activation under Th1 biasing conditions. However, under Th2 biasing conditions, it acquires de novo methylation as the locus is silenced (13–17). In contrast, the Il4 locus is largely methylated in naïve CD4 T cells and remains so under Th1 biasing conditions. It is only under Th2 biasing conditions that the Il4 locus becomes progressively demethylated, favoring enhanced expression of IL-4, IL-5, and IL-13 (18–21). Such an inhibitory effect of genomic methylation on cytokine expression is supported by the observation that 5-aza-2-deoxycytidine, which inactivates DNMT1 and leads to progressive loss of DNA methylation, enhances expression of both IFN-γ and IL-4 in cultured T cells (19). Furthermore, T cell specific deletion of DNMT1, which results in profound global genomic DNA hypomethylation, leads to increased and promiscuous T cell cytokine expression (22). Mice that lack MBD2, whose gene product binds to methylated DNA and helps partition methylated genes into silent chromatin, share a similar phenotype with T cells that co-express IFN-γ and IL-4 (23).
A potential role for the de novo DNMTs, DNMT3a and DNMT3b, in regulation of T cell cytokine expression remains incompletely defined. DNMT3a is reported to localize at the Ifng and Il18r1 promoters in T cells (13, 24). In other cell types it is reported to localize at the Fgf-1, Ssx2ip, Hmga1, and Wrnip genes (25, 26). Association at these sites is correlated with increased methylation and decreased target gene expression. Mice globally deficient in DNMT3a, DNMT3b, or both have been reported; DNMT3a KO mice are runted after birth and do not survive past 4 weeks of age, while the DNMT3b and double KO mice are embryonic lethal (27). Hematopoetic stem cell precursors from such mice are capable of generating T cells in lethally irradiated recipients, however functional characterization of such T cells has not been described (28).
Given the dynamic changes in methylation identified at T cell cytokine gene loci and the apparent correlation between methylation and decreased gene expression, it was of interest when we observed that expression of one DNMT family member is regulated by TCR signaling. Our laboratory previously reported a microarray analysis that compared unstimulated T cells with cells stimulated with anti-CD3 alone or in combination with a variety of TCR signaling inhibitors to better understand regulation of T cell fate after activation (29). These experiments demonstrated a selective 38-fold upregulation of DNMT3a transcription after TCR signaling. This pattern of TCR-regulated de novo methyltransferase activity made DNMT3a a strong candidate for participation in the acquired methylation described in activated and differentiated T cells (5). By utilizing T cells lacking DNMT3a expression, we demonstrate that TCR-induced DNMT3a is required for establishing normal patterns of DNA methylation at the Ifng locus under Th2 biasing conditions. Such methylation appears dispensable for directing the appropriate patterns of IFN-γ and IL-4 expression in Th1 and Th2-skewed cells. However, when DNMT3a−/−Th2 cells are placed into Th1 biasing conditions, they are able to produce increased amounts of IFN-γ compared to WT Th2 cells. These data support DNMT3a as a critical participant in the stability of T cell gene expression via DNA methylation.
Materials and Methods
Mice
B10.A mice and B10.A/AiTac-[Tg]TCRCyt5CC7-I-[KO]Rag2 mice were purchased from Taconic. DNMT3a−/− mice and DNMT3a 2loxP mice were provided by E. Li (Novartis Pharmaceuticals, Cambridge, Massachusetts). DNMT3a−/− mice were bred from C57.B6/129 onto 6.5 TCR-transgenic Thy-1.1+ B10.D2 mice, a gift from C. Drake (Johns Hopkins University, Baltimore, Maryland). DNMT3a 2loxP mice were bred onto CD4 Cre 6.5 TCR-transgenic Thy-1.1+ B10.D2 mice, also a gift from C. Drake (Johns Hopkins University, Baltimore, Maryland). All animal protocols were approved by the Institutional Animal Care and Use Committee of Johns Hopkins University (Baltimore, Maryland).
Cells, antibodies and reagents
A.E7, a CD4+ T helper type 1 clone specific for PCC(81–104), was grown as described (30). Naïve CD4 T cells were isolated from pooled spleen and lymph nodes of 5C.C7 Rag2−/− mice using the CD4 T cell Isolation Kit (Miltenyi Biosciences). Previously activated CD4 T cells were generated by culturing 5C.C7 splenocytes with 5 μM PCC (Sigma) for 48 hours, then resting cells for 5 to 7 days in media containing 1 ng/ml IL-2 (Invitrogen). These antigen-experienced cells were then purified over a Ficoll gradient (GE Healthcare), yielding >98% CD4+CD44+CD62L− T cells. For activation, 107 A.E7 or 5C.C7 cells per well were cultured in a six-well plate coated with anti-CD3 (2C11; BD PharMingen) at 1 μg/ml. Indicated cultures were supplemented with anti-CD28 (37.51; BD PharMingen) at 10 μg/ml. Culture of CD4 T cells lacking DNMT3a was performed by pooling spleen and lymph nodes from 2½ to 3½-week-old DNMT3a−/− mice or age matched littermates, purifying CD4 T cells by negative selection (Miltenyi Biosciences), and culturing 2×106 cells with 10-fold excess of irradiated syngeneic splenocytes and 0.3 μg/ml of anti-CD3 (BD Biosciences) in 2 ml of media. After 48 hours of culture, samples were expanded into 8 ml of fresh media supplemented 1 ng/ml IL-2 and cultured an additional 5 days. Cells were harvested on day 7 for analysis, and a portion was recultured as above. Non-biased cultures of naïve T cells used 2×106 FACS sorted CD4+ CD62L− CD44- naïve T cells from 2loxP or 2loxP;CD4-Cre mice cultured as above. For Th1 and Th2 biasing cultures, 2×106 FACS sorted CD4+ CD62L+ CD44− naïve T cells from 2loxP or 2loxP;CD4-Cre mice were cultured with 5 μg/ml plate-bound anti-CD3 and 10 μg/ml soluble anti-CD28 in 2 ml of media. Th1 cultures included 10 ng/ml IL-12 (eBioscience) and 10 μg/ml Anti IL-4 (11B11; NCI Repository, Frederick). Th2 cultures included 10 ng/ml IL-4 (eBioscience) and 10 μg/ml Anti IFN-γ (BD Biosciences). After 48 hours of stimulation, cells were collected and added to 8 ml of fresh media supplemented with 1 ng/ml IL-2 and cultured an additional 5 days. For subsequent restimulation, 2×106 cells were recultured with 1 μg/ml plate-bound anti-CD3 and 10 μg/ml soluble anti-CD28 plus biasing cytokines and antibodies as above for 48 hours then collected and added to 8 ml of media with 1 ng/ml IL-2 and cultured an additional 5 days. Anti DNMT3a (H-295, Santa Cruz Biotechnology), Anti Actin (Sigma), and Anti-rabbit IgG HRP (GE Healthcare) were used for Western Blot. Anti CD4 PE, Anti CD4 PerCP, Anti CD8 FITC, Anti CD8 APC, Anti IL-4 APC, and Anti IFN-γ PE (BD Biosciences) were used for FACS.
Software
Statistical analysis and graphing was performed with Prism v5.0b (Graphpad Software).
Microarray analysis
Generation and analysis of the microarray data set has been described (29).
Real-time RT-PCR
Total RNA was isolated from cultured T cells using Trizol Reagent (Invitrogen). RT was performed on 1 μg of total RNA with the Superscript III Reverse Transcriptase Kit (Invitrogen). Real-time PCR was performed using Ready Made Primer/Probe Sets for DNMT3a1, DNMT3a2, 18S Endogenous Control, and 2X PCR Master Mix (Applied Biosystems) on ABI 7900 and 7500 machines. Data were analyzed using SDS 2.3 and SDS 1.5 software (Applied Biosystems). RNA abundance was calculated using delta Ct method normalized to control 18S RNA and is plotted as 2−DCt unless otherwise indicated.
