Preface
Eukaryotic protein modification by ubiquitin-like proteins (Ubls) controls an enormous range of physiological processes. Ubl attachments principally regulate interactions with other macromolecules, such as proteasome-substrate binding or recruitment of proteins to chromatin. Different Ubl systems use related enzymes to attach specific Ubls to proteins (or other molecules), and most Ubl attachments are transient. Mounting evidence suggests that Ubl-protein modification evolved from prokaryotic sulfurtransferase systems or related enzymes. Surprisingly, proteins similar to Ubl-conjugating and Ubl-deconjugating enzymes appear to have already become widespread by the time of the last universal common ancestor, suggesting that Ubl-protein conjugation is not a eukaryotic invention.
Introduction
The array of proteins in a eukaryotic cell or organism is subject to an impressive variety of post-translational covalent modifications; these alterations greatly extend the functional diversity and dynamics of the proteome. Proteins can be attached to small molecules such as phosphate, methyl, or acetyl groups, or they can be covalently modified, usually only transiently, by certain other proteins1,2,3. Among these latter protein modifiers, the first to be described and most thoroughly understood is ubiquitin. Ubiquitin is a small protein that is extremely well conserved among the eukarya but is absent from eubacteria and archaea. It can be transiently attached to thousands of different proteins.
An intricate enzymatic pathway catalyzes ubiquitin modification of substrate proteins (Fig. 1 and Box 1). In a similar fashion, distinct but evolutionarily related enzyme cascades catalyze the attachment of ubiquitin-like proteins (Ubls) to proteins or other molecules4. The Ubls, including ubiquitin itself, share the same basic three-dimensional core structure, the β-grasp fold. Thus, it is clear that these different Ubl modification systems share a common ancestry. In addition to ubiquitin, at least nine Ubls that covalently modify proteins (or in one case, a phospholipid) are currently known.
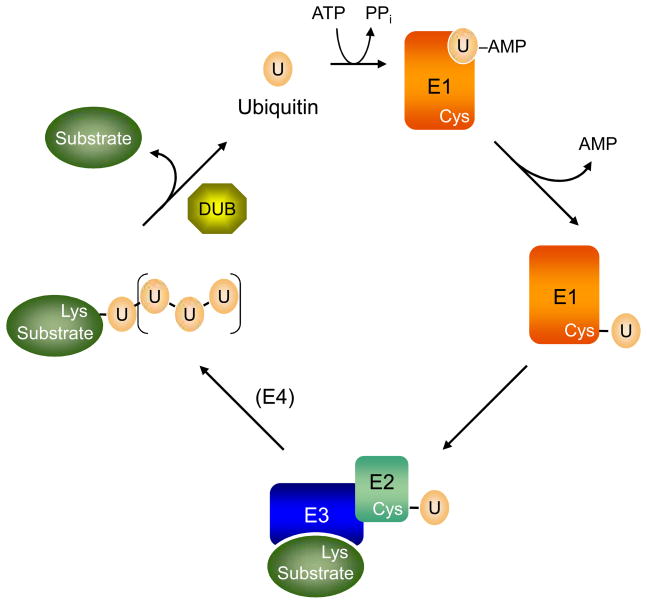
Figure 1.
The ubiquitin (U)-protein conjugation cycle. For ubiquitin (and at least some other Ubls), an E3 ligase is usually necessary to stimulate ubiquitin transfer from the E2 to a substrate, generally to a lysine ε-amino group. Additional ubiquitin molecules can be added either to other lysine side chains on the substrate or to ubiquitin itself, the latter leading to polymeric ubiquitin chains. Additional E3s can help assemble ubiquitin chains on substrates; when they act in this way, they are sometimes called “E4s”. Ubiquitin-substrate modifications are transient and can be removed by deubiquitylating enzymes or DUBs (or more generally, Ubl-specific proteases or ULPs). In addition, ubiquitin and most Ubls are synthesized in precursor forms, and the C-terminal extensions are also removed by DUBs or ULPs (not depicted).
Box 1. Basics of ubiquitin enzymology.
Ubiquitin-protein conjugation illustrates the general mechanisms used by cells to attach and remove Ubls from their substrates (Fig. 1). The 76-residue ubiquitin polypeptide is activated and attached to substrate proteins by a series of enzymes. The trio of E1 ubiquitin-activating, E2 ubiquitin-conjugating, and E3 ubiquitin-ligase enzymes perform an impressive array of ubiquitin modification reactions, including assembly of polyubiquitin chains. All eukaryotic species express multiple E2 and E3 isozymes, which can range up to several dozen E2s and many hundreds of E3s. This allows for the highly specific modification of many different proteins by ubiquitin, and such modifications are often under strict temporal and spatial control.
Ubiquitin is usually joined to proteins by an amide linkage between the C-terminus of ubiquitin and primary amino groups of the acceptor proteins2, 52. The amine is most often a lysine ε-amino group, but it can also be the N-terminal Nα-amino group70. In addition, recent work has shown in vivo ubiquitin attachment to cysteines, serines, and threonines in proteins71, 72, 73. When ubiquitin forms polymers, the ubiquitin molecules are linked through the lysine side chain of one ubiquitin with the C-terminal carboxyl of the next ubiquitin. Ubiquitin has seven lysines, and all can contribute to such linkages.
The C-terminal glycine of ubiquitin must be activated before it can form a covalent bond with another protein2, 52 (Fig. 1). Initially, the C-terminus is adenylated by E1, with the ubiquitin-AMP adduct remaining bound to the enzyme. An E1 cysteine side chain then attacks the ubiquitin C-terminus, yielding an E1-ubiquitin thioester intermediate. The activated ubiquitin is subsequently passed to the active site cysteine of an E274. E2 proteins catalyze substrate ubiquitylation in conjunction with an E3 ligase. Ubiquitin E3s play a paramount role in substrate recognition, although not all Ubl pathways necessarily require one. In the ubiquitin pathway, a different E3 may sometimes help add ubiquitins to a protein already modified by one or a few ubiquitins. Such E3s are sometimes called E4s, particularly when they are thought to extend a polyubiquitin chain. Due to the activity of deubiquitylating enzymes (DUBs), ubiquitin-modified proteins are only transiently modified61, 75. Dynamic modification of proteins by ubiquitin and other Ubls creates reversible switches between different functional states.
Ubiquitin is not only attached to proteins as a monomer but also in the form of polyubiquitin chains. Ubiquitin molecules in these chains are linked to one another in the same way that they are usually linked to substrate proteins. In particular, the C-terminus of the more distal ubiquitin molecule is attached to the ε-amino group of a lysine in the preceding ubiquitin molecule, forming an isopeptide (amide) bond. Often when a substrate protein is coupled to a polyubiquitin chain, it binds to the 26S proteasome, a large, multisubunit protease complex, which degrades the substrate into small peptides and recycles the ubiquitin tag (Fig. 2). The 26S proteasome is composed of two major subcomplexes: the 20S proteasome or core particle (CP), which bears the proteolytic sites in a sequestered interior chamber, and the 19S regulatory particle (RP), a complex of at least 18 different polypeptides, including a half-dozen different AAA+ ATPases, several polyubiquitin-binding subunits, and deubiquitinating enzymes (DUBs) that cleave the ubiquitin chain from the substrate so that the ubiquitin can be reused.