Immunoblot analysis
Whole cell extracts were made with RIPA lysis buffer containing 1 mM orthovanadate, 1mM sodium fluoride, 5 μg/ml PMSF, and 1× protease inhibitor mixture (Sigma), separated by SDS-PAGE, and transferred to PVDF membrane (Invitrogen). Western blot by standard methods was detected with the ECLPlus Detection Kit (GE Healthcare).
ELISA, Flow Cytometry, and Intracellular Cytokine Staining
IL-2, IFN-γ, and IL-4 were measured in culture supernatants by ELISA (eBioscience). ICS was performed on 2×105 cells stimulated for 4 hours with PMA and ionomycin in the presence of GolgiStop. Cells were stained using the Cytofix/Cytoperm kit (BD Biosciences). Samples were run on a FACSCalibur cytometer and data analyzed using CellQuest software (BD Biosciences).
5-Methylcytosine Level Determination by Mass Spectrometry
Total 5-methylcytosine content in genomic DNA, as a percentage of total cytosine content, was determined by an high-performance liquid chromatography/mass spectrometry as decribed previously (31).
Methylation Sensitive PCR
Genomic DNA prepared with the DNEasy Purification Kit (Qiagen) was quantified and diluted to 10 ng/μl in water. For Il4 methylation analysis, 200 ng of DNA was incubated in enzyme buffer alone or enzyme buffer plus HpaII (NEB) for IL-4 PCR and digested for 1 hour followed by heat inactivation for 20 minutes at 80°C. Mock digested DNA and HpaII digested DNA were amplified using primers targeting DNAse Hypersensitive Site V flanking an HpaII site (methylation primers) (forward 5′-GAAGGAGGAGCAATTTTGCTAATCC-3′, reverse 5′-CGTCTTTTCCAGTGAATCTCTCAAA-3′) or control primers lacking an HpaII site from Hypersensitive Site Va (forward 5′-GAGATGTGAATTCAGGTCCTGA-3′, reverse 5′-TGCACACATGCTCTAAATATACAGAT-3′) that lie 3′ of the IL-4 gene locus. PCR products were detected by ethidium bromide staining after agarose gel electrophoresis. For Ifng methylation analysis, 84 ng of DNA was incubated in enzyme buffer with or without 5 U of SnaBI (NEB), digested for 1 hour, and heat inactivated. Mock and SnaBI digested DNA were amplified using 2xSYBR Green Master Mix (Applied Biosystems) and primers from the promoter (forward 5′-CCCACCTATCTGTCACCATCTTAA-3′, reverse 5′-CCTCTCTGGCTTCCAGTTTTATACC-3′) which flank a SnaBI site at −53 bp from the transcription start site (methylation primers) or control primers from exon 1 (forward 5′-GCCAGGCTGCCGTCTCT-3′, reverse 5′-GTCAATAAACACCTCTCCAGCAAA-3′) using Real-time PCR. Reactions were quantified against a standard curve of undigested genomic DNA. Fractional methylation is calculated as (quantity of SnaBI digested DNA/quantity of mock digested DNA).
Real-time PCR to Detect Non-deleted 2loxP Alleles
Primer sequences to detect 2loxP (142 forward 5′-CTGTGGCATCTCAGGGTGATGAGCA-3′, 144 reverse 5′-AAGCCTCAGGCCCTCTAGGCAAGAT-3′) and 1loxP (142 forward shown above, 145 reverse 5′-TGAGTGGTGAGGCCCAGCTTATCGA-3′) were provided by Dr. En Li. PCR was performed on 5 ng of input genomic DNA per well in triplicate wells using 100 nM primers and SYBR Green 2X PCR Master Mix (Applied Biosystems) under default cycling conditions. Standard curves were prepared using DNA purified from CD4 T cells of Cre- mice for 2loxP PCR and Cre+ mice for 1loxP PCR over a range of 50 ng to 50 pg in 10-fold dilutions. Control reactions from Cre- samples demonstrated a sensitivity of detection of 100 pg input genomic DNA using 2loxP primers and <50 pg input genomic Cre+ DNA using 1loxP primers based on standard curves. Percent non-deleted was calculated as 100*(amount of 2loxP DNA amplified)/(amount of 2loxP DNA amplified + amount of 1loxP DNA amplified).
Results
DNMT3a is upregulated by TCR stimulation
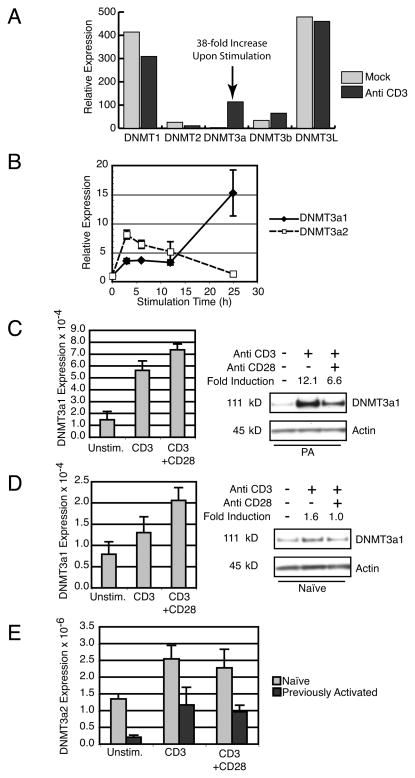
Prior work from our laboratory used microarray analysis to identify genes upregulated by TCR signaling using the murine CD4 T cell clone A.E7 (29). We noted that one member of the DNMT family, DNMT3a, was selectively upregulated 38-fold by TCR stimulation (Fig. 1A). This was confirmed by Real-Time PCR of samples from both A.E7 cells (unpublished data) as well as primary CD4 T cells from 5C.C7 TCR transgenic Rag2−/− mice that were previously activated by antigen/APC and rested to model antigen experienced cells. DNMT3a has two alternative promoters that result in a longer DNMT3a1 and a shorter DNMT3a2 transcript (32), and both are regulated by TCR signaling (Fig. 1B). DNMT3a1 transcript levels progressively increase in 5C.C7 T cells during 24 hours of stimulation to 15-fold above baseline at 24 hours. DNMT3a2 transcripts peak at 8-fold induction after 6 hours of stimulation, and almost return to baseline by 24 hours.
Figure 1.
DNMT3a expression is regulated by TCR stimulation. (A) Microarray analysis of DNMT family members in A.E7 T cells cultured without stimulation (Mock) or with plate-bound anti-CD3 mAb. (B) Real-time PCR of previously activated 5C.C7 TCR Tg T cells stimulated for indicated times with plate-bound anti-CD3. Expression of DNMT3a1 (black diamonds) and DNMT3a2 transcripts (white squares) is shown as the fold induction compared to unstimulated cells. Error bars depict standard deviation of triplicate samples. Results are representative of three experiments. (C) Left, Real-Time PCR for DNMT3a1 from previously activated 5C.C7 T cells cultured overnight under indicated conditions. Error bars depict standard deviation of triplicate samples. Results are representative of three experiments. Right, Western blot of DNMT3a1 from whole cell lysates of previously activated 5C.C7 T cells cultured overnight under indicated conditions. Fold Induction is normalized to Actin expression. Results are representative of three experiments. (D) Left, Real-Time PCR for DNMT3a1 from naïve 5C.C7 T cells cultured overnight under indicated conditions. Error bars depict standard deviation of triplicate samples. Results are representative of two experiments. Right, Western blot of DNMT3a1 from whole cell lysates of naïve 5C.C7 T cells cultured overnight under indicated conditions. Fold Induction is normalized to Actin expression. Results are representative of three experiments. (E) Real-Time PCR for DNMT3a2 from naïve (light grey) and previously activated (dark grey) 5C.C7 T cells cultured overnight under indicated conditions. Error bars depict standard deviation of triplicate samples. Data are representative of 2 (naïve) and 3 (previously activated) independent experiments.
Having demonstrated TCR-regulated DNMT3a transcription in primary CD4 T cells, we wanted to determine the effects of TCR stimulation with and without costimulation in previously activated (CD62L−, CD44+) and in naïve CD4 T cells (CD62L+, CD44−) at the RNA level. Previously activated T cells show low basal levels of DNMT3a1 message with 3 to 4-fold induction after overnight CD3 stimulation that was only slightly increased by CD28 costimulation (Fig. 1C, left panel). Unstimulated, naïve 5C.C7 T cells demonstrate similar basal DNMT3a1 transcript expression to previously activated cells, however induction with anti-CD3 was a more modest 1.5-fold, with CD28 costimulation enhancing this to a 2-fold increase from baseline (Fig. 1D, left panel). DNMT3a2 transcription (Fig. 1E) was about 2 orders of magnitude lower than DNMT3a1 transcription. However, it was similarly induced 4 to 5-fold in previously activated cells and about 2-fold in naïve cells. CD28 costimulation did not affect DNMT3a2 transcript levels.
Interestingly, regulation of DNMT3a protein expression was found to differ from mRNA. DNMT3a amino-terminal specific polyclonal antiserum preferentially binds DNMT3a1 protein due to the greater amount of the antigen present in the larger polypeptide (unpublished observation). This antiserum detected constitutive expression of DNMT3a1 protein in resting previously activated 5C.C7 T cells with dramatic induction by anti-CD3, about 12-fold. Upregulation of DNMT3a protein expression in previously activated cells is consistently attenuated by the presence of CD28 costimulation, to about half that of cells activated with anti-CD3 alone (Fig. 1C, right panel). This is in spite of a modest augmentation in RNA level by CD28 costimulation. DNMT3a1 expression in resting naïve cells is similar to resting previously activated cells but induction by anti-CD3 is more modest (1.6-fold). As in previously activated cells, induction of DNMT3a was attenuated by CD28 costimulation (Fig. 1D, right panel). Similar results were obtained using either whole cell lysate with RIPA buffer (shown) or fractionated nuclear extracts (unpublished data). Thus, DNMT3a1 protein expression in T cells differs from that predicted by mRNA expression, in that CD28 costimulation appears to exert a negative post-transcriptional effect on DNMT3a1 protein levels.
Similar T cell phenotype with reduced cellularity in DNMT3a KO mice
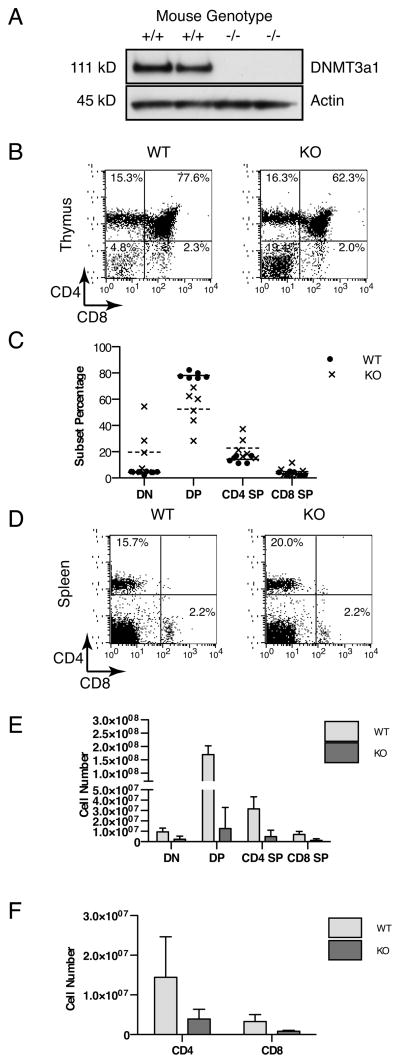
DNMT3a KO mice have abnormal post-natal development with severe runting and death by 4 weeks of age (27). While these authors noted thymic expression of DNMT3a, they did not characterize the immune phenotype of the runted KO mice. Therefore, we obtained DNMT3a KO mice and examined the number and phenotype of T cells isolated from runted 2½ to 3½-week-old KO animals in comparison to their littermates. As expected, DNMT3a1 protein was readily detected in thymocytes of WT littermates, but not in DNMT3a KO thymocytes (Fig. 2A). The immunophenotype of thymocytes from DNMT3a KO mice (Fig. 2B) demonstrated a variable increase in the percentage of double negative cells (mean±SD) from 4.3±1.1% in WT to 19.8±19.5% in KO and a corresponding decrease in the percentage of double positive cells from 78.2±2.5% in WT to 52.5±14.7% in KO, with a modestly increased percentage of CD4 single positive (WT 14.2±2.6%, KO 22.8±8.6%) and similar percentage of CD8 single positive cells (WT 3.4±1.4%, KO 4.9±3.8%) (Fig. 2C). Despite these differences in thymocyte subset percentages, in the spleen, percentages of CD4 and CD8 T cells were similar between WT and KO mice (Fig. 2D). Consistent with their overall runted phenotype, the total cellularity of thymocytes and splenocytes in KO mice is variably decreased with average cellularity about 10% of their WT littermates (Fig. 2E and Fig. 2F). There was considerable variation noted in cellularity between individual animals, with lower cellularity appearing to correlate with animals that are more severely runted (unpublished observation). Lower T cell numbers in KO mice does not appear to be due to an intrinsic defect in proliferation, as DNMT3a KO splenocytes proliferate similarly to WT in vitro to anti-CD3 (WT 37350±6956 CPM, KO 42060±1059 CPM at 24 hours, N=2 mice per group, p=0.5722 by unpaired two-tailed t-test). Thus, T cells lacking DNMT3a expression are able to mature through all stages of thymic development and circulate in the periphery of KO mice in similar proportions to WT T cells. Furthermore, their proliferative capacity in vitro remains similar to DNMT3a expressing T cells. Overall decreases in T cell numbers are consistent with the decrease in size of the runted KO mice.
Figure 2.
DNMT3a KO mice are capable of generating peripheral T cells at similar frequencies to WT. (A) Western blot for DNMT3a1 (111 kD) or Actin (45 kD) prepared from whole cell lysates of thymocytes from individual DNMT3a WT or KO mice. (B) Flow cytometry depicting CD4 versus CD8 staining of thymocytes from a representative pair of DNMT3a WT and KO mice. (C) Percentages of thymocyte subsets (DN, double negative; DP, double positive; CD4 SP, CD4 single positive; CD8 SP, CD8 single positive) from N=6 mice per group with solid horizontal bar indicating mean value for WT and dashed horizontal bar indicating mean value for KO. (D) Flow cytometry depicting CD4 versus CD8 staining of splenocytes from a representative pair DNMT3a WT and KO mice. Results are representative of at least six independent experiments. (E) Absolute number of each thymocyte subset for DNMT3a WT (light grey) and KO (dark grey) mice from N=6 mice per group with bars depicting mean and error bars depicting standard deviation. (F) Total cellularity of pooled spleen and lymph nodes from DNMT3a WT (light grey) and DNMT3a KO (dark grey) mice with N=6 mice per group. Bars depict mean value and error bars depict standard deviation.