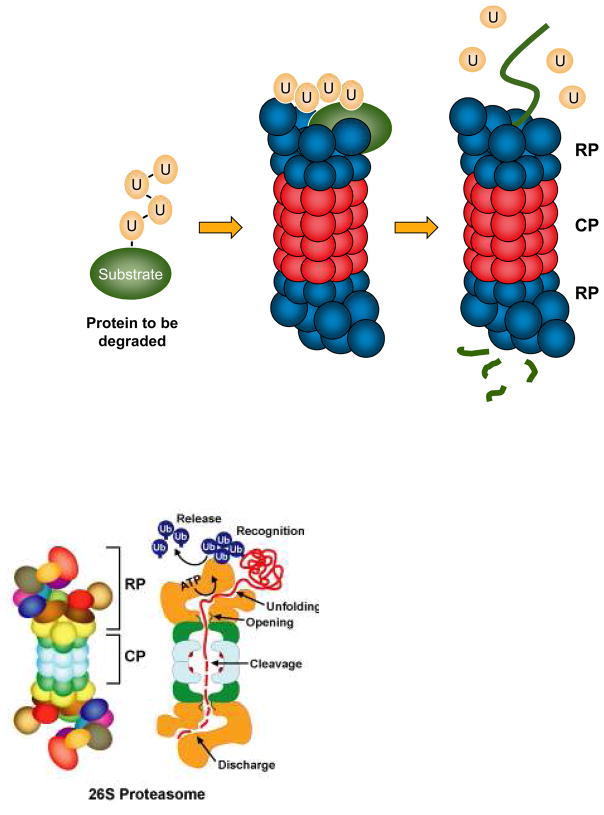
Figure 2.
Polyubiquitin-tagged proteins are often targeted for proteasome-mediated degradation. The ubiquitin-proteasome pathway is responsible for the degradation of hundreds, and probably thousands, of different proteins. Many of these substrates are regulatory proteins, such as transcription factors or cell cycle regulators, while others are misfolded or otherwise aberrant proteins that must be eliminated to prevent their aggregation or toxicity. A polyubiquitin-modified protein is the form most commonly targeted to the proteasome. Ubiquitin receptors either in the proteasome regulatory particle (RP, blue) of the 26S proteasome or adaptor proteins that associate reversibly with both polyubiquitylated proteins and specific proteasomal subunits (not shown) allow binding of the proteolytic substrate to the proteasome. ATPases within the RP unfold the substrate and translocate it into the 20S proteasome core particle (CP, red), which houses the proteolytic sites in an interior chamber. The substrate is cleaved to small peptides. Ubiquitin itself is normally recycled by DUBs that bind to or are intrinsic to the RP.
Even when considering just ubiquitin-protein modification, the diversity of the processes that are regulated is extraordinary4,5. The consequences of the modification also depend on whether the attached ubiquitin is monomeric or in the form of a polymer; polyubiquitin chains can have different ubiquitin-ubiquitin linkages, which also help dictate the fate of the modified substrate (Fig. 3). Ubiquitin-protein attachments that lead to proteasomal degradation include substrates that must be eliminated for proper cell-cycle progression, transcriptional regulation, protein quality control, signal transduction, and circadian rhythms. Ubiquitylation is also employed in nonproteolytic regulatory mechanisms, such as membrane protein endocytosis and intracellular trafficking, chromatin-mediated regulation of transcription, DNA repair, and assembly of signaling complexes. In this light, it is not surprising that the list of diseases implicating misregulation of the ubiquitin system is growing steadily. These disorders include many different cancers, certain severe forms of mental retardation such as Angelman’s Syndrome, neurodegenerative disorders such as Parkinson’s, Huntington’s and Alzheimer’s disease, and diabetes6.
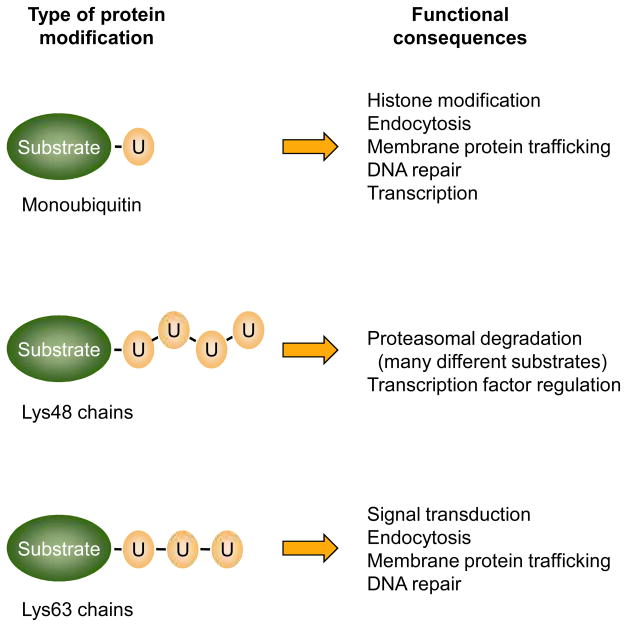
Figure 3.
Cellular processes that depend on ubiquitin conjugation. Protein attachment to a single ubiquitin allows recognition by a subset of ubiquitin-binding domains (UBDs) in target proteins, and this is important in the indicated general processes. Often a single, specific lysine is modified. Different polyubiquitin chains are thought to have different structures that allow discrimination among other UBDs, although other contextual cues, such as the cellular location where the modification occurs, may also help dictate the physiological consequences of the polyubiquitin attachment. Lys48-linked chains are most commonly associated with proteasomal binding and degradation. Not shown here are ubiquitin chains with mixed linkages or multi-site ubiquitylation of the substrate.
For the first few decades following the discovery of ubiquitin in 1975, the evolutionary precursors to the ubiquitin-conjugation system had gone largely undetected. Ubiquitin itself was regarded as perhaps the most highly conserved of all eukaryotic proteins7, but until relatively recently, sequence-comparison algorithms were not sufficiently sensitive to detect any bacterial proteins with primary sequence similarity to it. This situation has changed dramatically over the past few years. First, sequence comparison methods have become more sophisticated, revealing unexpected similarities between ubiquitin pathway components, including ubiquitin, and an assortment of bacterial proteins8. Second, structure determinations showed that a ubiquitin-like fold is adopted by many eukaryotic and prokaryotic proteins or protein domains even where the primary sequence similarity to ubiquitin is difficult to discern. Third, mechanistic analysis of the biosynthesis of secondary metabolites and enzyme cofactors such as thiamin (vitamin B1) has uncovered surprising parallels to Ubl activation and conjugation9. Together, these studies imply that the ubiquitin system has been cobbled together from a variety of pre-existing parts and pathways that had already undergone a great deal of diversification in early prokaryotes1, 8.