Simultaneous IFN-γ and IL-4 expression in DNMT3a KO T cells
Differentiation of naïve CD4 T cells into Th1 or Th2 cells is associated with well-described changes in DNA methylation at the Ifng and Il4 genes (5, 13–21). In naïve T cells, the Ifng locus is hypomethylated and the Il4 locus is hypermethylated. With Th1 differentiation, this is maintained. Under Th2 differentiation Il4 becomes progressively hypomethylated and Ifng becomes methylated, de novo. To determine if TCR-induced DNMT3a plays a role in Th1 versus Th2 differentiation, we isolated DNMT3a WT and KO CD4 T cells purified by negative selection with magnetic beads and stimulated them using anti-CD3, irradiated syngeneic WT splenocytes, and IL-2 supplementation. KO T cells underwent similar expansion to WT and both could be cultured for several weeks by restimulation with anti-CD3 and fresh irradiated splenocytes once weekly. Secretion of IL-2 is similar between DNMT3a WT and KO T cells and remained stable following weekly restimulation through 5 weeks (Fig. 3A, top left panel). T cells from WT mice on a B10.D2 background spontaneously acquire a Th1 cytokine expression profile in such cultures that only includes IL-2 supplementation, with production of increasing amounts of IFN-γ but not IL-4 over time (Fig. 3A, top right and bottom panels). Initially, T cells from KO mice have slightly decreased IFN-γ production compared to WT littermates, but like WT counterparts, they do not secrete IL-4 (Fig. 3A, top right and bottom panels). Following at least two restimulations and increasing with each subsequent restimulation, however, we observed that KO T cells increase their IFN-γ secretion but also begin to spontaneously secrete IL-4 (Fig. 3A, bottom panel), in contrast to the Th1 phenotype acquired by WT T cells under these conditions.
Figure 3.
DNMT3a KO T cells spontaneously secrete IFN-γ and IL-4 in non-selected culture in vitro. (A) ELISA for IL-2 (top left), IFN-γ (top right), and IL-4 (bottom) of cell culture supernatant harvested from primary stimulation or after the indicated number of weeks of culture with weekly stimulation with anti-CD3, irradiated splenocytes, and IL-2 as described in Materials and Methods. Results are representative of two experiments. (B) Flow cytometry depicting IL-4 versus IFN-γ intracellular staining of CD4 T cells from DNMT3a WT (top) or KO (bottom) mice. The panel depicts cells harvested after 5 weeks of culture. Results are representative of three experiments.
The presence of both IFN-γ and IL-4 in the culture supernatants of DNMT3a KO T cells suggested that they were able to simultaneously differentiate into Th1 and Th2 cells in culture, a pattern normally mitigated by the inhibitory effect of IFN-γ on Th2 differentiation and IL-4 on Th1 differentiation. Therefore, we measured IFN-γ and IL-4 production by intracellular cytokine staining (ICS) performed on cells cultured for 3–5 weeks and expressing both cytokines by ELISA. While WT T cells cultured 5 weeks demonstrated expression of IFN-γ without IL-4 (Fig. 3B), DNMT3a KO T cells have a population of Th2 phenotype IL-4 positive, IFN-γ negative cells (17.7% KO vs. 0% WT), a very limited number of Th1 phenotype IFN-γ positive, IL-4 negative cells (2.4% KO vs. 85.8% WT), and a unique population that secrete both IFN-γ and IL-4 simultaneously (11.3% KO vs. 0.5% WT). Thus, stimulation of CD4 T cells that lack DNMT3a with anti-CD3, APC, and IL-2 results in the loss of normally mutually exclusive patterns of IFN-γ and IL-4 expression.
DNMT3a KO T cells express T-bet and GATA-3
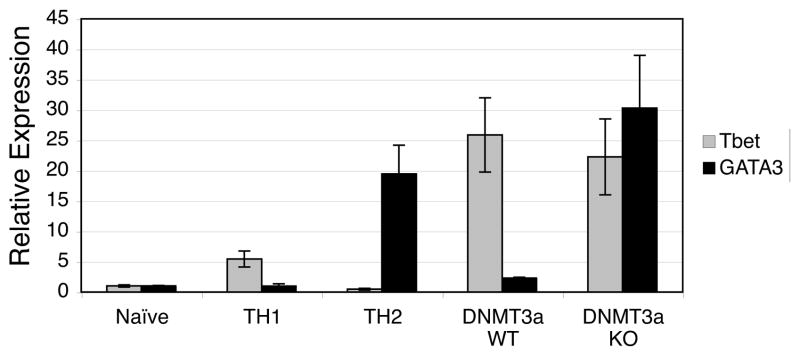
The normal pattern of exclusive IFN-γ expression in Th1 cells and IL-4 expression in Th2 cells is regulated both at the transcriptional level by cross-regulation of T-bet and GATA-3 and by changes in DNA methylation. In order to determine whether transcriptional regulation of cytokine expression was disrupted in DNMT3a KO T cells, we performed Real-Time PCR to measure T-bet and GATA-3 expression. RNA isolated from control freshly harvested (naïve) 5C.C7 CD4 T cells, control Th1 differentiated 5C.C7 T cells, and control Th2 differentiated 5C.C7 T cells demonstrating the expected pattern of low but detectable levels of each transcription factor in naïve cells with upregulation of T-bet alone in Th1 cells and GATA-3 alone in Th2 cells (Fig. 4). WT T cells cultured for 5 weeks that express only IFN-γ under our culture conditions also express only T-bet. DNMT3a KO T cells cultured for 5 weeks that express both IFN-γ and IL-4 express high levels of both T-bet and GATA-3 mRNA (Fig. 4). While these findings suggest possible co-expression of both GATA-3 and T-bet in KO T cells, formal determination awaits analysis of T-bet and GATA-3 expression at the single cell level.
Figure 4.
T-bet and GATA-3 are expressed in DNMT3a KO T cells. Real-time PCR was performed on cDNA prepared from the indicated samples. DNMT3a WT and KO cells had been cultured for 5 weeks resulting in cytokine expression as shown in Figure 3B. Relative expression is plotted with that of naïve T cells set to 1. The figure depicts mean±standard deviation of triplicate samples and is representative of two experiments.
Co-expression of IFN-γ and IL-4 by DNMT3a KO T cells is associated with simultaneous demethylation of both genes
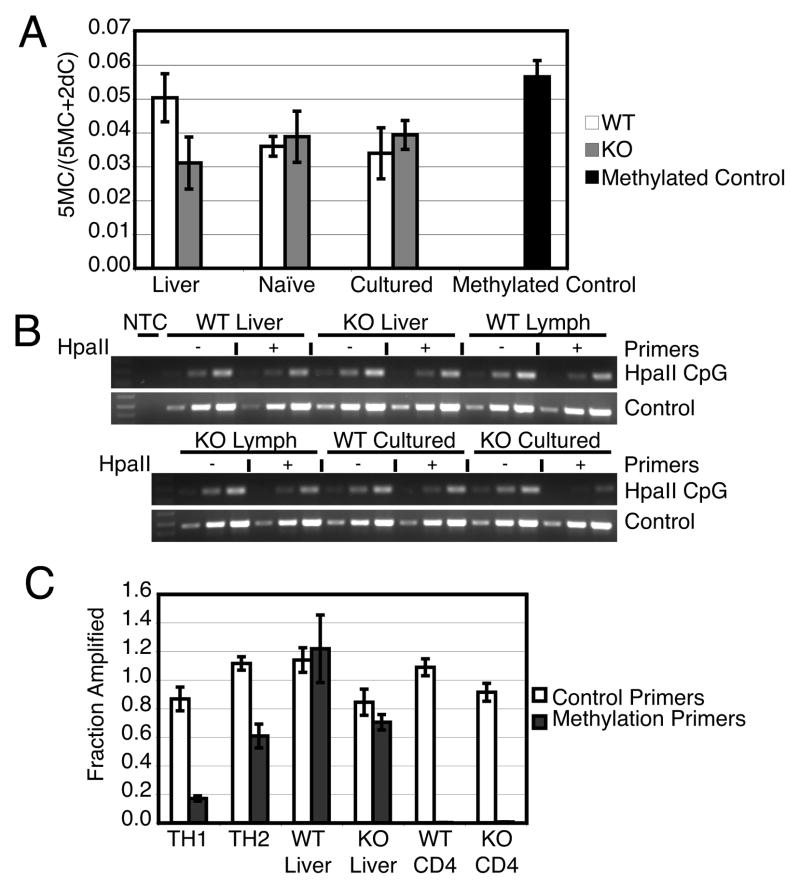
In order to better understand the abnormal regulation of IFN-γ and IL-4, we examined the methylation status of the Ifng and Il4 genes and the effects of exogenous cytokines on Th1 and Th2 skewing of KO T cells. We hypothesized that changes in methylation at the Ifng and Il4 loci in DNMT3a KO T cells would be associated with the abnormal cytokine expression patterns observed. We began by assessing DNMT3a KO T cells for a global derangement of methylation by measuring total methylcytosine content of liver and lymphocyte DNA. We observed that total methylcytosine levels were similar in liver, freshly isolated CD4 T cells, or T cells expanded for 5 weeks from WT and DNTM3a KO mice (Fig. 5A). Thus, overall levels of methylation in T cells do not appear to be altered by the absence of DNMT3a. This stands in contrast to the profound global hypomethylation noted by the Wilson laboratory in DNMT1 KO T cells (22).