In this review, I will discuss basic features of Ubl-protein conjugation and how such protein modifications contribute, in a general way, to cell regulatory mechanisms. A major emphasis will be on the likely evolutionary antecedents to eukaryotic Ubl-protein conjugation in prokaryotes, and on parallels between enzymes responsible for synthesis of specific small molecules, particularly certain sulfurtransferases, and enzymes of the ubiquitin system. The evolution of the proteasome will not be discussed, as it is well covered elsewhere10,11.
Similarities and differences in Ubl conjugation
Studies begun in the late 1980s identified an interferon-stimulated gene product of 15 kDa (ISG15) that shares significant sequence similarity with ubiquitin and also covalently modifies other proteins12,13. ISG15 turned out to be the first in a wave of Ubls found to act as protein modifiers (Table 1). Despite being the first Ubl identified, ISG15 function is still not well understood, and it took some time before its E1-like activating and E2-like conjugating enzymes (Box 1) were identified14, 15, 16, 17. These enzymes, like ISG15 itself, are strongly induced by type-I interferons. Recent results with mouse models indicate that ISG15-protein conjugation contributes to anti-viral responses, consistent with the interferon induction of ISG1518, 19, 20.
Table 1.
Known or suspected Ubls.
| Modifiera | Identity with Ub (%) | E1a | E2a | Comments |
|---|---|---|---|---|
| Ubiquitin (Ub) | 100 | Uba1& Uba6 | many | Multiple genes encode ubiquitin precursors |
| Rub1/NEDD8 | 55 | Uba3-Ula1 | Ubc12 | Substrates: cullins, p53 |
| Smt3/SUMO1–3 | 18 | Uba2-Aos1 | Ubc9 | Vertebrates have 3–4 distinct SUMO genes |
| Atg12 | NDc | Atg7 | Atg10 | ~20% identical to Atg8 |
| Atg8 | ND | Atg7 | Atg3 | 3 known human isoforms; β-grasp fold |
| Urm1 | ND | Uba4 | – | Related to MoaD, ThiS; β-grasp fold |
| ISG15 | 32/37b | Ube1L | UbcH8 | Induced by type I interferons |
| UFM1 | ND | Uba5 | Ufc1 | β-grasp fold |
| FUBI/MNSFβ | 38 | – | – | Derived from ribosomal protein precursor |
| FAT10 | 32/40b | Uba6 | – | Substrates unknown; β-grasp fold |
| Ubl-1 | 40 | – | – | Nematode ribosomal protein precursor |
| BUBL1, 2 | variable (up to 80%) | – | – | Putative autoprocessed proteins (ciliates) |
| SF3a120 | 30 | – | – | Ubl at C-terminus; no data for conjugation |
| Oligo(A) synthetase | 30/42b | – | – | Ubl at C-terminus; no data for conjugation |
Yeast names are listed for E1s and E2s except for the ISG15, FAT10, and UFM1 systems, which are not found in S. cerevisiae (Uba6 has a much more limited phylogenetic distribution); for the Ubls, the S. cerevisiae names are given if present in this organism (first six entries) and are listed first if a vertebrate ortholog is known and goes by a different name;
Two Ub-related domains;
Not detectable by standard BLAST searches.
At present, ten distinct Ubls, including ubiquitin, have been demonstrated to covalently modify other macromolecules, usually proteins (Table 1), and several others are suspected of having this ability (noted below the dashed line in Table 1). This list is likely to be incomplete. Ubiquitin has been reported to modify over a thousand different proteins in yeast21. Some Ubls, such as SUMO (small ubiquitin-related modifier), may approach ubiquitin in the number and diversity of substrates targeted, but others have a far more limited range of substrates4. For example, Atg12 (autophagy protein 12) appears to have a single target (Atg5), and Atg8 is attached to a specific phospholipid, phosphatidylethanolamine22.
As noted, most Ubl modification pathways utilize highly similar enzymatic mechanisms. The major pathways of protein conjugation appear to have evolved by repeated rounds of duplication and diversification of enzymes and protein modifiers derived from ancient biosynthetic pathways (see below). However, a few unusual Ubl conjugation mechanisms have been proposed for specific Ubl pathways. In one of these models, a ubiquitin hydrolase was proposed to be able not only to cleave ubiquitin from substrates but to work backwards, in effect, and ligate ubiquitin to a protein23. Another provocative model was suggested from sequence analysis of an unusual group of putative self-splicing polyproteins in ciliates24. These polyproteins consist of a tandem series of variant Ubl domains with interspersed self-splicing bacterial intein-like (BIL) domains; the corresponding genes probably arose from a polyubiquitin gene that had acquired a sequence encoding the BIL domain.
BIL domains have a Ser or Cys residue at their N-terminal end, and they are known to stimulate an N-O or N-S acyl rearrangement at the boundary with their upstream sequences and to catalyze cleavage and self-splicing reactions24. It was suggested that the BIL domains in the BIL-ubiquitin-like (BUBL) proteins might also trigger such (N-S) acyl shifts and catalyze nucleophilic attack on the resulting thioester by a protein (or other molecule) during autocatalytic processing, leading to ligation of the upstream Ubl sequence to the attacking molecule24. If the attacking group were a Lys side chain of a protein, the resulting product would be a Ubl-protein conjugate similar to those formed through the standard pathway (Box 1) even though no E1 or E2 (or ATP) is involved. Notably, there is no obvious requirement for the sequences upstream of the BIL domains in BUBL precursors to be Ubls. Thus, other protein modifiers might exist that are not related to ubiquitin but are fused upstream of domains with functional similarity to BIL domains.
There are also many ubiquitin-related proteins in which a ubiquitin-like domain (ULD) is part of a larger polypeptide but is usually neither processed nor covalently attached to other proteins1. Such ULDs confer properties on a protein similar to those from a transferable Ubl, such as the ability to bind specific target proteins. Certain ULDs may be cleaved under some specific conditions (and might even become competent for ligation to other proteins). For example, autocleavage at an internal ULD occurs in the USP1 deubiquitylating enzyme after UV damage to cells; this inactivates the enzyme and allows accumulation of a monoubiquitylated protein required for trans-lesion DNA synthesis25.
As noted, all structurally characterized Ubls and ULDs have a similar tertiary structure, which is generally referred to as the β-grasp fold26. The β-grasp fold is phyletically widespread and ancient, potentially having arisen as an RNA-binding module in a primitive protein translation system26. It has been adapted to a broad spectrum of functions, ranging from a scaffold for different enzymatic activities and iron-sulfur-cluster binding to an adaptor module for specific protein-protein interactions.
General functions of Ubl-protein conjugation
Early work on ubiquitin focused primarily on its role in proteolysis27, 28, 29. The 26S proteasome is responsible for the degradation of polyubiquitylated proteins, and direct binding between a polyubiquitin chain and the proteasome can fully account for the observed affinity of model polyubiquitylated proteins for this protease30. In short, the polyubiquitin chain provides a generic affinity tag that leads to tight binding of the proteolytic substrate to the proteasome. Multiple polyubiquitin receptors exist within the proteasome31, 32, 33. In addition, polyubiquitin-binding domains are found in shuttling factors that direct polyubiquitylated proteins to the proteasome34, 35, 36.