Figure 5.
Genomic DNA methylation in DNMT3a WT and KO T cells. (A) Total genomic DNA 5-methylcytosine content determined after hydrolysis using GC-MS. The fraction of methylated cytosine is depicted as the mean±standard error of triplicate samples. Differences in means between WT and KO samples of each tissue assessed by unpaired two tailed t-test (liver p= 0.1388, naïve p= 0.7440, cultured p= 0.5652) were not significant. (B) Il4 gene methylation as detected by methylation sensitive restriction digestion and semi-quantitative PCR. Indicated genomic DNA samples were incubated without (−) or with (+) HpaII. Semiquantitative PCR was performed in 3 reactions on serial diluted input (left to right equals low to high input). WT Cultured and KO Cultured samples are from cells cultured 5 weeks with cytokine expression as depicted in Figure 3B. Results are representative of three experiments. (C) Ifng promoter methylation determined by methylation sensitive restriction digestion and quantified by Real-time PCR. WT CD4 and KO CD4 cells were cultured for 5 weeks with cytokine expression as depicted in Figure 3B. Bars depict mean±standard deviation of triplicate samples and are representative of three experiments.
We next looked for selective changes in methylation of Il4 and Ifng in regions previously noted to be marked by methylation during Th1 and Th2 differentiation. Several of the described methylation sites are recognized by methylation sensitive restriction enzymes (13, 20), therefore we used restriction digestion followed by PCR flanking these sites to assess their methylation status in WT and KO T cells. Semiquantitiative PCR of the Il4 locus after restriction digestion with HpaII found DNA from both DNMT3a WT and KO livers is resistant to digestion, indicating methylation at Il4 (Fig. 5B, top panel). DNA from freshly isolated lymphocytes from both WT and KO mice is likewise resistant to digestion (Fig. 5B, top panel at right, bottom panel at left), consistent with basal Il4 methylation in both liver and T cells arising independently of DNMT3a. DNA from cultured DNMT3a WT T cells, displaying a Th1 phenotype and not secreting IL-4, remains resistant to digestion indicating methylation. DNA from KO T cells that spontaneously secrete IL-4 becomes sensitive to restriction digestion indicating loss of methylation (Fig. 5B bottom panel center and right). Thus Il4 remains methylated in the liver, freshly isolated T cells, and cultured T cells from WT mice and in the liver and freshly isolated T cells from KO mice; however, cultured KO T cells which gain IL-4 expression demonstrate loss of Il4 methylation.
In contrast to Il4, the Ifng promoter is hypomethylated in naïve T cells but becomes methylated in Th2 cells (13–15). Previously, it has been shown that methylation at a CpG dinucleotide −53 bp from the transcription start site is well correlated with silencing of IFN-γ expression in Th2 cells. This region of the Ifng promoter is bound by DNMT3a under Th2 but not Th1 conditions, as measured by chromatin immunoprecipitation (13). This site corresponds to a SnaBI methylation-sensitive restriction site; thus, this enzyme was used to interrogate the methylation status of the promoter by amplifying with primers flanking the SnaBI site (methylation primers) or an adjacent region lacking any SnaBI site (control primers). The ratio of DNA amplified from a SnaBI digested sample divided by that amplified from a mock digested sample should be unity for control primers. The same ratio of signal resulting from amplification with methylation primers represents the fraction of alleles methylated at the SnaBI site. When we examined the Ifng promoter, DNMT3a is not required for developmental methylation of Ifng in the liver, as both WT and KO samples were hypermethylated (Fig. 5C). Freshly isolated WT, control Th1 differentiated WT, and spontaneously generated WT IFN-γ secreting cells are sensitive to restriction digestion (hypomethylated) but control Th2 differentiated WT cells acquire resistance to digestion (methylation) (Fig. 5C). In contrast, DNMT3a KO T cells that spontaneously acquire the ability to produce IL-4 do not have de novo methylation of the IFN-γ promoter after expansion in vitro (Fig. 5C).
DNMT3a KO T cells correctly bias into Th1 and Th2 subsets in vitro
Given the unusual pattern of IFN-γ and IL-4 co-expression and the hypomethylation of both genes that we observed in KO CD4 T cells, we wanted to determine if KO CD4 T cells were able to correctly bias into Th1 or Th2 cells under strong skewing culture conditions with exogenous cytokines and neutralizing antibodies. If methylation were essential to the initiation of Th1/Th2 skewing, then KO cells might produce IFN-γ/IL-4 dual producing cells under either Th1 or Th2 culture conditions; i.e. the intrinsic capacity of naïve cells to express low levels of both IFN-γ and IL-4 might simply turn both cytokine genes on regardless of the local cytokine milieu. If, however, signaling and transcriptional activators were taking advantage of a permissive chromatin structure in KO T cells, then the inclusion of neutralizing antibodies in the biasing conditions might be expected to abolish expression of the neutralized cytokine and allow normal skewing to occur.