Specific interactions between ubiquitin and ubiquitin-binding domains (UBDs) are not limited to proteasome targeting. A general theme that has emerged over the past decade is that many functions of ubiquitin and Ubls are mediated by such associations (Fig. 4). At least 16 structurally distinct UBD classes have been characterized to date, and they vary considerably in both length (~30–150 residues) and structure37. Evolution of this array of UBDs allowed the remarkable diversification of ubiquitin signaling functions found in modern eukaryotes.
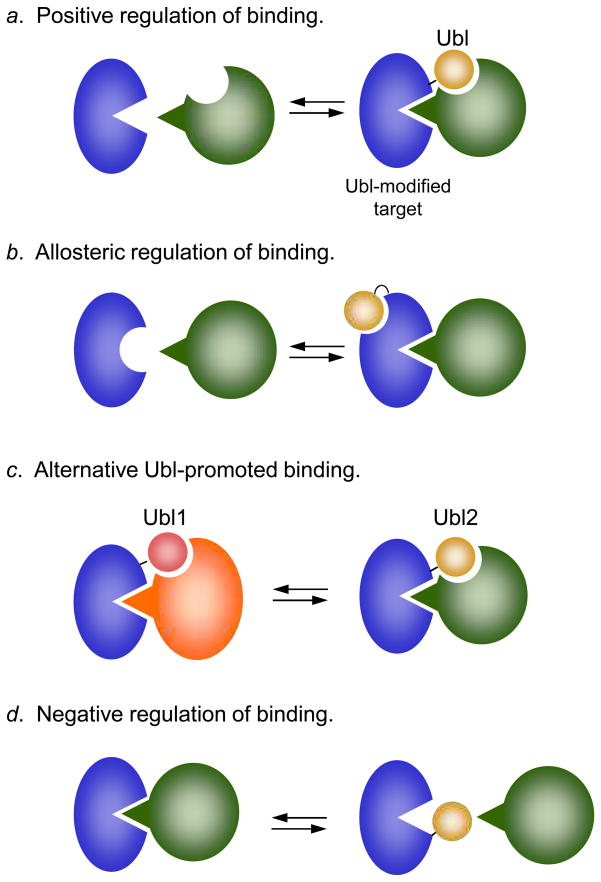
Figure 4.
General functions of Ubl tagging. a. Ubl conjugation facilitates protein association by providing an additional binding site. The classic example of this type of regulation is polyubiquitin modification of proteins, which can then bind specific UBD receptors in the proteasome33. b. Ubl conjugation causes a conformational change that enhances binding (as shown) or inhibits binding to a target site. For instance, SUMO attachment to thymine-DNA glycosylase (TDG) triggers a conformational change in TDG that lowers its affinity for DNA67. c. Modification by one Ubl helps recruit a factor that is different from the protein that would be recruited were the substrate modified by another type of Ubl. These modifications may be mutually exclusive and can potentially involve the same attachment site. The modification of proliferating cell nuclear antigen (PCNA), a DNA replication and recombination protein, by SUMO, ubiquitin, or ubiquitin polymers causes PCNA to bind distinct factors68. d. Ubl conjugation directly blocks an interaction between two proteins. A potential example of this is the sumoylation of the vaccinia A40R protein, which prevents association and aggregation between A40R monomers69. Other possible variations on these basic mechanisms are not shown.
UBD binding to ubiquitin is usually weak, with apparent dissociation constants ranging from ~20 μM to over 500 μM, depending on the UBD, although tighter binding is occasionally observed37. Despite these generally low affinities, mutational studies support their physiological relevance. Linking multiple ubiquitin moieties into a chain can have dramatic effects on the affinity or avidity for target proteins. A nice example comes from analysis of polyubiquitin-proteasome binding30. Using competitor ubiquitin chains of various lengths, Thrower et al.30 measured the inhibition of degradation of model substrates. Tetrameric chains displayed very strong inhibition (Ki ~ 170 nM), i.e., high affinity, whereas inhibition was extremely weak with trimeric chains, and undetectable with dimeric chains. The lack of a simple dependence of affinity on the length of the chain implies that formation of a tetrameric chain creates a unique binding determinant that is not present in shorter chains or monoubiquitin38.
Besides ubiquitin, SUMO has been the most extensively studied Ubl, and analyses of SUMO-protein interactions confirm and extend many of the ideas about ubiquitin-protein interactions sketched above. Interestingly, genetic, biochemical, and biophysical studies have converged on a single noncovalent SUMO-binding element, called the SUMO-interaction motif (SIM)39. SIM peptides as short as nine residues, which is much shorter than the domains that typically bind ubiquitin, have dissociation constants between 5 and 10 μM, which is substantially tighter binding than that seen with most known UBDs. The SIM consensus sequence contains a central group of 3–4 hydrophobic residues usually flanked on one or the other side by a cluster of acidic residues. The SIM forms a β strand that sits in a hydrophobic surface depression between the β2 strand and α1 helix of SUMO and extends the SUMO β sheet; acidic residues at the ends of the SIM interact with basic residues on the SUMO surface40, 41. This is the opposite face of theβ-grasp fold from that recognized by most UBDs on ubiquitin, which is a hydrophobic patch centered on Ile44. Thus, different Ubls can function analogously as adaptor modules but the exact ways in which they bind to target proteins need not be the same.
As implied by the above discussion, Ubl conjugation frequently functions by promoting protein-protein interactions. When such modifications enhance an interaction with another macromolecule, they usually do so by participating directly in the formation of the binding interface with the target molecule (Fig. 4a). In principle, an interaction can also be modulated by an allosteric change in a target-binding site induced by the attached Ubl (Fig. 4b). Many examples of Ubl regulation fall into the former category, and only a handful in the latter42, 43. Even if only a small fraction of a particular protein were modified, its new activity could suffice to effect a change in physiological state. As mentioned, such noncovalent Ubl-protein interactions tend to be weak. Specific binding can be greatly enhanced either by polymerization of the Ubl signal (see above) or by combining the weak binding from the Ubl with additional weak binding sites in the conjugated protein (Fig. 4a, c). An example of such multivalent binding is the association of the SUMO-RanGAP1 conjugate with the nuclear pore complex, which is observed for the conjugate, but not for either contributing polypeptide44.
Given their bulk, another way by which Ubls could function would be by steric hindrance: the attached Ubl simply blocks the binding of a substrate to another protein (or another part of the same protein) (Fig. 4d). There are relatively few well-established in vivo examples of this intermolecular inhibitory mechanism. One likely reason is that for such a mechanism to operate effectively, a large fraction of the protein would need to be modified by the Ubl. However, for many proteins, only a miniscule amount is observed in the conjugate form. In principle, such an inhibitory mechanism could still operate if the small fraction of modified protein were localized to some functionally privileged cellular site or if a transient modification were sufficient to put the protein in a new state (see Fig. 4 legend).
Origins of Ubl-protein conjugation
An early clue to the antecedents of Ubl-protein conjugation came from the E1 ubiquitin-activating enzyme sequence; the protein displays weak but significant similarity to MoeB, an E. coli protein required for the biosynthesis of the molybdenum cofactor (Moco)45. At the time E1 was sequenced, the biochemical function of MoeB was unknown, so this similarity was not particularly informative. During the late 1990s, however, the protein sequences and catalytic mechanisms of the enzymes used to synthesize Moco (and thiamin) began to be deciphered, and intriguing similarities to ubiquitin activation were noted46, 47, 48. The thiamin synthesis pathway is nearly universally distributed among bacterial species and Moco synthesis enzymes are also found very broadly, so these enzymatic systems are thought to be far older than ubiquitin conjugation8.
Ubl-related sulfur carrier proteins
Moco and thiamin synthesis involves insertion of sulfur atoms into precursors of these cofactors. Remarkably, a small sulfur-carrier protein, named MoaD and ThiS, respectively, donates the necessary sulfur(s). Sulfur is taken from a thiocarboxylate group formed at the C-terminus of these proteins (see Fig. 5). MoaD and ThiS sequences are related and, like ubiquitin, end with a pair of glycines. Most interestingly, conversion of the C-terminal glycine carboxylate to a thiocarboxylate in these proteins is preceded by C-terminal adenylation by an E1-related enzyme: MoeB for MoaD and ThiF for ThiS47, 48, 49. Both MoaD and ThiS were later shown to share the ubiquitin/β-grasp fold despite minimal sequence similarity to ubiquitin50, 51. Therefore, ubiquitin, MoaD, and ThiS are all structurally related proteins whose C-termini are activated through adenylation by homologous E1-like enzymes52, 53.
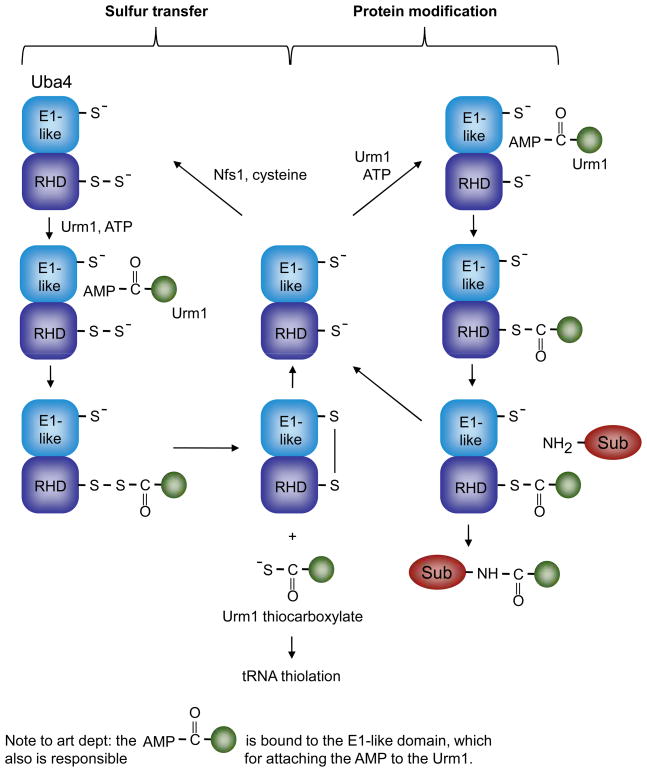
Figure 5.
Uba4 and Urm1, at the crossroads of Ubl-protein modification and sulfur transfer. Two modes of Uba4 activity are depicted: In the cycle on the left, Uba4 functions as a sulfurtransferase, transferring sulfur from a persulfide formed on its rhodanese-homology domain (RHD) to the C-terminus of Urm1, yielding a Urm1 thiocarboxylate (bottom, middle). The sulfur is ultimately transferred to a specific subset of tRNAs. On the right, Uba4 catalyzes transfer of Urm1 to protein substrates (Sub) through a hypothetical Uba4-Urm1 thioester intermediate (middle right). Although it is possible that the Urm1-Uba4 acyl disulfide (bottom left) functions in Urm1-protein modification, it is difficult to explain the requirement for the E1-like domain cysteine residue in such a scheme. Nfs1 is a cysteine desulfurase that mobilizes sulfur from cysteine56.
Further insight into the potential evolutionary link between these sulfur-transfer systems and Ubl activation came with the discovery of a MoaD/ThiS-related protein in the yeast Saccharomyces cerevisiae, called ubiquitin-related modifier-1 (Urm1; Fig. 5)54. Although S. cerevisiae lacks any Moco-containing enzymes and uses a different mechanism for thiamin synthesis, it does express an E1-related protein, Uba4, which has sequence similarity to ThiF and MoeB. Uba4 binds Urm1 and stimulates covalent addition of Urm1 to cellular proteins54, 55. These findings demonstrated that Urm1 and Uba4 function as part of a Ubl-protein conjugation system despite bearing much closer sequence relatedness to bacterial sulfur-transfer enzymes. However, as discussed below, Uba4 was recently shown to have the ability to function in sulfur transfer as well, so Uba4 is potentially a bifunctional enzyme56. The Urm1-Uba4 system may thus represent a “molecular fossil” that retains features of the more ancient sulfur transfer pathway yet is also able conjugate the Ubl to proteins.
Urm1: both sulfur carrier and protein modifier?
Enzymatic activation of the C-termini of Ubls by ATP-dependent E1-related enzymes is analogous to other adenylation reactions such as the activation of amino acids by aminoacyl-tRNA synthetases2. For the Ubls, the energy of ATP hydrolysis is conserved by formation of the E1-Ubl thioester linkage. However, despite being chemically activated, all but one of the Ubls listed in Table 1 (Urm1) are known to first undergo an energetically neutral transesterification to the thiol of a second enzyme, E2 (Fig. 1). The one apparent exception to the use of an E1-E2 relay, the Urm1 pathway, has some distinct features, which it shares with bacterial sulfur-transfer pathways. Uba4 (the Urm1 E1) includes a rhodanese-homology domain (RHD), unlike the E1s for other Ubls1, 56 (Fig. 5). Rhodanese and a number of RHD proteins are sulfurtransferases that transfer sulfur to their targets via an intermediate persulfide (-S-S-H) on their active site cysteine57. Many MoeB family proteins have a domain organization similar to that of Uba4, with an E1-like domain followed by an RHD. Based on these and other considerations, it had been proposed that thiocarboxylate formation in MoaD proceeds through an RHD-MoaD intermediate and that, by analogy, Urm1 transfer from Uba4 to substrate also utilizes the RHD in Uba41. In particular, the Uba4 RHD was suggested to form a transient thioester intermediate with Urm1, which is then transferred directly to the substrate, bypassing the requirement for a separate E2 (Fig. 5). The RHD cysteine is required for Urm1-protein ligation in yeast42.
Remarkably, recent work demonstrated that the RHD in Uba4 is also capable of forming a persulfide and can transfer the terminal sulfur to the C-terminus of Urm1 in vitro, creating an Urm1-thiocarboxylate56. The E1-domain cysteine is not required for this, and these authors found no evidence for an Urm1-Uba4 thioester. Instead, they inferred that the adenylated Urm1 is attacked by a persulfide on the RHD, forming an acyl disulfide intermediate with Urm1, with subsequent release of the thiocarboxylate (also see Ref. 42). Schmitz et al. suggested that this could also be an intermediate in Urm1-protein conjugation56.