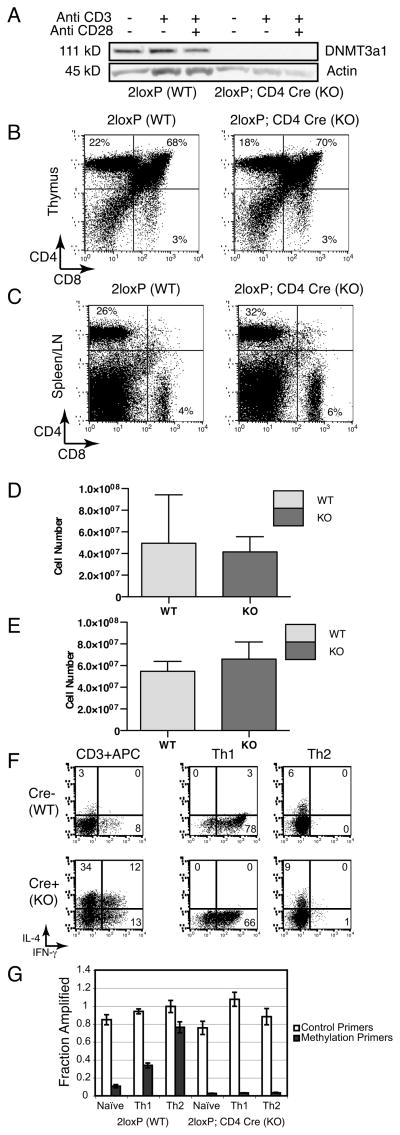
Due to the limited number of naïve T cells that we could obtain from individual DNMT3a KO mice and the challenges associated with their perinatal mortality, we obtained mice bearing a floxed allele of DNMT3a that allow conditional deletion of DNMT3a (33) so that we might collect large enough numbers of naïve CD4 T cells to permit isolation after sorting by flow cytometry. We crossed these mice with CD4-Cre transgenic mice, and obtained animals with no detectable DNMT3a protein in total CD4 T cells before or after TCR stimulation/costimulation (Fig. 6A). However, unlike runted KO mice globally deficient in DNMT3a, the T cell specific KO offspring were healthy with normal body size and cellularity. The immunophenotype of the thymocytes (Fig. 6B) and pooled spleen and lymph node cells (Fig. 6C) from conditional KO mice was similar to their WT Cre- littermates and cellularity of the thymus (Fig. 6D) and peripheral T cells (Fig. 6E) were not statistically different (WT N=3, KO N=4, unpaired t-test p=0.09 for thymus and p=0.53 for spleen/lymph node). Naïve CD4+ CD62L+ CD44 − T cells were isolated by FACS sorting from 2loxP; Cre- (WT) and 2loxP; Cre+(KO) mice and cultured under non-biasing conditions with anti-CD3 plus irradiated APC or in APC-free Th1 or Th2 culture conditions. As we observed with CD4 T cells from mice constitutively deficient in DNMT3a, conditional KO CD4 T cells spontaneously secrete both IFN- γ and IL-4 after expansion with anti-CD3 plus irradiated APC while WT cells maintain distinct populations of either IFN- γ or IL-4 secreting cells (Fig. 6F). Thus, this second, independently generated mouse line confirms our observation of dysregulated IFN-γ and IL-4 co-expression observed earlier in T cells from mice deficient in DNMT3a in all tissues. However, under biasing culture conditions, DNMT3a conditional KO CD4 T cells show appropriate mutually exclusive expression of either IFN- γ or IL-4 that is indistinguishable from WT cells (Fig. 6F). IL-13 expression was similarly restricted to Th2 biased WT and KO T cells (unpublished observation). CD4-Cre mediated deletion of floxed genes has been reported to be incomplete (22); however, this phenomenon does not explain the similar cytokine expression pattern in our biasing WT and KO cultures. Non-deleted 2loxP alleles were undetectable in fourteen of eighteen 2loxP;CD4-Cre samples tested using quantitative Real-time PCR as described in Materials and Methods, corresponding to >98% deletion based on the sensitivity of the PCR assay. This includes the KO Th1 and Th2 samples depicted in Fig. 6C. In the four KO samples where 2loxP alleles were detectable, the fraction present varied: 31% (anti-CD3 + APC in Fig. 6C), 7% (Th1 biased), 11% (Th2 biased), and 4% (Th1 biased). Most importantly, regardless of the precise efficiency of 2loxP deletion, the “gain of function” IFN-γ+ IL4+ dual expressing KO T cells that recapitulate the global KO mice were consistently suppressed under biasing culture conditions. Thus, this normal patterning of cytokine expression in biased culture suggests that proximal cytokine signaling and critical transcriptional events necessary for Th1 and Th2 differentiation remain intact in the conditional KO T cells, despite the absence of DNMT3a expression. In these experiments, WT Th2 cells had significant de novo methylation of the Ifng promoter, but Ifng methylation was virtually absent in conditional KO cells despite the successful Th2 biasing based on their pattern of intracellular cytokine staining (Fig. 6G). Thus, DNMT3a expression is not required to establish normal patterns of cytokine expression under Th1 and Th2 biasing culture conditions, but it is essential for the de novo methylation of the Ifng promoter observed in Th2 cells.
Figure 6.
T cell conditional DNMT3a KO mice have normal lymphocyte cellularity, distributions, and CD4 T cell polarization to Th1 and Th2 cells but they fail to methylate the Ifng promoter. (A) Western blot for DNMT3a1 (111 kD) or Actin (45 kD) prepared from whole cell lysates of DNMT3a 2loxP; CD4-Cre − (WT) and CD4-Cre+ (KO) CD4 T cells left unstimulated or cultured overnight with anti-CD3 alone or anti-CD3 + anti-CD28. Data are representative of two experiments. (B) Flow cytometry depicting CD4 and CD8 staining from representative DNMT3a 2loxP (WT) and 2loxP; CD4 Cre (KO) mouse thymocytes. Data are representative of three independent experiments. (C) Flow cytometry depicting CD4 and CD8 staining of pooled spleen and lymph node cells. Data are representative of three independent experiments. (D) Mean cellularity of the thymus from DNMT3a 2loxP (WT) and 2loxP;CD4 Cre (KO) mice with error bars depicting standard deviation. N=3 WT and 4 KO mice per group. (E) Mean cellularity of pooled spleen and lymph nodes from DNMT3a 2loxP (WT) and 2loxP;CD4 Cre (KO) mice with error bars depicting standard deviation. N=3 WT and 4 KO mice per group. (F) Flow cytometry depicting IL-4 versus IFN-γ intracellular staining of CD4 T cells from DNMT3a 2loxP WT (top) or 2loxP; CD4-Cre KO (bottom) mice. Cells were cultured under the indicated conditions for 2 weeks prior to measurement of cytokine secretion. Results are representative of three experiments. (G) Ifng promoter methylation determined by methylation sensitive restriction digestion and quantified by Real-time PCR as in Figure 5C. Samples are prepared from DNMT3a 2loxP (WT) or DNMT3a 2loxP; CD4-Cre (KO) mice cultured under the indicated conditions for 2 weeks. Results are shown as the mean±standard deviation of triplicate samples and are representative of three experiments.
DNMT3a conditional KO Th2 cells are able to express higher amounts of IFN-γ after culture under Th1 biasing conditions
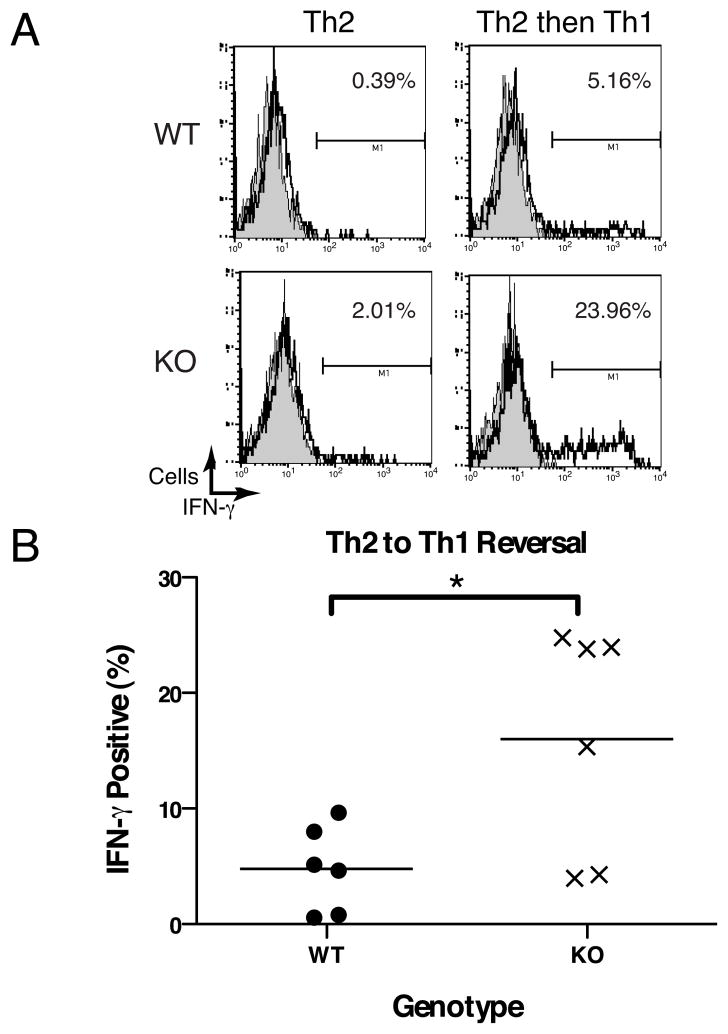
Since DNMT3a conditional KO T cells are able to correctly differentiate into Th2 cells but the Ifng promoter of these cells is devoid of methylation, we hypothesized that such cells would possess an increased ability to express IFN-γ after transfer to culture in Th1 biasing conditions. It has been reported that CD4 Th1 and Th2 cells possess some plasticity in their cytokine phenotype after up to 1 week in biasing cultures, but that this plasticity is extinguished by 3 weeks in culture under biasing conditions (34). Therefore we cultured WT or conditional DNMT3a KO CD4 T cells under Th2 skewing conditions for 2 weeks, followed by culture for 1 week under either Th2 or Th1 skewing conditions. Although the percentage of IFN-γ producing cells after culture of Th2 cells in Th1-biasing conditions varied between individual experiments, percentages of conditional KO Th2 cells expressing IFN-γ were consistently higher than paired WT samples (representative individual experiment in Fig. 7A, pooled data from 5 experiments with 6 pairs of mice in Fig. 7B). When we examined samples used for the reversal experiment depicted in Fig. 7A, we were unable to detect non-deleted 2loxP alleles. Thus, the selective loss of methylation marks at the Ifng promoter of DNMT3a KO Th2 cells correlates with an increased ability to recover IFN-γ expression after culture of Th2 cells under Th1 skewing conditions.