A newly discovered function for the Urm1 pathway is the substitution of sulfur for oxygen in position 2 of the wobble uridine in the anticodon of certain tRNAs, which modulates their decoding specificity58, 59. Urm1 is required for these thiolation reactions, likely functioning as a sulfur carrier in the form of an Urm1-thiocarboxylate. The degree to which tRNA modification by the Urm1 pathway accounts for the pleiotropic physiological roles of Urm1 remains to be determined. Protein urmylation may also stimulate tRNA thiolation59, although a minimal hypothesis would require only the formation of an Urm1-Uba4 acyl disulfide that resolves into the Urm1-thiocarboxylate, from which the sulfur is ultimately transferred to the tRNA.
Although the conserved E1 domain cysteine in Uba4 is dispensable for Urm1-thiocarboxylate formation in vitro, it is necessary for protein conjugation to Urm1 in vivo54. This cysteine could function in reductive cleavage of the Urm1-RHD linkage56, but this should stimulate formation of an Urm1 C-terminal thiocarboxylate rather than an Urm1-protein amide linkage. Potentially, the E1-like cysteine undergoes a persulfide exchange with the RHD, freeing the RHD thiol for attack on the Urm1-adenylate (not shown in Fig. 5), or the Urm1-adenylate is directly attacked by a substrate lysine, although this would not explain the requirement for the two Uba4 active-site cysteines. Reductive cleavage of the Urm1-RHD disulfide by the E1 domain active site sulfhydryl group would allow the RHD thiol to be regenerated, potentially making it competent for Urm1 thioester formation. Whether the Urm1-protein ligation mechanism reflects an early precursor to other Ubl conjugation mechanisms is not known. Conceivably, during the evolution of Ubl-conjugation systems, a distinct E2-like factor could at some point have been co-opted, leading to loss of the RHD from the E1 and thereby eliminating the persulfide/thiocarboxylate “side reaction.”
Radiation of E2s and Ubl-specific proteases
When did the E2-like enzymes arise and when did they first associate with E1-like proteins to form the now nearly universal E1-E2 relay used for Ubl-protein conjugation? Earlier sequence searches had not revealed any E2-like proteins in bacteria, but recent surveys have revealed an astonishing number of E2-related sequences in the same DNA neighborhoods (i.e., in presumptive operons or co-regulated genes) or in the form of domain fusions with Ubl-related, E1-like, or JAMM (JAB1/MPN/Mov34) metalloprotease coding sequences8, 60. In eukaryotes, specific JAMM-class proteases act as deubiquitylating enzymes (DUBs) or Ubl-specific proteases (ULPs)61.
A striking radiation of E2-like proteins therefore appears to have occurred in bacteria concomitant with the diversification of Ubl and E1-like proteins. One subfamily that is most closely related to classical Ubl-conjugating E2 enzymes has been proposed to be ancestral to the eukaryotic Ubl E2s60. Although none of these prokaryotic E2-like proteins has yet been shown to catalyze Ubl-substrate modification in conjunction with an E1, these contextual associations suggest that at least some of them will.
As shown in Fig. 1, ULPs or DUBs are often necessary for C-terminal processing of Ubl precursors and for removing Ubls from their targets. While multiple JAMM proteins have now been linked by contextual sequence analysis to potential Ubl-modification systems in prokaryotes, some of these JAMM enzymes participate in sulfur transfer mechanisms rather than Ubl-protein conjugation. For instance, Mycobacterium tuberculosis has an unusual cysteine biosynthetic pathway that involves thiocarboxylate derivatization of CysO, a β-grasp protein, by the E1-related MoeZ protein62. The gene for a JAMM enzyme clusters with the gene for CysO, and it is likely to hydrolyze cysteine from CysO in the final step of cysteine biosynthesis. Synthesis of the sulfur-containing thioquinolobactin siderophore (an iron-chelating compound) in Pseudomonas fluorescens requires a sulfur-carrier protein called QbsE that is related to MoaD and ThiS. QbsE, however, is made in precursor form with two additional amino acids following the diglycine motif 63. A JAMM protease expressed in the same thioquinolobactin biosynthetic operon cleaves these last two residues from QbsE. Therefore, proteases of the type that in eukaryotes remove Ubls from protein conjugates might originally have been part of bacterial βgrasp protein-based biosynthetic pathways, just like the Ubls and E1-like (and possibly E2-like) enzymes.
E1-like activation of a non-Ubl substrate
It is worth pointing out that the E1-like superfamily of adenylating enzymes catalyzes a spectrum of biochemical reactions that goes beyond C-terminal activation of β-grasp proteins. The best evidence for this comes from enterobacteria that synthesize and secrete the small antibiotic microcin C7 (MccC7). MccC7 is a modified heptapeptide encoded by a large E. coli plasmid64. An isoasparaginyl moiety at the C-terminus of MccC7 has a phosphoramidate linkage to a modified adenylate, and attachment of this modified AMP requires the plasmid-encoded mccB product. MccB is a member of the E1-like enzyme superfamily. Therefore, the substrate of this E1-like enzyme is not a β-grasp protein, and the C-terminal modification chemistry is different from the sulfurtransferases described above, even though the initial adenylation of a C-terminal α-carboxylate by the E1-related enzymes is similar.
Outlook
The fundamental biochemical consequence of protein modification by a Ubl is usually a change in association with other proteins or macromolecules (Fig. 4). Eukaryotes have elaborated multiple variants of the same basic enzymatic mechanism for Ubl attachment, and one system even modifies a lipid rather than a protein. Ten eukaryotic Ubl-modification systems have been documented to date, and for nine of these, at least one enzyme in the pathway for substrate conjugation is known. Ubiquitin and SUMO, and potentially other Ubls, can attach to proteins in the form of polymers, and topological variants of these chains impart differences in function as well. The basic E1–E2 relay, which probably arose early in the evolution of Ubl-conjugation systems, has now diversified to include multiple E2 (and E3) isozymes, especially in the ubiquitin pathway. Ubl-specific proteases make most of the Ubls into highly dynamic and closely regulated protein modifiers.
How much of the diversity of Ubl-protein modification or analogous conjugation systems is yet to be discovered? Some Ubls are difficult to recognize by primary sequence comparison, so additional β-grasp/Ubl modifiers might still have been overlooked. The exciting possibility of a multitude of prokaryotic Ubl-related protein modification systems, none of which has been analyzed experimentally, was made apparent when contextual sequence analyses suggested the existence of a bevy of bacterial regulons that bring together genes encoding novel β-grasp proteins, E1-like enzymes, E2-like proteins, and hydrolases related to those in known Ubl systems. Moreover, not all E1-like enzymes act on β-grasp/Ubl proteins, so further insights into the ability of such enzymes to modify specific proteins or peptides (or other molecules) can be anticipated.
Conversely, there may be novel intracellular protein-protein conjugation mechanisms that involve neither E1-like adenylating enzymes nor β-grasp proteins, such as intein-mediated protein trans-splicing. Along these lines, a 64-residue protein in M. tuberculosis called Pup (prokaryotic ubiquitin-like protein) was recently shown to modify specific proteins in vivo and, remarkably, to target them for degradation by the mycobacterial proteasome65. Pup is not a βgrasp/Ubl protein, and Pup attachment involves linkage of a substrate lysine to what had originally been a glutamine residue at the Pup C-terminus. The terminal Pup glutamine is converted to a glutamate either during or before substrate conjugation. Similar amide bond-forming reactions are seen with transglutaminases and γ-glutamylcysteine synthetases (involved in glutathione synthesis). In fact, the M. tuberculosis PafA protein, which is required for substrate pupylation65, was recently shown to have distant sequence similarity to γ-glutamylcysteine synthetases and glutamine synthetases66. Mass spectrometry-based proteomic studies could yield further surprises about protein modification and ligation in vivo. Viewed through this wider lens, protein-protein conjugation can be seen as a multifaceted and nearly universally employed means of cellular regulation, and we probably still do not know the half of it.
Acknowledgments
I thank V.J. Rubenstein, Andrew Kusmierczyk, and Joanna Bloom for comments on the manuscript. Work from my laboratory is funded by grants from the NIH.
References
- 1.Hochstrasser M. Evolution and function of ubiquitin-like protein-conjugation systems. Nat Cell Biol. 2000;2:E153–E157. doi: 10.1038/35019643. [DOI] [PubMed] [Google Scholar]
- 2.Pickart CM. Mechanisms Underlying Ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 3.Xu P, Peng J. Dissecting the ubiquitin pathway by mass spectrometry. Biochim Biophys Acta. 2006;1764:1940–7. doi: 10.1016/j.bbapap.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 5.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–5. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz AL, Ciechanover A. Targeting Proteins for Destruction by the Ubiquitin System: Implications for Human Pathobiology. Annu Rev Pharmacol Toxicol. 2008 doi: 10.1146/annurev.pharmtox.051208.165340. [DOI] [PubMed] [Google Scholar]
- 7.Sharp PM, Li WH. Molecular evolution of ubiquitin genes. 1987;2:328–332. doi: 10.1016/0169-5347(87)90108-X. [DOI] [PubMed] [Google Scholar]
- 8.Iyer LM, Burroughs AM, Aravind L. The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like beta-grasp domains. Genome Biol. 2006;7:R60. doi: 10.1186/gb-2006-7-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Begley TP. Cofactor biosynthesis: an organic chemist’s treasure trove. Nat Prod Rep. 2006;23:15–25. doi: 10.1039/b207131m. [DOI] [PubMed] [Google Scholar]
- 10.Volker C, Lupas AN. Molecular evolution of proteasomes. Current Topics in Microbiology & Immunology. 2002;268:1–22. doi: 10.1007/978-3-642-59414-4_1. [DOI] [PubMed] [Google Scholar]
- 11.Cavalier-Smith T. Rooting the tree of life by transition analyses. Biol Direct. 2006;1:19. doi: 10.1186/1745-6150-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haas AL, Ahrens P, Bright PM, Ankel H. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J Biol Chem. 1987;262:11315–23. [PubMed] [Google Scholar]
- 13.Loeb KR, Haas AL. The interferon-inducible 15-kDa ubiquitin homolog conjugates to intracellular proteins. J Biol Chem. 1992;267:7806–13. [PubMed] [Google Scholar]
- 14.Yuan W, Krug RM. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. Embo J. 2001;20:362–71. doi: 10.1093/emboj/20.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao C, et al. The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-alpha/beta-induced ubiquitin-like protein. Proc Natl Acad Sci U S A. 2004;101:7578–82. doi: 10.1073/pnas.0402528101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim KI, Giannakopoulos NV, Virgin HW, Zhang DE. Interferon-inducible ubiquitin E2, Ubc8, is a conjugating enzyme for protein ISGylation. Mol Cell Biol. 2004;24:9592–600. doi: 10.1128/MCB.24.21.9592-9600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durfee LA, Kelley ML, Huibregtse JM. The basis for selective E1–E2 interactions in the ISG15 conjugation system. J Biol Chem. 2008 doi: 10.1074/jbc.M804069200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lenschow DJ, et al. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc Natl Acad Sci U S A. 2007;104:1371–6. doi: 10.1073/pnas.0607038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okumura A, Pitha PM, Harty RN. ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc Natl Acad Sci U S A. 2008;105:3974–9. doi: 10.1073/pnas.0710629105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malakhova OA, Zhang DE. ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J Biol Chem. 2008;283:8783–7. doi: 10.1074/jbc.C800030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng J, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–6. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 22.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–6. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT., Jr The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson’s disease susceptibility. Cell. 2002;111:209–18. doi: 10.1016/s0092-8674(02)01012-7. [DOI] [PubMed] [Google Scholar]
- 24.Dassa B, Yanai I, Pietrokovski S. New type of polyubiquitin-like genes with intein-like autoprocessing domains. Trends Genet. 2004;20:538–42. doi: 10.1016/j.tig.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 25.Huang TT, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–47. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 26.Burroughs AM, Balaji S, Iyer LM, Aravind L. Small but versatile: the extraordinary functional and structural diversity of the beta-grasp fold. Biol Direct. 2007;2:18. doi: 10.1186/1745-6150-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hershko A, Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- 28.Goldberg AL, Rock KL. Proteolysis, Proteasomes and Antigen Presentation. Nature. 1992;357:375–379. doi: 10.1038/357375a0. [DOI] [PubMed] [Google Scholar]
- 29.Hochstrasser M. Ubiquitin-dependent protein degradation. Ann Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 30.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. Embo J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S Protease Subunit That Binds Ubiquitin Conjugates. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- 32.Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart CM. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature. 2002;416:763–7. doi: 10.1038/416763a. [DOI] [PubMed] [Google Scholar]
- 33.Husnjak K, et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–8. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L, Madura K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol Cell Biol. 2002;22:4902–13. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Funakoshi M, Sasaki T, Nishimoto T, Kobayashi H. Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:745–50. doi: 10.1073/pnas.012585199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim I, Mi K, Rao H. Multiple interactions of rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol Biol Cell. 2004;15:3357–65. doi: 10.1091/mbc.E03-11-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–72. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varadan R, et al. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem. 2004;279:7055–63. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- 39.Kerscher O. SUMO junction-what’s your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007;8:550–5. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song J, Zhang Z, Hu W, Chen Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J Biol Chem. 2005;280:40122–9. doi: 10.1074/jbc.M507059200. [DOI] [PubMed] [Google Scholar]