Figure 7.
DNMT3a KO Th2 cells are able to express more IFN-γ when cultured under Th1 biasing conditions. (A) Intracellular cytokine staining of CD4 T cells from a pair of DNMT3a WT and KO mice cultured for 3 weeks under Th2 conditions or 2 weeks under Th2 conditions followed by 1 week under Th1 conditions. Histograms depict isotype control (filled grey) or IFN-γ (solid line). (B) Aggregate data depicting IFN-γ secretion for 6 pairs of DNMT3a WT and KO CD4 T cells cultured for 2 weeks under Th2 conditions followed by 1 week under Th1 conditions demonstrating percentage of IFN-γ expression measured by ICS as in top panel. Bars depict mean value, N=6 mice per group, *p=0.0145 by paired, 2-tailed Student’s t-test.
Discussion
The acquired restriction of cytokine expression that is observed as naïve T cells are recruited to an immune response is controlled both by the selective expression of specific transcriptional activators and also by the accumulation of histone modifications and dynamic changes in DNA methylation at many effector genes. It was therefore of great interest when we observed that expression of one member of the DNA methyltransferase family, DNMT3a, is regulated by TCR stimulation. Specifically, we observed a 5-fold increase in DNMT3a RNA and 12-fold increase in DNMT3a1 protein expression after reactivation of antigen experienced T cells with a 1.5 to 2-fold increase in RNA and a 1.6 fold increase in protein in naïve T cells.
Notably, the upregulation of DNMT3a protein in both naïve and memory phenotype cells is attenuated by CD28 costimulation. The mechanism of DNMT3a post-transcriptional regulation remains under investigation, but recently published data would suggest members of the miR-29 family of microRNA might contribute (35). The DNMT3a 3 ′ UTR contains a miR-29 binding site, and transfection of miR-29 family members both suppressed expression of a luciferase reporter with the DNMT3a 3 ′ UTR and reduced cellular DNA methylation levels. The consequences of enhanced DNMT3a expression in the absence of costimulation remain under investigation, but the basis of our original microarray screen that identified DNMT3a induction by TCR signaling was, in fact, to identify candidate genes that participate in T cell anergy (29). A recent report of increased genomic methylation at the Il2 locus in anergic cells provides support for such a hypothesis (36). Characterization of the response of DNMT3a KO T cells to anergizing stimuli may help clarify the potential role of de novo methylation in the stability of the anergic phenotype.
DNMT3a is a de novo methyltransferase, that is, it is able to add new methylation marks to unmethylated cytosine bases during differentiation (6). This distinguishes it from DNMT1 which acts to copy existing methylation marks on hemimethylated DNA from parental to daughter strands following DNA replication. These differences in substrate specificity are apparent in the phenotypic differences reported between our two models of T cells lacking DNMT3a and earlier reports from the Wilson lab describing the phenotype of T cells deficient in DNMT1 (22, 37). While the DNMT1 deficient T cells have global defects in DNA methylation owing to the dilution of DNA methylation marks following each round of proliferation, our mice demonstrated preservation of total DNA methylation but defective addition of DNA new methylation marks at the Ifng locus that we examined. Cytokine expression of the DNMT1 deficient T cells, while modulated by local cytokines, is promiscuous even under skewing Th culture conditions (37). In contrast, we find DNMT3a deficient T cells are able to maintain faithful Th cytokine gene expression under skewing conditions but fail to maintain this pattern when skewing conditions are subsequently changed. Such plasticity in the form of increased IFN-γ expression in the DNMT3a KO T cells is associated with decreased de novo promoter methylation at the Ifng locus under Th2 culture conditions.
Although the observed methylation data support that changes in cytokine expression are due to defective silencing at the level of the cytokines themselves, they cannot exclude additional defects in methylation that affect more proximal elements regulating cytokine expression. For example, GATA-3 induction during Th2 differentiation typically interferes with the ability of IL-12 to promote Th1 differentiation, T-bet expression, and IFN-γ expression due to the down-regulation of STAT4 (38). Precise roles for DNA methylation in this process have not been defined, but it has been reported that culture of human T cells with a DNMT inhibitor is able to increase STAT4 expression (39). Thus, KO T cells may remain responsive to IL-12 even after they begin IL-4 mediated GATA-3 upregulation if such cells fail to methylate the STAT4 locus. Alternatively, the promoter of the Tbx21 gene encoding T-bet has also been reported to acquire limited de novo methylation in Th2 cells (40). Either of these processes would be predicted to promote GATA-3 and T-bet co-expression and potentially lead to the dual cytokine expression that is observed when DNMT3a deficient T cells are cultured under non-biased conditions with anti-CD3 plus APC. It is also possible that DNMT3a KO T cells may accumulate multiple sites of failed de novo methylation and that each contributes to the atypical cytokine expression phenotype. Ongoing efforts to characterize the defective targets of de novo methylation in DNMT3a KO cells will help to clarify the potential contributions of each of these mechanisms to the observed changes in cytokine gene expression.
Acknowledgments
We are grateful to Dr. En Li of the Novartis Institute for Biological Research for generously providing us the DNMT3a KO and DNMT3a 2loxP mice. We are also grateful for the technical support of Krystal Matthews and Angela Alme.
This research was supported by NIH T32CA60441 (C.G.), a Hyundai Scholar Award (C.G.) and ACS RSG0605301LIB (J.P.).
Footnotes
DNMT-DNA Methyltransferase; 5C.C7-B10.A/AiTac-[Tg]TCRCyt5CC7-I-[KO]Rag2 mouse.
Disclosures
The authors have no conflicts of interest to disclose.
References
- 1.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 2.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 3.Ivanov, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 4.Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 5.Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24:369–379. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Jeltsch A. Molecular enzymology of mammalian DNA methyltransferases. Curr Top Microbiol Immunol. 2006;301:203–225. doi: 10.1007/3-540-31390-7_7. [DOI] [PubMed] [Google Scholar]
- 7.Gowher H, Liebert K, Hermann A, Xu G, Jeltsch A. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J Biol Chem. 2005;280:13341–13348. doi: 10.1074/jbc.M413412200. [DOI] [PubMed] [Google Scholar]
- 8.Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, Benes V, Jeltsch A, Gannon F, Salbert G. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 9.Mizuno S, Chijiwa T, Okamura T, Akashi K, Fukumaki Y, Niho Y, Sasaki H. Expression of DNA methyltransferases DNMT1, 3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood. 2001;97:1172–1179. doi: 10.1182/blood.v97.5.1172. [DOI] [PubMed] [Google Scholar]
- 10.Tao J, Yang M, Chen Z, Huang Y, Zhao Q, Xu J, Ren H, Zhao H, Chen Z, Ren Q, Yang R. Decreased DNA methyltransferase 3A and 3B mRNA expression in peripheral blood mononuclear cells and increased plasma SAH concentration in adult patients with idiopathic thrombocytopenic purpura. J Clin Immunol. 2008;28:432–439. doi: 10.1007/s10875-008-9223-2. [DOI] [PubMed] [Google Scholar]
- 11.Luo Y, Li Y, Su Y, Yin H, Hu N, Wang S, Lu Q. Abnormal DNA methylation in T cells from patients with subacute cutaneous lupus erythematosus. Br J Dermatol. 2008;159:827–833. doi: 10.1111/j.1365-2133.2008.08758.x. [DOI] [PubMed] [Google Scholar]
- 12.Balada E, Ordi-Ros J, Serrano-Acedo S, Martinez-Lostao L, Rosa-Leyva M, Vilardell-Tarres M. Transcript levels of DNA methyltransferases DNMT1, DNMT3A and DNMT3B in CD4+ T cells from patients with systemic lupus erythematosus. Immunology. 2008;124:339–347. doi: 10.1111/j.1365-2567.2007.02771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones B, Chen J. Inhibition of IFN-gamma transcription by site-specific methylation during T helper cell development. Embo J. 2006;25:2443–2452. doi: 10.1038/sj.emboj.7601148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winders BR, Schwartz RH, Bruniquel D. A distinct region of the murine IFN-gamma promoter is hypomethylated from early T cell development through mature naive and Th1 cell differentiation, but is hypermethylated in Th2 cells. J Immunol. 2004;173:7377–7384. doi: 10.4049/jimmunol.173.12.7377. [DOI] [PubMed] [Google Scholar]
- 15.Young HA, Ghosh P, Ye J, Lederer J, Lichtman A, Gerard JR, Penix L, Wilson CB, Melvin AJ, McGurn ME, et al. Differentiation of the T helper phenotypes by analysis of the methylation state of the IFN-gamma gene. J Immunol. 1994;153:3603–3610. [PubMed] [Google Scholar]
- 16.Chang S, Aune TM. Dynamic changes in histone-methylation ‘marks’ across the locus encoding interferon-gamma during the differentiation of T helper type 2 cells. Nat Immunol. 2007;8:723–731. doi: 10.1038/ni1473. [DOI] [PubMed] [Google Scholar]
- 17.Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, Stamatoyannopoulos JA, Wilson CB. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat Immunol. 2007;8:732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- 19.Bird JJ, Brown DR, Mullen AC, Moskowitz NH, Mahowald MA, Sider JR, Gajewski TF, Wang CR, Reiner SL. Helper T cell differentiation is controlled by the cell cycle. Immunity. 1998;9:229–237. doi: 10.1016/s1074-7613(00)80605-6. [DOI] [PubMed] [Google Scholar]
- 20.Lee DU, Agarwal S, Rao A. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity. 2002;16:649–660. doi: 10.1016/s1074-7613(02)00314-x. [DOI] [PubMed] [Google Scholar]
- 21.Kim ST, Fields PE, Flavell RA. Demethylation of a specific hypersensitive site in the Th2 locus control region. Proc Natl Acad Sci U S A. 2007;104:17052–17057. doi: 10.1073/pnas.0708293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, Cherry SR, Tsai JH, Tucker SM, Weaver WM, Kelso A, Jaenisch R, Wilson CB. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 23.Hutchins AS, Mullen AC, Lee HW, Sykes KJ, High FA, Hendrich BD, Bird AP, Reiner SL. Gene silencing quantitatively controls the function of a developmental trans-activator. Mol Cell. 2002;10:81–91. doi: 10.1016/s1097-2765(02)00564-6. [DOI] [PubMed] [Google Scholar]
- 24.Yu Q, V, Thieu T, Kaplan MH. Stat4 limits DNA methyltransferase recruitment and DNA methylation of the IL-18Ralpha gene during Th1 differentiation. Embo J. 2007;26:2052–2060. doi: 10.1038/sj.emboj.7601653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oka M, Rodic N, Graddy J, Chang LJ, Terada N. CpG sites preferentially methylated by Dnmt3a in vivo. J Biol Chem. 2006;281:9901–9908. doi: 10.1074/jbc.M511100200. [DOI] [PubMed] [Google Scholar]
- 26.Samuel MS, Lundgren-May T, Ernst M. Identification of putative targets of DNA (cytosine-5) methylation-mediated transcriptional silencing using a novel conditionally active form of DNA methyltransferase 3a. Growth Factors. 2007;25:426–436. doi: 10.1080/08977190801931081. [DOI] [PubMed] [Google Scholar]
- 27.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 28.Tadokoro Y, Ema H, Okano M, Li E, Nakauchi H. De novo DNA methyltransferase is essential for self-renewal, but not for differentiation, in hematopoietic stem cells. J Exp Med. 2007;204:715–722. doi: 10.1084/jem.20060750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Safford M, Collins S, Lutz MA, Allen A, Huang CT, Kowalski J, Blackford A, Horton MR, Drake C, Schwartz RH, Powell JD. Egr-2 and Egr-3 are negative regulators of T cell activation. Nat Immunol. 2005;6:472–480. doi: 10.1038/ni1193. [DOI] [PubMed] [Google Scholar]
- 30.Powell JD, Bruniquel D, Schwartz RH. TCR engagement in the absence of cell cycle progression leads to T cell anergy independent of p27(Kip1) Eur J Immunol. 2001;31:3737–3746. doi: 10.1002/1521-4141(200112)31:12<3737::aid-immu3737>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 31.Agoston AT, Argani P, Yegnasubramanian S, De Marzo AM, Ansari-Lari MA, Hicks JL, Davidson NE, Nelson WG. Increased protein stability causes DNA methyltransferase 1 dysregulation in breast cancer. J Biol Chem. 2005;280:18302–18310. doi: 10.1074/jbc.M501675200. [DOI] [PubMed] [Google Scholar]
- 32.Chen T, Ueda Y, Xie S, Li E. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J Biol Chem. 2002;277:38746–38754. doi: 10.1074/jbc.M205312200. [DOI] [PubMed] [Google Scholar]
- 33.Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 34.Murphy E, Shibuya K, Hosken N, Openshaw P, Maino V, Davis K, Murphy K, O’Garra A. Reversibility of T helper 1 and 2 populations is lost after long-term stimulation. J Exp Med. 1996;183:901–913. doi: 10.1084/jem.183.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas RM, Saouaf SJ, Wells AD. Superantigen-induced CD4+ T cell tolerance is associated with DNA methylation and histone hypo-acetylation at cytokine gene loci. Genes Immun. 2007;8:613–618. doi: 10.1038/sj.gene.6364415. [DOI] [PubMed] [Google Scholar]
- 37.Makar KW, Wilson CB. DNA methylation is a nonredundant repressor of the Th2 effector program. J Immunol. 2004;173:4402–4406. doi: 10.4049/jimmunol.173.7.4402. [DOI] [PubMed] [Google Scholar]
- 38.Usui T, Nishikomori R, Kitani A, Strober W. GATA-3 suppresses Th1 development by downregulation of Stat4 and not through effects on IL-12Rbeta2 chain or T-bet. Immunity. 2003;18:415–428. doi: 10.1016/s1074-7613(03)00057-8. [DOI] [PubMed] [Google Scholar]
- 39.Shin HJ, Park HY, Jeong SJ, Park HW, Kim YK, Cho SH, Kim YY, Cho ML, Kim HY, Min KU, Lee CW. STAT4 expression in human T cells is regulated by DNA methylation but not by promoter polymorphism. J Immunol. 2005;175:7143–7150. doi: 10.4049/jimmunol.175.11.7143. [DOI] [PubMed] [Google Scholar]
- 40.Hewitt SL, High FA, Reiner SL, Fisher AG, Merkenschlager M. Nuclear repositioning marks the selective exclusion of lineage-inappropriate transcription factor loci during T helper cell differentiation. Eur J Immunol. 2004;34:3604–3613. doi: 10.1002/eji.200425469. [DOI] [PubMed] [Google Scholar]