- 41.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–92. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hochstrasser M. In: Protein Degradation: The Ubiquitin-Proteasome System. Mayer RJ, Ciechanover A, Rechsteiner M, editors. Wiley-VCH Verlag; Weinheim: 2006. pp. 249–278. [Google Scholar]
- 43.Archer CT, et al. Physical and functional interactions of monoubiquitylated transactivators with the proteasome. J Biol Chem. 2008;283:21789–98. doi: 10.1074/jbc.M803075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 45.McGrath JP, Jentsch S, Varshavsky A. UBA1: an essential yeast gene encoding ubiquitin-activating enzyme. Embo J. 1991;10:227–36. doi: 10.1002/j.1460-2075.1991.tb07940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rajagopalan KV. Biosynthesis and processing of the molybdenum cofactors. Biochem Soc Trans. 1997;25:757–61. doi: 10.1042/bst0250757. [DOI] [PubMed] [Google Scholar]
- 47.Taylor SV, et al. Thiamin biosynthesis in Escherichia coli. Identification of this thiocarboxylate as the immediate sulfur donor in the thiazole formation. J Biol Chem. 1998;273:16555–60. doi: 10.1074/jbc.273.26.16555. [DOI] [PubMed] [Google Scholar]
- 48.Appleyard MV, et al. The Aspergillus nidulans cnxF gene and its involvement in molybdopterin biosynthesis. Molecular characterization and analysis of in vivo generated mutants. J Biol Chem. 1998;273:14869–76. doi: 10.1074/jbc.273.24.14869. [DOI] [PubMed] [Google Scholar]
- 49.Leimkuhler S, Wuebbens MM, Rajagopalan KV. Characterization of Escherichia coli MoeB and its involvement in the activation of molybdopterin synthase for the biosynthesis of the molybdenum cofactor. J Biol Chem. 2001;276:34695–701. doi: 10.1074/jbc.M102787200. [DOI] [PubMed] [Google Scholar]
- 50.Rudolph MJ, Wuebbens MM, Rajagopalan KV, Schindelin H. Crystal structure of molybdopterin synthase and its evolutionary relationship to ubiquitin activation. Nat Struct Biol. 2001;8:42–6. doi: 10.1038/83034. [DOI] [PubMed] [Google Scholar]
- 51.Wang C, Xi J, Begley TP, Nicholson LK. Solution structure of ThiS and implications for the evolutionary roots of ubiquitin. Nat Struct Biol. 2001;8:47–51. doi: 10.1038/83041. [DOI] [PubMed] [Google Scholar]
- 52.Huang DT, Walden H, Duda D, Schulman BA. Ubiquitin-like protein activation. Oncogene. 2004;23:1958–71. doi: 10.1038/sj.onc.1207393. [DOI] [PubMed] [Google Scholar]
- 53.Duda DM, Walden H, Sfondouris J, Schulman BA. Structural Analysis of Escherichia Coli ThiF. J Mol Biol. 2005;349:774–86. doi: 10.1016/j.jmb.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 54.Furukawa K, Mizushima N, Noda T, Ohsumi Y. A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J Biol Chem. 2000;275:7462–5. doi: 10.1074/jbc.275.11.7462. [DOI] [PubMed] [Google Scholar]
- 55.Goehring AS, Rivers DM, Sprague GF., Jr Attachment of the ubiquitin-related protein Urm1p to the antioxidant protein Ahp1p. Eukaryot Cell. 2003;2:930–6. doi: 10.1128/EC.2.5.930-936.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmitz J, et al. The sulfurtransferase activity of Uba4 presents a link between ubiquitin-like protein conjugation and activation of sulfur carrier proteins. Biochemistry. 2008;47:6479–89. doi: 10.1021/bi800477u. [DOI] [PubMed] [Google Scholar]
- 57.Mueller EG. Trafficking in persulfides: delivering sulfur in biosynthetic pathways. Nat Chem Biol. 2006;2:185–94. doi: 10.1038/nchembio779. [DOI] [PubMed] [Google Scholar]
- 58.Nakai Y, Nakai M, Hayashi H. Thio-modification of Yeast Cytosolic tRNA Requires a Ubiquitin-related System That Resembles Bacterial Sulfur Transfer Systems. J Biol Chem. 2008;283:27469–76. doi: 10.1074/jbc.M804043200. [DOI] [PubMed] [Google Scholar]
- 59.Huang B, Lu J, Bystrom AS. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. Rna. 2008;14:2183–94. doi: 10.1261/rna.1184108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burroughs AM, Jaffee M, Iyer LM, Aravind L. Anatomy of the E2 ligase fold: implications for enzymology and evolution of ubiquitin/Ub-like protein conjugation. J Struct Biol. 2008;162:205–18. doi: 10.1016/j.jsb.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 62.Burns KE, et al. Reconstitution of a new cysteine biosynthetic pathway in Mycobacterium tuberculosis. J Am Chem Soc. 2005;127:11602–3. doi: 10.1021/ja053476x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Godert AM, Jin M, McLafferty FW, Begley TP. Biosynthesis of the thioquinolobactin siderophore: an interesting variation on sulfur transfer. J Bacteriol. 2007;189:2941–4. doi: 10.1128/JB.01200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roush RF, Nolan EM, Lohr F, Walsh CT. Maturation of an Escherichia coli ribosomal peptide antibiotic by ATP-consuming N-P bond formation in microcin C7. J Am Chem Soc. 2008;130:3603–9. doi: 10.1021/ja7101949. [DOI] [PubMed] [Google Scholar]
- 65.Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322:1104–7. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iyer LM, Burroughs AM, Aravind L. Unraveling the biochemistry and provenance of pupylation: a prokaryotic analog of ubiquitination. Biol Direct. 2008;3:45. doi: 10.1186/1745-6150-3-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Steinacher R, Schar P. Functionality of human thymine DNA glycosylase requires SUMO-regulated changes in protein conformation. Curr Biol. 2005;15:616–23. doi: 10.1016/j.cub.2005.02.054. [DOI] [PubMed] [Google Scholar]
- 68.Ulrich HD. How to activate a damage-tolerant polymerase: consequences of PCNA modifications by ubiquitin and SUMO. Cell Cycle. 2004;3:15–8. [PubMed] [Google Scholar]
- 69.Palacios S, et al. Quantitative SUMO-1 Modification of a Vaccinia Virus Protein Is Required for Its Specific Localization and Prevents Its Self-Association. Mol Biol Cell. 2005;16:2822–35. doi: 10.1091/mbc.E04-11-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ciechanover A, Ben-Saadon R. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 2004;14:103–6. doi: 10.1016/j.tcb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 71.Cadwell K, Coscoy L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science. 2005;309:127–30. doi: 10.1126/science.1110340. [DOI] [PubMed] [Google Scholar]
- 72.Ravid T, Hochstrasser M. Autoregulation of an E2 enzyme by ubiquitin-chain assembly on its catalytic residue. Nat Cell Biol. 2007;9:422–7. doi: 10.1038/ncb1558. [DOI] [PubMed] [Google Scholar]
- 73.Wang X, et al. Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J Cell Biol. 2007;177:613–24. doi: 10.1083/jcb.200611063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee I, Schindelin H. Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell. 2008;134:268–78. doi: 10.1016/j.cell.2008.05.046. [DOI] [PubMed] [Google Scholar]
- 75.Nijman SM, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–86. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]